Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
Purification strategies developed for human Mk-E progenitors, as well as committed Mk and E progenitors.
MYB regulates the biphenotypic fate decision of human MEPs.
Abstract
Bipotent megakaryocyte/erythroid progenitors (MEPs) give rise to progeny limited to the megakaryocyte (Mk) and erythroid (E) lineages. We developed a novel dual-detection functional in vitro colony-forming unit (CFU) assay for single cells that differentiates down both the Mk and E lineages (CFU-Mk/E), which allowed development and validation of a novel purification strategy for the identification and quantitation of primary functional human MEPs from granulocyte colony-stimulating factor–mobilized peripheral blood and bone marrow. Applying this assay to fluorescence-activated cell sorter–sorted cell populations, we found that the Lin−CD34+CD38midCD45RA−FLT3−MPL+CD36−CD41− population is much more highly enriched for bipotent MEPs than any previously reported subpopulations. We also developed purification strategies for primary human lineage-committed Mk and E progenitors identified as CFU-Mk and burst forming unit-E. Comparative expression analyses in MEP, MkP, and ErP populations revealed differential expression of MYB. We tested whether alterations in MYB concentration affect the Mk-E fate decision at the single cell level in MEPs and found that short hairpin RNA-mediated MYB knockdown promoted commitment of MEPs to the Mk lineage, further defining its role in MEP lineage fate. There are numerous applications for these novel enrichment strategies, including facilitating mechanistic studies of MEP lineage commitment, improving approaches for in vitro expansion of Mk and E cells, and developing improved therapies for benign and malignant hematologic disease.
Introduction
Megakaryocyte/erythroid progenitors (MEPs) are bipotent cells that undergo a fate decision to become either megakaryocytes (Mk) or erythroid (E) cells. Detailed mechanistic knowledge of the human MEP fate decision is not only critical for our understanding of normal and perturbed hematopoiesis, but also has important therapeutic implications. Potential applications include refinement of regenerative approaches to produce platelets and red blood cells in vitro, providing insight into engraftment of these lineages in clinical hematopoietic transplantation, and development of therapeutic agents for treatment of benign and malignant hematologic disease.
Previous studies of the MEP fate decision have primarily used mouse bone marrow (BM),1,2 in vitro cell lines (of leukemic origin),3-6 and in vitro–expanded human CD34+ cells.7-9 The existence of bipotent MEPs in human BM was first reported in 1996; Debili et al10 identified bipotent MEPs within the CD34+CD38lo and CD34+CD38mid fraction of BM. Since that time, multiple publications’ strategies for MEP enrichment from CD34+CD38+Lin− cells have been described. Manz et al11 enriched MEPs using IL3RA−CD45RA− selection. Edvardsson et al12 replaced the IL3RA with thrombopoietin receptor (myeloproliferative leukemia [MPL], CD110),13,14 and showed that, in BM, the MPL+CD45RA− fraction of CD34+CD19− cells was restricted to Mk and E fates. They also found that other CD34+ cells did not stain for MPL, which was unexpected, as hematopoietic stem cells (HSCs) express MPL mRNA, and TPO promotes HSC self-renewal.15-18 This discrepancy was addressed in later studies19 showing that the BAH-1 clone20 of anti-MPL antibody used is not specific for MPL. Abbot et al,19 using more sensitive and specific anti-MPL antibodies (clones 1.6 and 1.7), showed that MPL is expressed on a larger percentage of CD34+ cells, as expected. They also showed that the BAH-1 clone has both false-positive and false-negative activity on MPL− and MPL+ cells, respectively. It is unknown if more specific MPL antibodies (eg, clone 1.6) are useful for purifying MEPs, and which hematopoietic stem and progenitor cells (HSPCs) subsets have surface expression of MPL. A third21 published approach to enrich primary human MEP is the FLT3−CD45RA− population, which was reported to contain almost entirely E potential, and to lack granulocyte/monocyte differentiation potential in methylcellulose colonies, but for which the Mk or E/Mk potential were not assessed. In summary, primary human MEP purification strategies described to date are inconsistent in the source of HSPCs and the assays used for quantifying biphenotypic potential. In addition, these strategies have not been applied to the enumeration of MEPs in mobilized peripheral blood (MPB), the predominant source of HSPCs used clinically.
A recently published study suggests that adult humans do not have MEPs and that megakaryocytes are derived directly from HSCs or multipotent progenitors (MPPs).22 Consistent with these findings, murine studies have revealed that HSPCs under stress conditions may commit to the Mk lineage without seeming to go through the MEP stage of differentiation. Strong molecular and functional data suggest that there are von Willebrand factor–expressing murine HSPCs that are biased toward the Mk lineage.23,24 Also, murine single cell transplantation of daughter cells produced in vitro provided evidence for a relatively long-term (∼6 months) self-renewing population of Mk-committed hematopoietic stem/progenitor cells.25
In vitro, a MEP should retain both Mk and E potential and should lack the potential to differentiate down other myeloid lineages (monocytes, granulocytes). Such MEPs are distinct from unipotent megakaryocyte and erythroid progenitors (MkPs and ErPs), as well as more multipotent progenitors, such as common myeloid progenitors (CMPs) and MPPs. To distinguish between assumed marker-pure subsets from functionally pure subsets, one needs an assay with single cell resolution, but with high capacity so that hundreds of cells can be assayed simultaneously to assess not only the quantification of this purified population, but also the molecular mechanisms on which their function relies.
Together, these data indicate that improved approaches for both purification and functional characterization of human MEPs are needed. In this report, we describe a novel dual detection approach for single cell colony-forming unit (CFU) capable of differentiation down both the Mk and E lineage, which we call CFU-Mk/E, allowing the identification and quantitation of functional MEPs. Applying this assay to fluorescence-activated cell sorter (FACS)-sorted populations, we find that the Lin−CD34+CD38midCD45RA−FLT3−MPL+CD36−CD41− population is much more highly enriched for bipotent MEPs than populations previously reported. We also developed sorting strategies for purifying primary human MkPs and ErPs, assessed as CFU-Mk and CFU-E.
These highly enriched populations can be used to better understand the Mk vs E lineage fate decision of MEPs. Transcription factors are major drivers of hematopoiesis, guiding a cascade of gene expression programs that determine the fate decision of different types of stem and progenitor cells.26 Comparative expression analyses in MEP, MkP, and ErP cells revealed differential expression of the transcription factor MYB, a known participant in Mk-E lineage choice. MYB targets genes important for E differentiation27,28; however, its role in the biphenotypic fate decision in human MEPs has been unknown. We tested whether alterations in MYB concentration affect the Mk-E fate decision at the single cell level in bipotent MEPs and found that MYB knockdown promoted commitment of MEPs to the Mk lineage, further defining its role in MEP lineage fate. Thus, the improved approaches for the identification and quantitation of primary functional human MEPs provide novel tools for studies of biphenotypic fate decisions in normal and altered hematopoiesis.
Materials and methods
Cell source, preparation, and FACS
Human granulocyte colony-stimulating factor (G-CSF) MPB was either CD34-enriched (CliniMACS; Miltenyi) and cryopreserved in 10% dimethyl sulfoxide for future use or was used fresh for sorting. Human BM was purchased from Lonza (Walkersville, MD). For non–CD34-selected cells, immunomagnetic lineage depletion was performed using biotinylated antibodies against CD2, CD11b, CD14, CD15, CD20, and CD56 (full list of antibodies in supplemental Table 1, available on the Blood Web site) followed by streptavidin-coated BD-IMAG beads (Becton Dickinson) according to the manufacturer’s instructions.
Cell sorting
Lineage-depleted or CD34-selected MPB or BM cells were stained with desired antibodies and sorted on a FACSAria. The sorted populations were collected in culture medium and counted for cell concentration and postsort viability (range, 86-95%).
Modified MegaCult assays
For the Dual Mk/E colony assay, 250 sorted cells were plated in 2 plates (125 cells per plate) of MegaCult C Medium plus Lipids (Stem Cell Technologies) with rhEPO (recombinant human erythropoietin) (3.0 U/mL), recombinant human interleukin rhIL-3 (10 ng/mL), rhIL-6 (10 ng/mL), and rhSCF (recombinant human stem cell factor) (25 ng/mL), with/without rhTPO (recombinant human thrombopoietin) (50 ng/mL). All cytokines were purchased from PeproTech (Rocky Hill, NJ) except rhEPO (Amgen). Fixed and dried MegaCult cultures were then stained using a dual immunohistochemistry assay adapted from published methods,29 using rabbit anti-GlyA antibody (1:1500, ABD Serotec, clone YTH89.1), horse anti-rabbit Impress-horseradish peroxidase secondary kit (Vector Labs) signal amplification, and nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate.30 For Mk staining, we used mouse anti-human CD41a antibody (BD), and then goat anti-mouse Impress-AP secondary (Vector Labs), ABC-AP, followed by detection with Vector Red substrate (Vector Labs). Alternatively, we also stained Mks with anti-vWF antibody clone F8/86 (Dako) and detected the signal with Vector SG (Vector Labs). Colonies were scored based on GlyA and CD41a staining as megakaryocyte only (CFU-Mk), erythroid only burst forming unit (BFU)-E, or megakaryocyte + erythroid (CFU-Mk/E). We also performed liquid culture of sorted MEPs in StemSpan Serum Free Expansion Medium (Stem Cell Technologies) with rhEPO (3.0 U/mL), rhIL-3 (10 ng/mL), rhIL-6 (10 ng/mL), and rhSCF (25 ng/mL) with/without rhTPO (50 ng/mL) for 48 hours.
Methylcellulose assay
Cells were plated in Methocult (Stem Cell Technologies) per the manufacturer’s instructions.
Statistical analysis
Average cloning frequencies for each colony assay were compared using Student t tests.
Gene expression analysis
RNA was extracted from sorted MEPs, MkPs, and ErPs using the RNAqueous-Micro Total RNA Isolation Kit (Ambion). Briefly, 1 to 10 ng total RNA was reverse transcribed using the iScript cDNA synthesis system (BioRad). Polymerase chain reaction (PCR) was performed in triplicate for each reverse transcriptase (RT) product, following the manufacturer’s protocol with Syber Green on a CFx96 Real Time System, and threshold cycles for GATA1, GATA2, ITGA2B, ITG3B, CD36, GFI1B, KLF1, CA1, FLI1, IL6R, NFE2, TFRC, VWF, and MAX were normalized to GAPDH expression. For MYB (Hs00920556_m1) mRNA analysis, RT-PCR was performed using TaqMan (Applied Biosystems), and threshold cycles were normalized to Ct values of corresponding 18S (4333760T) rRNA reactions. Relative gene expression was calculated as 2−ΔΔCT.
MYB vector cloning and production.
Lentiviral vector pLKO.8 was produced in the laboratory of Jun Lu (Yale University), by replacing the puromycin resistance gene in pLKO.231with enhanced green fluorescent protein (GFP) cDNA amplified by oligonucleotide pairs: agcggatcctcgccaccatggtgagcaag and aggggtaccagctagctactagctagtc from pMIRWAY-GFP.32,33 pLKO.8 was used with short hairpin RNA (shRNA) against MYB or against luciferase (shLuc). The sequences of the shRNAs against human MYB were obtained from the RNAi Consortium (http://www.broad.mit.edu/genome_bio/trc/): shMYB-1 and shMYB-2 clones are TRCN0000040058 and TRCN0000009853, respectively. For lentivirus production, 90% confluent 293T cells were transfected with pLKO.8 shMYB-1/2 (or pLKO.8 shLuc), Gag/VSVG, and Gag/Rev with FuGENE HD (Promega). Virus was collected 24, 36, 48, and 60 hours after transfection, and each collection was 0.45-µm filtered, concentrated using Amicon filters (Millipore, Billerica, MA), and frozen in aliquots at −80°C.
Transduction.
CD34+ cells were cultured at 5 × 105 cells/mL in expansion media (100 ng/mL hFLT3, 100 ng/mL SCF, 20 ng/mL IL-3, 20 ng/mL IL-6, and 20 ng/mL hTPO) in StemSpan Serum Free Expansion Medium (Stem Cell Technologies) for 16 hours, transduced with lentivirus at MOI9, and sorted for GFP+Lin−CD34+CD38midCD45RA−FLT3−MPL+CD36− cells 24 hours later.
Human subjects.
All work was conducted according to Declaration of Helsinki principles. Collection and use of human cells were approved by the Yale University institutional review board. Healthy donors who were already donating cells for allogeneic transplantation provided written informed consent prior to use of their extra G-CSF mobilized cells for research.
Results
Assay for quantitating biphenotypic MEPs
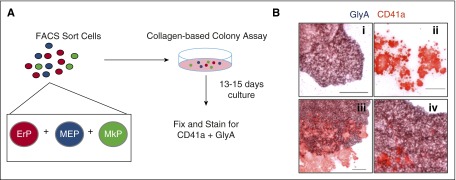
To date, no method for purifying primary human MEPs yields a homogeneous, bipotent population of cells. Instead, current methods yield a population of cells that is a mix of bipotent progenitors (MEPs), unipotent progenitors (MkPs and ErPs), and progenitors with broader myeloid potential. To determine the relative numbers of each progenitor type in these populations, we developed an assay combining modification of a commercial collagen-based colony assay (MegaCult) with dual immunohistochemistry for CD41a and GlyA (Figure 1A). In this Dual CFU-Mk/E assay, we cultured candidate MEP subpopulations with cytokines that promote simultaneous Mk and E differentiation. Three types of colonies were quantitated: (1) BFU-E with typical BFU-E morphology and GlyA staining; (2) CFU-Mk, which had typical CFU-Mk morphology and CD41a staining; and (3) dual CFU-Mk/E colonies, which had morphologic characteristics of CFU-Mk and BFU-E, plus dual staining for CD41a and GlyA (Figure 1B).
Figure 1.
Use of the dual CFU-Mk/E assay to quantify bipotent MEPs. (A) Schematic of the dual CFU-Mk/E assay to determine Mk/E potential of FACS-sorted populations. Candidate MEPs are plated in a collagen-based colony assay with EPO, TPO, SCF, IL-3, and IL-6 to promote growth of megakaryocyte and erythroid colonies. After 13 to 15 days of culture, colonies are fixed and stained for CD41a (Mk specific) and GlyA (E specific). (B) Representative images of different colony types are shown. (i) Typical BFU-E stains for GlyA (purple) only. (ii) CFU-Mk stains for CD41a (red) only. (iii-iv) Two representative CFU-Mk/E colonies show prominent CD41a+ Mk within a burst of GlyA+ cells. Scale bar, 100 µm.
Direct comparison of previous MEP purification methods
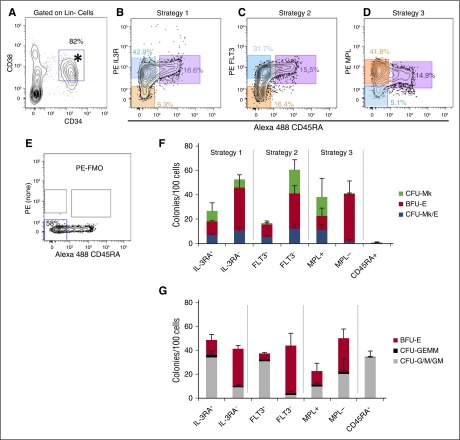
Three published methods were compared for enrichment of primary human MEPs from the CD34+CD38+Lin− population (Figure 2A). Each method identified granulocyte-monocyte progenitor (GMP) as CD45RA+. Thus, the primary difference in these methods is in how the CD45RA− populations were separated (Figures 2B-D) based on IL3RA,11 FLT3,21 or MPL.12 For all 3 strategies, we used fluorescence minus one (FMO) to gate for the negative populations (Figure 2E). In our hands, the staining patterns for IL3RA and FLT3 were consistent with the original publications. However, MPL staining (Figure 2D) was different, likely due to the use of antibody clone 1.6 (as discussed in the “Introduction”). Perhaps surprisingly, nearly all CD34+CD38+ cells expressed higher levels of MPL than the FMO, suggesting that in addition to HSCs, most hematopoietic progenitors express MPL. Therefore, we gated on MPLlo/− vs MPL+ cells.
Figure 2.
Comparison of 3 different MEP purification strategies for enrichment of CFU-Mk/E. Human MPB mononuclear cells were fractionated using 3 different approaches and analyzed for colony formation in the dual Mk/E assay and methylcellulose as described. (A) Starting from previously gated Lin− cells, CD34+CD38+ cells (*) were equivalently selected using each of 3 MEP purification strategies. (B-D) The candidate MEP population (orange) is CD45RA− (x-axis) and either IL-3Ra− (CD123, strategy 1), FLT3− (CD135, strategy 2), or MPL+ (CD110, strategy 3). The corresponding CMP (blue) and GMP (purple) populations are highlighted for each strategy. CMPs were also sorted for comparison. Percentages of total CD34+Lin− cells appear with gates. (E) FMO control used for strict gating of PE negative (bottom left gate) vs positive events shown in B-D. (F) For the dual Mk/E assay, colonies were enumerated based on dual immunohistochemistry for CD41a and GlyA. (G) For methylcellulose, colonies were enumerated based on typical morphology and validated with Wright-Geimsa–stained cytospins of selected colonies (data not shown). Results presented as an average for each colony type + standard deviation (SD) for ≥4 independent experiments except for strategy 1 (n = 3 for dual Mk/E and n = 2 for methylcellulose assays).
Sorted cells were analyzed using 2 different colony assays (Figure 2F-G). The first was a standard methylcellulose assay containing cytokines (EPO, IL-3, SCF, GM-CSF, G-CSF) that promote growth of CFU-granulocyte/monocyte (G/M) and CFU-granulocyte, erythrocyte, monocyte/macrophage, megakaryocyte (GEMM)/Mix, as well as BFU-E. Highly enriched candidate MEP populations, as well as MkP and ErP, should contain little to no G/M or GEMM potential, whereas CMPs and granulocyte-monocyte progenitors will contain CFU-GEMM and CFU-G/M or CFU-G/M, respectively. Second, the dual Mk/E assay was used to quantitate CFU-Mk/E, CFU-Mk, and BFU-E. Results showed that sorted GMP (CD45RA+) contained no Mk or E potential (Figure 2F) and purely CFU-G/M potential (Figure 2G). Populations enriched for CMP (FLT3+, IL3RA+) had mixed potential for all colony types, as expected. All 3 MEP-enriched populations tested (FLT3−, IL3RA−, and MPL+) had similar CFU-Mk/E cloning potential with ∼10% to 15% of cells being CFU-Mk/E (P > .05). However, the CFU-G/M, CFU-GEMM, BFU-E, and CFU-Mk contents varied. The FLT3− population had the lowest CFU-G/M contamination (P < .05 vs IL3R−), and the MPL+ population had the lowest BFU-E (P < .05 vs FLT3− or IL3R−), suggesting that MEP enrichment could be improved by combining approaches to achieve better BFU-E and CFU-G/M depletion. The MPLlo/− population contained little to no CFU-Mk/E or CFU-Mk potential, but retained BFU-E and G/M potential, implying that true MEP express MPL on their surface, which is lost after erythroid commitment. Note that there was no statistical difference between the BFU-E quantitated in Megacult vs methylcellulose assays (paired t test, P = .58, n = 68 conditions).
Two caveats prevent precise quantification of the frequency of MEP: (1) colonies with both Mk and E cells can result from MEP, CMP, or pre-CMP populations and (2) an MEP may not always grow as a colony with both mature Mk and E being apparent. Therefore, we can at least provide the current best approach for enriching cells that have both Mk and E potential and lack potential to form colonies with granulocytes and/or monocytes.
Improved MEP enrichment by selection for FLT3−MPL+ cells
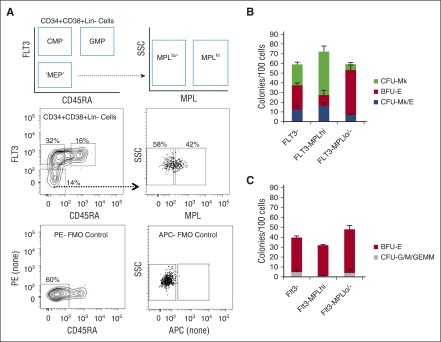
We tested whether MEP enrichment could be improved by combining sorting approaches. Based on results from Figure 2, we combined sorting for FLT3− and MPL+ cells. We did not further pursue IL3RA because the IL3R− and IL3R+ fractions had nearly equivalent CFU-Mk/E potential (P > .05). To be able to sort cells that were MPL+ and FLT3− cells, we modified the approach for simultaneous staining of FLT3 and MPL (Figure 3A). Compared with total Lin−CD34+CD38+FLT3− cells, the Lin−CD34+CD38+FLT3−MPL+ fraction had 36% improved enrichment for CFU-Mk/E and the FLT3-MPLlo/− fraction had a 44% decrease in CFU-Mk/E (Figure 3B). The FLT3−MPL+ population had a high cloning efficiency (72 ± 6.7 colonies/100 cells), 25% of which were CFU-Mk/E, with the rest being CFU-Mk and BFU-E. The FLT3−MPLlo/− fraction was mostly comprised of BFU-E. The FLT3−MPL+ fraction had only 0.73% CFU-G/M + CFU-GEMM (Figure 3C). Although there is inter-donor variation, in every experiment performed, there was enrichment for CFU-Mk/E in FLT3−MPL+ compared with total FLT3− (P = .02) or FLT3-MPLlo/− (P = .01) cells (data not shown).
Figure 3.
Improvement in MEP purification by combining FLT3 and MPL selection. (A) Schematic of FACS gating for further purifying FLT3− MPB cells into MPLhi and MPLlo/− fractions. Representative flow plots with associated FMO controls used to gate for positive and negative populations. (B) Results of dual Mk/E assay showing average colony counts per 100 seeded cells from 3 experiments. The FLT3-MPLhi population has the highest Mk/E potential (blue). (C) Methylcellulose assay results showing average CFU-G/M + CFU-GEMM (gray) and BFU-E (red) colony counts per 100 seeded cells for different subpopulations. CFU-G/M + CFU-GEMM (gray) colonies likely arise from CMP contamination in the sorted MEP-enriched population. The FLT3−MPLhi population has the lowest G/M contamination (P = .02 in comparison with FLT3− population).
MEPs are primarily in the CD38mid subpopulation
When we assessed the separate CD38−, CD38lo, CD38mid, and CD38hi populations of Lin−CD34+CD45RA−FLT3−MPL+ cells, we found that CFU-Mk/E were enriched, and CFU-G/M together with CFU-GEMM were depleted from the CD38mid population (Figure 4), excluding the possibility of containing CMPs or MPPs. Analysis of the proportion of CFU-Mk/E in the different populations reveals that 70% of CFU-Mk/E are in the MPL+ population (supplemental Figure 1A), and within the MPL+ population, 63% of CFU-Mk/E are in the CD38mid population (supplemental Figure 1B). Similarly, >60% of CFU-Mk are in the MPL+ population. In contrast, the majority of BFU-E are in the MPL− fraction (supplemental Figure 1A). We observed a higher proportion of CFU-Mk in the CD38− fraction; however, this population also gives rise to a high number of other myeloid colonies (CFU-G/M and especially CFU-GEMM), so we cannot rule out their MPP/CMP potency (Figure 4B-C).
Figure 4.
Majority of MEPs are in the CD38mid fraction. (A) Sorting strategy used to evaluate MPL+ and MPL− cells within CD38 negative, low, mid, and high subsets of Lin−CD34+ MPB cells followed by exclusion of FLT3+ and CD45RA+ cells. (B) Dual Mk/E and (C) methylcellulose colony assay results. Results are presented as average + SD from 3 independent experiments.
Simultaneous enrichment approaches for MEPs, MkPs, and ErPs
Importantly, we observed that gating on CD38hi cells provides a significant increase in enrichment for CD36+ cells and for BFU-E in both the dual CFU-Mk/E and methylcellulose assays even when the cells are MPL+ (Figure 4B-C). Based on this observation, we developed strategies to enrich for ErPs (Lin−CD34+CD38highFLT3−CD45RA−MPL−) and MkPs (Lin−CD34+CD38midFLT3−CD45RA−MPL+CD36−CD41+) as shown (Figure 5A). Although the resultant populations are not 100% pure, they result in the highest purifications to date of primary human MEPs, MkPs, and ErPs. The ErP population is greatly enriched (80% purity) for BFU-E with a small percentage of CFU-Mk/E and CFU-Mk. The absolute CFU-Mk/E content of the MEP population (Lin−CD34+CD38midFLT3−MPL+CD36−CD41−) varies from 45% to 60% of cells plated, and this percentage is consistently higher than in the MkP population, which is comprised of >70% CFU-Mk. Although the fractions of CFU-Mk/E, BFU-E, and CFU-Mk are not identical from donor to donor, the relative enrichment of each in the 3 sorted populations is consistent (n = 5 donors). We next tested whether these sorting strategies for MEPs, MkPs, and ErPs are also applicable in primary human BM. We performed CFU assays on sorted BM subpopulations and found that they were enriched for CFU-Mk/E, CFU-Mk, and BFU-E in proportions very similar to MPB (Figure 5B-E).
Figure 5.
Improved strategies for isolating MEPs, MkPs, and ErPs in MPB and BM. (A) Improved strategy to enrich for MEPs (Lin−CD34+CD38midCD45RA−FLT3−MPL+CD36−CD41−), MkPs (Lin−CD34+CD38midFLT3−CD45RA−MPL+CD36−CD41+), and ErPs (Lin−CD34+CD38hiFLT3−CD45RA−MPL−). Dual Mk/E assay results for (B) MPB and (C) BM indicate similar enrichment levels in MPB and BM. Methylcellulose assay from the same populations in (D) MPB and (E) BM showing low contamination with other myeloid colonies (CFU-G/M/GM).
To compare our findings to 2 recent papers that rely on CD71 expression to distinguish cells with erythroid potential22,34 and also on CD105 as a marker for commitment to BFU-E, we assessed CD71 and CD105 expression in our newly established MEP, MkP, and ErP populations and also in subpopulations separated according to differential CD38 expression (CD38−, CD38low, CD38mid, CD38high). In our hands, there is no difference in CD105 expression between any of the populations analyzed; however, there is a strong correlation between CD38 and CD71 expression, as CD38high cells (ErP) have the highest intensity of CD71 compared with MEPs and MkPs, as well as CD38mid, CD38low (supplemental Figure 2).
Purification of MEPs, MkPs, and ErPs provides an excellent system for elucidating how exogenous factors may influence the fate decision. In preliminary studies, we performed the dual CFU-Mk/E assay with sorted MEPs in the presence and absence of TPO to test whether this cytokine affects the Mk vs E fate decision. We observed that TPO plays a role in the proliferation/survival of all colony types (Mk/E, Mk, and E), but does not affect the relative distribution of colony types (supplemental Figure 3A). Dual CFU-Mk/E assays performed in the absence of EPO grew only Mk colonies, consistent with the role of EPO in erythroid maturation (data not shown).
Use of purified MEP to assess the role of MYB in the MEP fate decision
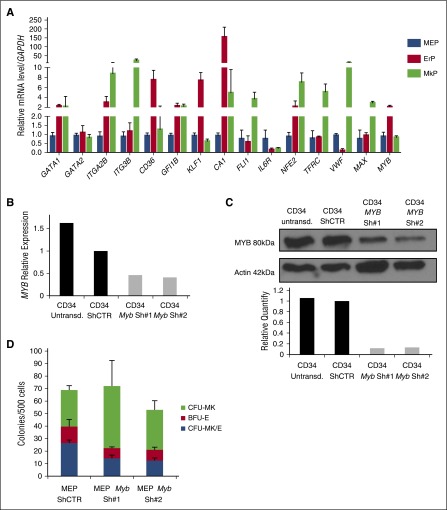
We next compared the gene expression profiles of highly purified MEPs, MkPs, and ErPs. The data show differential expression with MkP, expressing the high levels of ITGA2B (CD41), ITG3B (CD49C), NFE2, and VWF. The unique ErP signature includes expression of CD36, CA1, and KLF1 and elevated levels of MYB compared with MEPs and MkPs (Figure 6A). This is consistent with published data28,35,36 that MYB expression promotes erythroid differentiation while inhibiting Mk differentiation. Also, these data suggest that MEPs are not biphenotypic, with known unique markers of both Mk and E cells that are lost on differentiation.
Figure 6.
MYB expression controls the MEP fate decision. MYB is differentially expressed in MEPs, MkPs, and ErPs, and MYB downregulation in MEPs promotes megakaryocytic commitment. (A) Quantitative reverse transcriptase-PCR analysis of multiple genes in purified MEP, MkP, and ErP subsets (GAPDH used as an internal control). Knockdown of MYB in CD34+ cells showing decreased (B) mRNA and (C) protein expression 24 hours after transduction. (D) Dual Mk/E assay results show increased CFU-Mk as the expense of both CFU-Mk/E and BFU-E when MYB is knocked down (P < .05). Results are presented as average + SD from 3 independent experiments.
No studies have directly shown that MYB can influence lineage choice in human MEPs. We knocked down MYB expression in MEPs isolated as described above with 2 different shRNAs against MYB (Figure 6B-C). Inhibition was >60% for both MYB mRNA and protein. To assess the effect of MYB knockdown on the Mk vs E fate decision, 24 hours after transduction, GFP+ (transduced) MEPs were sorted as Lin−CD34+CD38midFLT3−MPL+CD36−CD41− cells and plated in the CFU Mk/E assay. Knockdown of MYB increased CFU-Mk at the expense of CFU-Mk/E and BFU-E (shCTR vs shMyb#1, P = .01 and shCTR vs shMyb#2, P = .0009), confirming that MYB plays a role in the MEP fate decision inhibiting Mk commitment and promoting E commitment (Figure 6D). Consistent with the lack of effect of TPO on CFU distribution, there is no statistically significant difference in MYB expression level between MEP cultured in the presence vs in the absence of TPO (supplemental Figure 3B).
Discussion
The process of hematopoiesis is one of the best-studied models of lineage choice and cell fate determination. However, our understanding of this process is far from complete. Transcriptome-based studies demonstrate the complexity of this process, even between closely related progenitor populations.37 Remarkably, strategies to functionally identify or to purify homogeneous populations of human MEPs, a critical cell in Mk and E fate choice, have been lacking. To determine the efficacy of different MEP purification strategies, we developed a method to quantitate functional MEPs as single cells capable of Mk and E differentiation exclusively. By assessing the same populations in standard methylcellulose assays, we excluded the presence of more multipotent progenitors, such as CMP and multipotent progenitors, which would differentiate down the granulocytic and monocytic lineages in addition to Mk and E.38
Using this assay, we performed the first direct comparison of MEP purification methods from MPB. This allowed us to develop an improved strategy for enriching MEPs from MPB and BM in the Lin−CD34+CD38midFLT3−MPL+CD36−CD41− population. The bipotent MEPs are distinct from unipotent MkPs and ErPs, which were also purified in this study. Although our approach is a significant improvement over prior methods, there is room for improvement in precisely defining human MEPs. Combining our dual detection with other emerging technologies may allow further refinement in the MEP population.
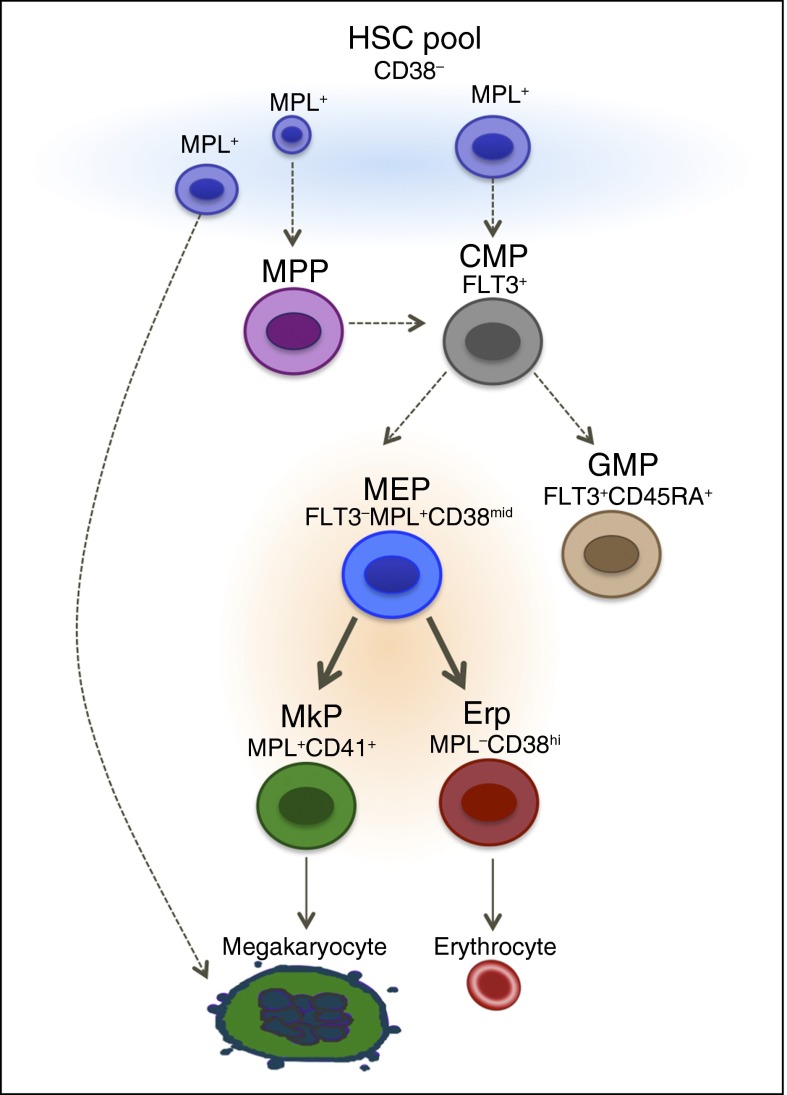
Alternate hierarchical models of hematopoiesis have been proposed, wherein a pool of long-term self-renewing stem cells or cells that have the surface phenotype of HSCs and MPPs are already Mk committed39 or have the ability to commit directly to the Mk lineage.40 In addition to murine studies cited in the introduction and above, a recently published study suggests that in adult humans, Mk/E activity together with multipotency are restricted to the stem cell compartment with no intermediate bipotent progenitor.22 However, there are numerous differences between our studies that may explain why these investigators22 did not identify primary MEPs in adult humans. (1) They present results from Lin−CD34+CD38hi for the presumed MEP gating strategy. We show that most of the cells from CD38hi population are erythroid committed, lacking both Mk and Mk/E potential, corroborating one of the first studies to identify human MEP using single cell assays, where they showed that 90% of CD34+CD38+ cells give rise to erythroid colonies, whereas the Mk and Mk/E potential was observed in both CD34+CD38mid and CD34+CD38low/− fractions.10 (2) When they identified cells that give rise to both erythroid and megakaryocyte colonies in the CD34+CD38lo population, they identified these cells as being multipotent progenitors. We also found significant Mk activity in the CD38− fraction, and it is likely that some MEP are CD38lo. However, this population is also enriched for CFU-G/M and CFU-GEMM, thus decreasing its utility in an MEP purification strategy. (3) Perhaps the most significant difference between our studies is the antibodies used. The BAH.1 antibody used by Notta et al22 is known not to target MPL.19 Cell lines that are known to lack MPL expression (eg, TF1 cells) stain positively with the BAH.1 clone, and some cells that are known to be MPL+ (eg, 32D cells transduced with human cMPL) do not stain with this monoclonal antibody. Thus, the cell populations analyzed in this report are likely to be different from the ones sorted herein using clone 1.6, which has been confirmed to be directed against human MPL.19 Overall, our findings do not exclude the possibility of megakaryocytes being generated directly from cells in the hematopoietic stem/multipotent progenitors pool, especially under stress conditions. It is likely that there are multiple routes in vivo to generate cells that are extremely important for survival such as platelets (Figure 7). (4) Another difference between the studies is the single cell assays used. We used CFUs in semisolid medium, whereas Notta et al22 cultured single cells on an MS5 feeder layer for 3 weeks and assayed the progeny by FACS analysis.
Figure 7.
Proposed model for human hematopoiesis. The classical model of hematopoiesis was based in hierarchical and sequential differentiation from hematopoietic stem cells to mature cells. However, recent studies provide evidence that mature cells such as megakaryocytes could be derived directly from HSCs. We propose a model that there is more than one origin of megakaryocytes, likely dependent on need. We observed that the heterogeneous mixture of cells within the Lin−CD34+CD38− population includes cells that can give rise to megakaryocytes directly. This is in addition to a strictly bipotent MEP population that gives rise to megakaryocytes and erythrocytes. Solid arrows, validated in the manuscript.
Our improved MEP, MkP, and ErP purification strategies have similar FACS gating patterns and dual CFU-Mk/E assay results in MPB and BM. In addition, we provide new data showing wide expression of MPL on hematopoietic stem and progenitor cells. An example of the utility of our methodologies is our observation, for the first time, of the role of MYB in lineage choice in human MEPs. Decreasing MYB expression in MEPs resulted in increased Mk colonies at the expense of erythroid and Mk/E colonies. This observation complements studies of MYB knockdown by siRNA in CD34+ cells, which lead to increased megakaryocytic differentiation.28 MYB favors erythropoiesis via binding to and activation of KLF1 and LMO. More recent studies show MYB induces erythropoiesis and restrains megakaryopoiesis through the transactivation of miR-486-3p expression followed by downregulation of MAF.27 Thus, these methodologies will have broad applicability across studies of normal and perturbed hematopoiesis, hematopoietic stem cell transplantation, and cell fate determination.
Authorship
Contribution: C.S. and J.X.-F. designed and performed experiments and wrote the paper; Y.-C.L. performed experiments and helped write the manuscript; E.K., E.M., P.-X.Z., S.Z., M.Z., and G.Z. performed experiments; P.G.G. helped interpret data and helped write the manuscript; and D.S.K. oversaw the project, provided intellectual input, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Juliana Xavier-Ferrucio, Yale University School of Medicine, Department of Laboratory Medicine, Yale Stem Cell Center, 10 Amistad St, New Haven, CT 06511; e-mail: juliana.xavierferrucio@yale.edu.
Acknowledgments
The authors thank the Yale Flow Sorting Core staff for performing cell sorting. The authors also thank Sara T. Olalla Saad, Emanuela Bruscia, Jun Lu, and Natalia Ivanova for helpful discussions and reagents. The authors also thank members of the laboratory of James Palis at the University of Rochester for assistance in developing the Dual Mk/E Assay.
This work was supported by National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases grants DK086267 and DK094934 and the CT Stem Cell Fund (D.S.K.), the Yale Cooperative Center of Excellence in Hematology (1U54DK106857), and 5T32HL007974 and 5T32DK007556 fellowship funding (C.S.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
References
- 1.Stachura DL, Chou ST, Weiss MJ. Early block to erythromegakaryocytic development conferred by loss of transcription factor GATA-1. Blood. 2006;107(1):87–97. doi: 10.1182/blood-2005-07-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doré LC, Chlon TM, Brown CD, White KP, Crispino JD. Chromatin occupancy analysis reveals genome-wide GATA factor switching during hematopoiesis. Blood. 2012;119(16):3724–3733. doi: 10.1182/blood-2011-09-380634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin P, Papayannopoulou T. HEL cells: a new human erythroleukemia cell line with spontaneous and induced globin expression. Science. 1982;216(4551):1233–1235. doi: 10.1126/science.6177045. [DOI] [PubMed] [Google Scholar]
- 4.Greenberg SM, Rosenthal DS, Greeley TA, Tantravahi R, Handin RI. Characterization of a new megakaryocytic cell line: the Dami cell. Blood. 1988;72(6):1968–1977. [PubMed] [Google Scholar]
- 5.Sato T, Fuse A, Eguchi M, et al. Establishment of a human leukaemic cell line (CMK) with megakaryocytic characteristics from a Down’s syndrome patient with acute megakaryoblastic leukaemia. Br J Haematol. 1989;72(2):184–190. doi: 10.1111/j.1365-2141.1989.tb07681.x. [DOI] [PubMed] [Google Scholar]
- 6.Lozzio CB, Lozzio BB. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975;45(3):321–334. [PubMed] [Google Scholar]
- 7.Bouilloux F, Juban G, Cohet N, et al. EKLF restricts megakaryocytic differentiation at the benefit of erythrocytic differentiation. Blood. 2008;112(3):576–584. doi: 10.1182/blood-2007-07-098996. [DOI] [PubMed] [Google Scholar]
- 8.Doré LC, Crispino JD. Transcription factor networks in erythroid cell and megakaryocyte development. Blood. 2011;118(2):231–239. doi: 10.1182/blood-2011-04-285981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elagib KE, Mihaylov IS, Delehanty LL, et al. Cross-talk of GATA-1 and P-TEFb in megakaryocyte differentiation. Blood. 2008;112(13):4884–4894. doi: 10.1182/blood-2008-03-145722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debili N, Coulombel L, Croisille L, et al. Characterization of a bipotent erythro-megakaryocytic progenitor in human bone marrow. Blood. 1996;88(4):1284-1296. [PubMed]
- 11.Manz MG, Miyamoto T, Akashi K, Weissman IL. Prospective isolation of human clonogenic common myeloid progenitors. Proc Natl Acad Sci USA. 2002;99(18):11872–11877. doi: 10.1073/pnas.172384399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edvardsson L, Dykes J, Olofsson T. Isolation and characterization of human myeloid progenitor populations--TpoR as discriminator between common myeloid and megakaryocyte/erythroid progenitors. Exp Hematol. 2006;34(5):599–609. doi: 10.1016/j.exphem.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 13.Kaushansky K, Lok S, Holly RD, et al. Promotion of megakaryocyte progenitor expansion and differentiation by the c-Mpl ligand thrombopoietin. Nature. 1994;369(6481):568–571. doi: 10.1038/369568a0. [DOI] [PubMed] [Google Scholar]
- 14.Debili N, Wendling F, Katz A, et al. The Mpl-ligand or thrombopoietin or megakaryocyte growth and differentiative factor has both direct proliferative and differentiative activities on human megakaryocyte progenitors. Blood. 1995;86(7):2516–2525. [PubMed] [Google Scholar]
- 15.Fox N, Priestley G, Papayannopoulou T, Kaushansky K. Thrombopoietin expands hematopoietic stem cells after transplantation. J Clin Invest. 2002;110(3):389–394. doi: 10.1172/JCI15430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Laval B, Pawlikowska P, Petit-Cocault L, et al. Thrombopoietin-increased DNA-PK-dependent DNA repair limits hematopoietic stem and progenitor cell mutagenesis in response to DNA damage. Cell Stem Cell. 2013;12(1):37–48. doi: 10.1016/j.stem.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 17.Qian H, Buza-Vidas N, Hyland CD, et al. Critical role of thrombopoietin in maintaining adult quiescent hematopoietic stem cells. Cell Stem Cell. 2007;1(6):671–684. doi: 10.1016/j.stem.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 18.Yoshihara H, Arai F, Hosokawa K, et al. Thrombopoietin/MPL signaling regulates hematopoietic stem cell quiescence and interaction with the osteoblastic niche. Cell Stem Cell. 2007;1(6):685–697. doi: 10.1016/j.stem.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 19.Abbott C, Huang G, Ellison AR, et al. Mouse monoclonal antibodies against human c-Mpl and characterization for flow cytometry applications. Hybridoma (Larchmt) 2010;29(2):103–113. doi: 10.1089/hyb.2009.0095. [DOI] [PubMed] [Google Scholar]
- 20.Deng B, Banu N, Malloy B, et al. An agonist murine monoclonal antibody to the human c-Mpl receptor stimulates megakaryocytopoiesis. Blood. 1998;92(6):1981–1988. [PubMed] [Google Scholar]
- 21.Doulatov S, Notta F, Eppert K, Nguyen LT, Ohashi PS, Dick JE. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat Immunol. 2010;11(7):585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- 22.Notta F, Zandi S, Takayama N, et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science. 2016 doi: 10.1126/science.aab2116. 351(6269):aab2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adolfsson J, Månsson R, Buza-Vidas N, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121(2):295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Månsson R, Hultquist A, Luc S, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26(4):407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto R, Morita Y, Ooehara J, et al. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell. 2013;154(5):1112–1126. doi: 10.1016/j.cell.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 26.Iwasaki H, Mizuno S, Arinobu Y, et al. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006;20(21):3010–3021. doi: 10.1101/gad.1493506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bianchi E, Bulgarelli J, Ruberti S, et al. MYB controls erythroid versus megakaryocyte lineage fate decision through the miR-486-3p-mediated downregulation of MAF. Cell Death Differ. 2015;22(12):1906–1921. doi: 10.1038/cdd.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bianchi E, Zini R, Salati S, et al. c-myb supports erythropoiesis through the transactivation of KLF1 and LMO2 expression. Blood. 2010;116(22):e99–e110. doi: 10.1182/blood-2009-08-238311. [DOI] [PubMed] [Google Scholar]
- 29.Tober J, Koniski A, McGrath KE, et al. The megakaryocyte lineage originates from hemangioblast precursors and is an integral component both of primitive and of definitive hematopoiesis. Blood. 2007;109(4):1433–1441. doi: 10.1182/blood-2006-06-031898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jamieson CH, Gotlib J, Durocher JA, et al. The JAK2 V617F mutation occurs in hematopoietic stem cells in polycythemia vera and predisposes toward erythroid differentiation. Proc Natl Acad Sci USA. 2006;103(16):6224–6229. doi: 10.1073/pnas.0601462103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muellner MK, Uras IZ, Gapp BV, et al. A chemical-genetic screen reveals a mechanism of resistance to PI3K inhibitors in cancer. Nat Chem Biol. 2011;7(11):787–793. doi: 10.1038/nchembio.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu J, Guo S, Ebert BL, et al. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell. 2008;14(6):843–853. doi: 10.1016/j.devcel.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng J, Guo S, Chen S, et al. An extensive network of TET2-targeting MicroRNAs regulates malignant hematopoiesis. Cell Reports. 2013;5(2):471–481. doi: 10.1016/j.celrep.2013.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mori Y, Chen JY, Pluvinage JV, Seita J, Weissman IL. Prospective isolation of human erythroid lineage-committed progenitors. Proc Natl Acad Sci USA. 2015;112(31):9638–9643. doi: 10.1073/pnas.1512076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McClinton D, Stafford J, Brents L, Bender TP, Kuehl WM. Differentiation of mouse erythroleukemia cells is blocked by late up-regulation of a c-myb transgene. Mol Cell Biol. 1990;10(2):705–710. doi: 10.1128/mcb.10.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosson D, O’Brien TG. Constitutive c-myb expression in K562 cells inhibits induced erythroid differentiation but not tetradecanoyl phorbol acetate-induced megakaryocytic differentiation. Mol Cell Biol. 1995;15(2):772–779. doi: 10.1128/mcb.15.2.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L, Kostadima M, Martens JH, et al. BRIDGE Consortium. Transcriptional diversity during lineage commitment of human blood progenitors. Science. 2014;345(6204):1251033. doi: 10.1126/science.1251033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404(6774):193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 39.Haas S, Hansson J, Klimmeck D, et al. Inflammation-induced emergency megakaryopoiesis driven by hematopoietic stem cell-like megakaryocyte progenitors. Cell Stem Cell. 2015;17(4):422-434. [DOI] [PubMed]
- 40.Mercier FE, Scadden DT. Not all created equal: lineage hard-wiring in the production of blood. Cell. 2015;163(7):1568–1570. doi: 10.1016/j.cell.2015.12.013. [DOI] [PubMed] [Google Scholar]