Abstract
Background
Pectin-rich wastes, such as citrus pulp and sugar beet pulp, are produced in considerable amounts by the juice and sugar industry and could be used as raw materials for biorefineries. One possible process in such biorefineries is the hydrolysis of these wastes and the subsequent production of ethanol. However, the ethanol-producing organism of choice, Saccharomyces cerevisiae, is not able to catabolize d-galacturonic acid, which represents a considerable amount of the sugars in the hydrolysate, namely, 18 % (w/w) from citrus pulp and 16 % (w/w) sugar beet pulp.
Results
In the current work, we describe the construction of a strain of S. cerevisiae in which the five genes of the fungal reductive pathway for d-galacturonic acid catabolism were integrated into the yeast chromosomes: gaaA, gaaC and gaaD from Aspergillus niger and lgd1 from Trichoderma reesei, and the recently described d-galacturonic acid transporter protein, gat1, from Neurospora crassa. This strain metabolized d-galacturonic acid in a medium containing d-fructose as co-substrate.
Conclusion
This work is the first demonstration of the expression of a functional heterologous pathway for d-galacturonic acid catabolism in Saccharomyces cerevisiae. It is a preliminary step for engineering a yeast strain for the fermentation of pectin-rich substrates to ethanol.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-016-0544-1) contains supplementary material, which is available to authorized users.
Keywords: Ethanol, d-galacturonic acid, Saccharomyces cerevisiae, Citrus pulp, Metabolic engineering
Background
Citrus pulp and sugar beet pulp are pectin-rich wastes that are produced in considerable amounts in various countries. Citrus pulp results from the production of orange juice concentrate. Its production is concentrated mainly in Brazil and the USA, which have a share of 54 and 26 % of the global market, respectively [1]. In the 2014/2015 harvest, 1.4 million tons of orange juice concentrate (65° Brix) were produced in these two countries [1], and this amount would result in the production of about 1.9 million metric tons of citrus pulp (dry matter). Likewise, large amounts of sugar beet pulp are generated during the extraction of sugar from sugar beets in temperate countries, especially Russia, the USA and several European countries [2]. In the 2013 harvest, a total of 246 million metric tons of sugar beet were produced worldwide [2], resulting in 12.3 million metric tons of beet pulp (dry matter).
Citrus pulp and sugar beet pulp are usually sold for incorporation in cattle feed, but the costs of drying these wastes make this application barely profitable [3]. On the other hand, these wastes are potentially important sources of carbohydrates that can be used as raw materials in biorefineries for the production of bio-based chemicals and biofuels. In fact, there is a rising demand for ethanol, especially in Brazil and the USA, and biorefineries using citrus wastes or sugar beet pulp could meet part of this demand [4, 5]. However, hydrolysates obtained from pectin-rich wastes have high contents of d-galacturonic acid, which represents 18 % (w/w) of citrus waste hydrolysates [6] and 16 % (w/w) of sugar beet pulp hydrolysates [7]. Unfortunately, this uronic acid is not catabolized by Saccharomyces cerevisiae, which is the microorganism of choice for ethanol production.
Conversely, many microorganisms that are able to use d-galacturonic acid do not produce ethanol in appreciable amounts. Escherichia coli, for example, has the isomerase pathway for d-galacturonic acid catabolism but, due to low ethanol and inhibitor tolerance, it is not the preferred organism for ethanol production. Alternatively, ethanol production with S. cerevisiae is already a highly productive and robust process. This yeast is preferred for commercial scale ethanol production for several reasons, including the resistance to contaminants, bacteriophages, inhibitors and low pH [8]. It also tolerates higher osmotic pressures, enabling the use of a concentrated culture medium, and greater concentrations of ethanol. Considering this, the expression of a heterologous pathway for catabolism of d-galacturonic acid into S. cerevisiae, instead of engineering bacteria for ethanol production, is the preferred path for industrial ethanol production from hydrolysates containing d-galacturonic acid.
Different approaches have been used to engineer S. cerevisiae for this purpose. Huijes et al. [9], for instance, introduced by integration into the chromosomes the five genes of the bacterial isomerase pathway (uxaC, uxaB, uxaA, kdgK and kdgA), which converts d-galacturonic acid to pyruvate and glyceraldehyde-3-phosphate. Although Huijes et al. [9] demonstrated by qPCR that all genes were transcribed, only two enzymes of the pathway showed detectable activity after integration in S. cerevisiae. In another approach, Souffriau [10] expressed the fungal pathway for d-galacturonic acid catabolism in S. cerevisiae by cloning the four genes of the pathway (gar1, lgd1, lga1 and gld1, from Trichoderma reesei) in two plasmids with the gap-DH-ADH1 bidirectional promoter. In this work the activity of all the heterologous enzymes were detected in the cell lysate. Despite this, the recombinant yeast strain did not consume d-galacturonic acid.
In the present work, we extended the work of Soufriau [10] by integrating to the yeast chromosomes the four genes of the reductive fungal d-galacturonic catabolic pathway; in addition, we also integrated of the recently discovered d-galacturonic acid transporter from Neurospora crassa, in order to engineer a yeast that is able to use d-galacturonic acid as a carbon source. This work represents an important step in the construction of a S. cerevisiae strain able to produce ethanol from d-galacturonic acid.
Results
Pathway assembly and gene expression
The fungal pathway for catabolism of d-galacturonic acid was expressed in Saccharomyces cerevisiae strain CEN.PK111-61A, with the genes being selected from three different filamentous fungi, Aspergillus niger, Trichoderma reesei and Neurospora crassa.
As a first step, four genes encoding the catabolic d-galacturonate pathway in A. niger (gaaA, gaaB, gaaC and gaaD) were integrated into the yeast chromosomes, with each one being expressed under strong and constitutive yeast promoters (PGK1 or TPI1). The corresponding enzyme activities of gaaA, gaaC and gaaD were detected in the cell lysate. The corresponding activity of gaaB, namely l-galactonate dehydratase, was not detectable, even when a codon-optimized gaaB ORF was used. This result was unexpected since gaaB activity was demonstrated when expressed in a multi copy expression vector [11]. Considering this, lgd1, an l-galactonate dehydratase encoding gene from T. reesei, was used instead. The resulting strain (H4531), having the complete pathway (gaaA, gaaC, gaaD and lgd1) integrated, was then re-tested for the activity of all four enzymes. The enzymatic activities obtained from H4531 are listed in Table 1.
Table 1.
Enzyme activities assayed from S. cerevisiae H4531 cell lysate
| Enzyme | Gene | Spec. Act. (nkat/mg) |
|---|---|---|
| EC 1.1.1.365 | gaaA | 0.246 |
| EC 4.2.1.146 | lgd1 | 0.018 |
| EC 4.1.2.B7 | gaaC | 0.274 |
| EC 1.1.1.372 | gaaD | 1.139 |
During the course of the study, a d-galacturonic acid transporter (coded by gat1) was described in N. crassa [12]. Although a previous study reported that native S. cerevisiae is able to import d-galacturonic acid when grown at acidic pH values [13], the introduction of a transporter might improve the intake, especially at higher pH values. For this reason, gat1-gfp fusion protein gene was integrated into H4531, resulting in the strain H4535. The location of GAT1 was confirmed by fluorescence microscopy, with the green fluorescence of GFP being observed in the plasma membrane.
d-galacturonic acid consumption
Both engineered strains, H4531 and H4535, were cultivated for 5 days under aerobic conditions in YP medium, supplemented with 12 g L−1 of d-galacturonic acid. The S. cerevisiae strain CEN.PK113-1A, which does not have auxotrophies, was used as a control. Even after adaptive laboratory evolution, the recombinant strains, as well as the control strain grew poorly and did not consume d-galacturonic acid in this condition.
A second fermentation was therefore done, with the difference that the medium was supplemented with 80 g L−1 of d-fructose (Fig. 1). This time, 20 % of the d-galacturonic acid was consumed by the yeast strain H4535, which expressed the four genes of the reductive pathway and the transporter for d-galacturonic acid. Notably, most of this consumption was observed in the first 24 h, and corresponds to the complete utilization of the d-fructose by the yeast. Consumption of d-galacturonic acid was negligible by the control strain and by the recombinant strain expressing the reductive pathway, but lacking the transporter. We could also exclude that d-galacturonic acid was simply converted to l-galactonate or other galacturonate pathway intermediates since these metabolites were not detected in neither in the culture medium nor in the cell extract. Besides the clear difference in the d-galacturonic acid consumption, every other parameter measured, such as ethanol, biomass, pH and d-fructose, did not differ significantly among the strains.
Fig. 1.
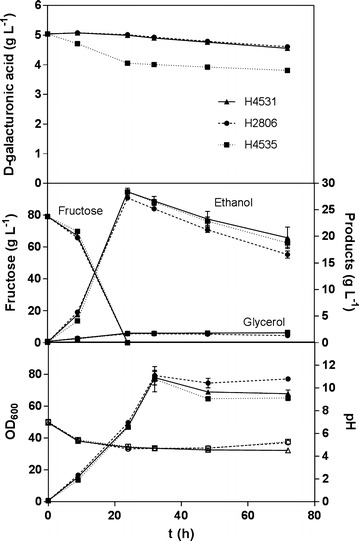
Cultivation of yeast strains in d-galacturonic acid and d-fructose. The control is H2806 (CEN.PK 113-1A), H4531 contains the genes gaaA, lgd1, gaaC and gaaD and H4535 contains the genes gaaA, lgd1, gaaC, gaaD and gat1. a d-galacturonic acid consumption. b Ethanol and glycerol production, fructose consumption. c Biomass production (closed markers) and pH (open markers). Error bars represent the standard error of the mean
To further analyse the role of d-fructose as a co-substrate that enables d-galacturonic acid consumption, an additional fermentation was performed. Here d-fructose was added in two batches of 40 g L−1, being the second at 72 h (Fig. 2). With this experiment was possible to observe that an additional galacturonic acid consumption followed the addition of fructose in 72 h.
Fig. 2.
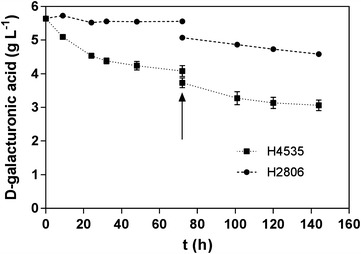
Cultivations in d-galacturonic acid and d-fructose, with a second addition of d-fructose. The control is H2806 (CEN.PK 113-1A) and H4535 contains the genes gaaA, lgd1, gaaC, gaaD and gat1. The fermentation was carried out with d-fructose as co-substrate, with a second load of d-fructose being added at 72 h of fermentation. d-galacturonic acid consumption was monitored over time. Error bars represent the standard error of the mean
Finally, to prove that the galacturonic acid was definitely being catabolized by the engineered yeast, and not only converted to e.g. l-galactonate, the fate of this sugar was monitored by NMR, using the uniformly labeled d-[UL-13C6] galacturonate as substrate. For that, a fermentation was carried out with 40 g L−1d-fructose and 4 g L−1 of d-galacturonic acid, with the labeled substrate corresponding to half of the total amount of d-galacturonic acid. Again, a control strain without the pathway was fermented in the same conditions. Figure 3a, b shows the NMR spectrum for the supernatant of this fermentation at 24 h, with a corresponding to the spectrum of the supernatant from the control strain and B from the engineered strain with the fungal pathway and the transporter. In Fig. 3b, there is a clear signal of uniformly labeled glycerol showing the 13C-13C scalar coupling fine structure, meaning that the carbon backbone was originally from the labeled d-galacturonic acid. In Fig. 3a this fully labeled glycerol is not detected in the spectrum. Glycerol is a product of the d-galacturonic acid pathway and of sugar catabolism in yeast, and was detected by HPLC for both control and engineered strains in the previous experiments. The detection of uniformly 13C-labeled glycerol already confirms that d-galacturonic acid is metabolized by the engineered strain, and not by the control. However, to further assure that the d-galacturonic acid is being metabolized, the yeast cells were collected after 96 h of fermentation and hydrolyzed. The spectrum of the resulting amino acids was analyzed for both the control strain (Fig. 3c) and the engineered strain (Fig. 3d). Also here we observed the 13C-13C scalar coupling fine structure in the sample of the engineered strain, proving that the labeled carbon is coming from the d-galacturonic acid. The alfa carbon of several amino are 13C-labeled and have 13C-labeled neighbors in the engineered strain, but not in the control strain, which is a proof that d-galacturonic acid is catabolized and ends up in the biomass of the engineered strain.
Fig. 3.
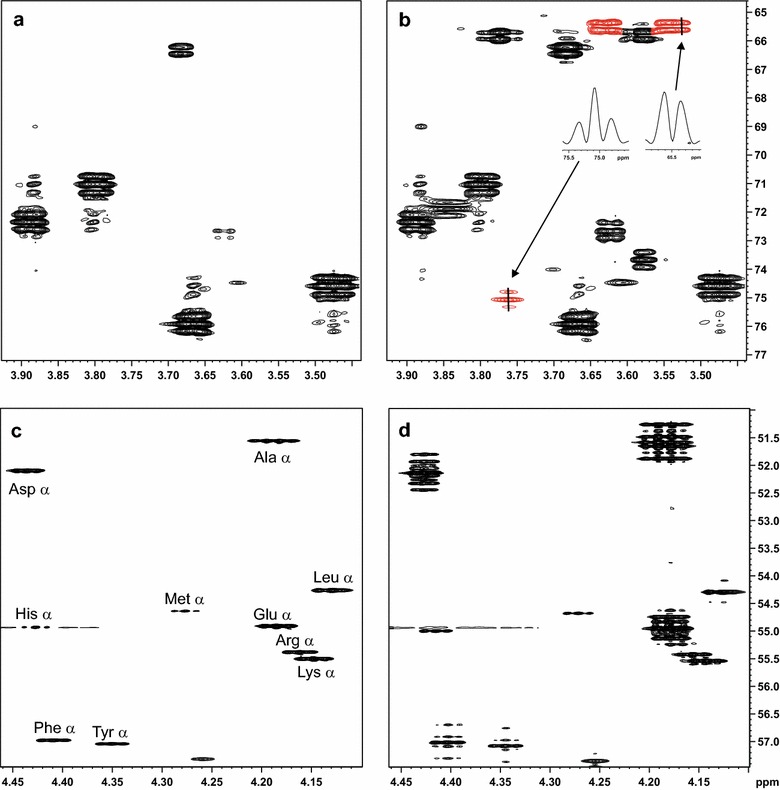
NMR analysis of the 13C tracing fermentations. Expansions of HSQC spectra of a the culture supernatant of the control strain H2806 (CEN.PK 113-1A) and b the strain H4535 containing the genes gaaA, lgd1, gaaC, gaaD and gat1. Signals of glycerol produced by H4535 are highlighted in red and the inserts show cross sections of these signals along F1, revealing the 13C-13C scalar coupling fine structure. These signals were not detected for the control strain. Part of the alpha carbon region of HSQC spectra of biomass hydrolysate of c the control strain and d the engineered strain, showing the 13C-13C scalar coupling fine structure in most signals
Discussion
In this work, we demonstrate for the first time a functional catabolic pathway for d-galacturonate consumption by S. cerevisiae. For this purpose, the fungal d-galacturonic acid pathway and the d-galacturonic acid transporter were integrated into the yeast chromosomes and the corresponding enzyme activities were shown in the cell lysate, and with the d-galacturonic acid transporter being located in the cell membrane. The fermentation trials with d-fructose as co-substrate showed that the engineered strain was, furthermore, able to catabolize d-galacturonic acid. The tracing of the 13C from uniformly labeled d-galacturonic acid showed that these carbons end up in products of the fermentation, particularly glycerol, and in the amino acids of the biomass.
The fungal pathway for d-galacturonic acid catabolism was recently discovered by Hilditch et al. [14] and confirmed by Martens-Uznova et al. [15]. It consists of four enzymes, with two reduction steps (Fig. 4). The first enzyme of the pathway, d-galacturonate reductase (EC 1.1.1.365), coded by gaaA in A. niger, converts d-galacturonate into l-galactonate, using NADPH or NADH as the electron donor. l-galactonate dehydratase (EC 4.2.1.146), coded by gaaB in A. niger and by lgd1 in T. reesei, converts l-galactonate into 2-keto-3-deoxygalactonate. The third enzyme, 2-keto-3-deoxygalactonate aldolase (EC 4.1.2.B7), coded by gaaC in A. niger, cuts 2-keto-3-deoxygalactonate between carbons 3 and 4, producing pyruvate and l-glyceraldehyde. The pyruvate can enter the citric acid cycle, while the l-glyceraldehyde is converted to glycerol by glyceraldehyde reductase (EC 1.1.1.372, encoded by gaaD in A. niger) using NADPH as the electron donor.
Fig. 4.
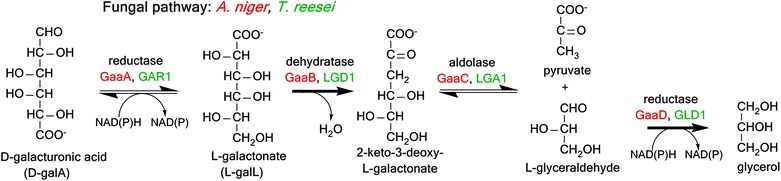
Fungal pathway for the catabolism of d-galacturonic acid
Recently, Benz [12] described the d-galacturonic acid transporter, which was suggested to act as an H+/d-galacturonic acid symporter. The corresponding gene was discovered in Neurospora crassa, and to further confirm its functionality, the transporter was also co-expressed in yeast with the first enzyme of the fungal pathway, galacturonate reductase (gaaA).
Souffriau [10], similarly to what we have done, also expressed the genes of the fungal pathway from Trichoderma reesei (gar1, lgd1, lga1 and gld1) in S. cerevisiae. Differently from our approach, the four genes were expressed from two plasmids with the gap-DH-ADH1 bidirectional promoter. Despite the fact that the activity of all the enzymes was shown in the cell lysates, the recombinant yeast strain did not grow on d-galacturonic acid. Further adaptive laboratory evolution, in a medium containing both glycerol and d-galacturonic acid, did not lead to mutants that were able to use d-galacturonic acid.
Similarly to Souffriau [10], we chose the reductive pathway for introduction into S. cerevisiae, since previous work showed that the individual enzymes could be expressed in this yeast [8, 15–17]. However, our approach differs from that of Souffriau [10] in four ways: First, we integrated the genes into the yeast genome, instead of using plasmid vectors; second, we used genes from both T. reesei and A. niger; third, we also integrated a d-galacturonic acid transporter [12]; and, fourth, we used d-fructose, instead of glycerol, as a co-substrate for growth of the recombinant yeast. Interestingly, both Souffriau [10] and we detected the activities of the d-galacturonic acid pathway enzymes in the cell lysate, but our use of hexose sugar as a co-substrate, rather than the glycerol used by Souffriau [10] and also the use of a d-galacturonic acid transporter may be the reasons why we were able to demonstrate d-galacturonic consumption. Additionally, the use of the A. nigerd-galacturonate reductase GAAA, which accepts both NADH and NADPH as a cofactor, instead of the T. reesei reductase, which strictly uses only NADPH, could also have enabled the observed d-galacturonic acid consumption.
Although the engineered strain was able to catabolize d-galacturonic acid, the strain is still not optimized for industrial processes using pectin-rich waste hydrolysates. Due to the fact that the fungal pathway requires reducing power that is also needed for ethanol production, further engineering of the strain and optimization of the cultivation conditions are still needed to address the redox balance of this process.
Conclusion
In this work, we describe for the first time a strategy for the introduction of a functional pathway that resulted in d-galacturonic acid catabolism in Saccharomyces cerevisiae. This strategy was based on the integration into the yeast chromosomes of not only the four enzymes of the fungal pathway of d-galacturonic acid catabolism, but also the recently described d-galacturonic acid transporter. This represents the first step in the construction of a strain of S. cerevisiae that would be able to produce ethanol from d-galacturonic acid. Such a strain would find application in a citrus/sugar beet waste biorefinery, in which pectin-rich wastes would be hydrolyzed and then fermented to produce ethanol.
Methods
Strains
The S. cerevisiae strain CEN.PK111-61A was used as a parental strain for the engineered strains. The prototrophic CEN.PK113-1A was used as a control in the fermentations. All the plasmids were produced in E. coli TOP10 cells. Table 2 shows the original and engineered S. cerevisiae strains used in this work.
Table 2.
Strains used in this work
| Strains | Description | Parent strain |
|---|---|---|
| Escherichia coli TOP10 | E. coli for electroporation hsdR, mcrA, lacZΔM15, recA | |
| CEN.PK111-61A | S. cerevisiae CEN.PK 111-61A MATα, ura3-52, his3-Δ1, leu2-3112, TRP1, MAL28c, SUC2 | |
| CEN.PK113-1A | S. cerevisiae CEN.PK 113-1A MATα, URA3, HIS3, LEU2, TRP1, MAL28c, SUC2 | |
| S. cerevisiae ATCC 90845 | MATα, his3Δ 200, ura3-52, leu2 Δ 1, lys2-Δ 202, trp1-Δ63 | |
| A. niger ATCC 1015 | Template DNA for gene gaaC | |
| Schizosaccharomyces pombe | Template DNA for gene his5 | |
| H4362 | S. cerevisiae CEN.PK 111-61A+ gaaC (MATα, ura3-52, HIS5, leu2-3112, TRP1, MAL28c, SUC2) | S. cerevisiae CEN.PK 111-61A |
| H4410 | S. cerevisiae CEN.PK 111-61A + gaaC + gaaD (MATα, ura3-52, HIS5, LEU2, TRP1, MAL28c, SUC2) | H4362 |
| H4425 | S. cerevisiae CEN.PK111-61A + gaaC + gaaD + gaaA (MATα, URA3, HIS5, LEU2, TRP1, MAL28c, SUC2) | H4410 |
| H4531 | S. cerevisiae CEN.PK111-61A + gaaC + gaaD + gaaA + lgd1 (MATα, URA3, HIS5, LEU2, TRP1, MAL28c, SUC2, can1−) | H4425 |
| H4535 | S. cerevisiae CEN.PK111-61A + gaaC + gaaD + gaaA + lgd1 + gat1-gfp (MATα, URA3, HIS5, LEU2, TRP1, MAL28c, SUC2, can1− , HO −) | H4531 |
Media and culture conditions
For plasmid multiplication, bacterial strains were cultivated in Luria Broth medium [18], supplemented with 100 μg mL−1 of ampicillin, at 37 °C and 250 rpm. For yeast transformations, several media were used. SCD: synthetic complete medium supplemented with 20 g L−1d-glucose [19]. SCD-URA: uracil deficient synthetic complete medium, supplemented with 20 g L−1d-glucose. SCD-HIS: histidine deficient synthetic complete medium, supplemented with 20 g L−1d-glucose. SCD-LEU: leucine deficient synthetic complete medium, supplemented with 20 g L−1d-glucose. SCD-URA/-HIS/-LEU: uracil, histidine and leucine deficient synthetic complete medium, supplemented with 20 g L−1d-glucose. YPD: 10 g L−1 yeast extract, 20 g L−1 peptone, 20 g L−1d-glucose [20]. YPD + G418: YPD supplemented with 200 μg mL−1 geneticin. YPD+ nourseothricin: YPD supplemented with 100 μg mL−1 nourseothricin.
Plasmid construction and gene integrations
Plasmids are listed in Table 3. Primers used for plasmid construction are listed in Table 4. All the PCR reactions were performed using Phusion High Fidelity (Finnzymes, Finland) and Phusion High Fidelity buffer or GC Buffer (Finnzymes, Finland). For bacterial and yeast colony PCR, DyNAzyme (Finnzymes, Finland) was used instead. For yeast colony PCR, cell disruption was carried out using Zymolyase 100 T (Seikagaku Biobusiness, Japan). Ligations were done using T4 DNA ligase (Promega, USA). Recombination in yeast for plasmid assembly or genome integration was done using Lab Transformation kit (Molecular Research Reagents Inc., USA), following the protocol of Gietz et al. [21]. Recombination in vitro was done using Gibson Assembly® Master Mix (New England Biolabs, USA).
Table 3.
Plasmids used in this work
| Plasmid | Description |
|---|---|
| B1181 | YEplac195 with PGK1 promoter, URA3, Amp R |
| pRS426 | Yeast integration vector, URA3, Amp R |
| B5430 | B1181 ligated to gaaC. Contains fragment 3 (PGK1/gaaC) of the gaaC integration vector in between bglII restriction sites |
| B5517 | Yeast integration vector created by homologous recombination in yeast. Contains the gene gaaC under PGK1 promoter, the integration locus HIS3 and marker HIS5 (S. pombe) in between NotI restriction sites |
| B5470 | pXY212 expressing the gene gaaD under TPI1 promoter |
| pRS405 | Yeast integration vector, LEU2, Amp R |
| B5555 | Yeast integration vector pRS405 expressing LEU2 and gaaD under TPI1 promoter in between AflII restriction sites |
| B2159 | pXY212, containing TPI1 promoter region, Amp R |
| B5706 | Genescript pUC57 plasmid containing codon-optimized gaaA in between EcoRI and BamHI restriction sites |
| B5696 | B2159 ligated to gaaA under TPI promoter |
| pRS406 | Yeast integration vector containing URA3, Amp R |
| B5697 | Yeast integration vector containing URA3 and gaaA under TPI1 promoter in between AflII restriction sites |
| B3033 | Yeast integration vector containing CAN1 locus, KnMX and lgd1 under PGK1 promoter in between LoxP sites and SaclL and Kpnl restriction sites |
| pSH66 | Deletion vector containing Cre, Amp R |
| B6367 | B2159 containing gat1-gfp under TPI1 promoter |
| B3531 | Yeast integration vector containing KnMX marker, HO locus |
| B6382 | B3531 yeast integration vector containing gat1-gfp under TPI1 promoter |
Table 4.
Primers used in this work
| Primer | Sequence | Description |
|---|---|---|
| P1 | ATGCCTTTTACCCCGCTCCG | For gaaC amplification from A. niger genome, for colony PCR and sequencing (forward) |
| P2 | CTAAGCAATATCCGGCAACG | For gaaC amplification from A. niger genome, for colony PCR and sequencing (reverse) |
| P3 | CGGGGGATCCACTAGTTCTAGAGCGGCCGCGTGAGGGTCAGTTATTTCAT | For fragment 1 (−1000 bp HIS3 locus) amplification from S. cerevisiae (forward) |
| P4 | TATTTCTTTCTACAAAAGCCCTCCTACCCATCTTTGCCTTCGTTTATCTTG | For fragment 1 (−1000 bp HIS3 locus) amplification from S. cerevisiae (reverse) |
| P5 | TAACTCGAAAATTCTGCGTTCGTTAAAGCTAGCTGCAGCATACGATATAT | For fragment 4 (+1000 bp HIS3 locus) amplification from S. cerevisiae (forward) |
| P6 | AAGCTGGAGCTCCACCGCGGTGGCGGCCGCGGAGCCATAATGACAGCAGT | For fragment 4 (+1000 bp HIS3 locus) amplification from S. cerevisiae (reverse) |
| P7 | AATGAGCAGGCAAGATAAACGAAGGCAAAGATGGGTAGGAGGGCTTTTGT | For fragment 2 (his5) amplification from S. pombae (forward) |
| P8 | TTCAGTTTTGGATAGATCAGTTAGAAAGCTATTAAGGGTTCTCGAGAGCT | For fragment 2 (his5) amplification from S. pombae (forward) |
| P9 | GGAAGATATGATCTACGTATGGTCATTTCTTC | For TPI1/gaaD amplification from B5470 (forward) |
| P10 | GGAGATCTCGAATTGGAGCTAGAGAAAG | For TPI1/gaaD amplification from B5470 (reverse) |
| P11 | GATCTACGTATGGTCATTTCTTC | For colony PCR and sequencing gaaD ORF (forward) |
| P12 | TCGAATTGGAGCTAGACAAAG | For colony PCR and sequencing gaaD ORF (reverse) |
| P13 | ATGGCTCCCCCAGCTGTGTT | For colony PCR and sequencing gaaA ORF (forward) |
| P14 | CTACTTCAGCTCCCACTTTC | For colony PCR and sequencing gaaA ORF (reverse) |
| P15 | CCTCGCACCCATGTACATTGG | For colony PCR and sequencing gat1 ORF (forward) |
| P16 | TTATATTGGCCTTTATGTCCGC | For colony PCR and sequencing gat1 ORF (reverse) |
| P17 | TATATACCCGGGGTGCCACCTGACGTCTAAGA | For amplification of TPI1/gat1-gfp from B6367 (forward) |
| P18 | TATATACCCGGGAGACCGAGATAGGGTTGAGT | For amplification of TPI1/gat1-gfp from B6367 (reverse) |
gaaC integration
The gaaC integration cassette was designed to have the gaaC gene under the PGK1 promoter (fragment 3), the flanking regions of S. cerevisiae HIS3 gene (fragments 1 and 4) used as homologous regions for genome integration, and the S. pombe HIS5 gene (yeast marker, fragment 2) assembled circularly in the pRS426 vector (fragment 5), which contains a URA3 marker.
gaaC was directly amplified from A. niger ATCC 1015 genome. It was inserted into the PGK1 promoter region of B1181 plasmid using Gibson Assembly. The resulting plasmid, B5430, contains the PGK1/gaaC component (fragment 3) of the gaaC integration cassette and was digested with BglII to obtain fragment 3. Fragment 1, which corresponds to the-1000 bp downstream region of S. cerevisiae his3, and fragment 4, which corresponds to the upstream +1000 bp region of his3, were amplified from the S. cerevisiae (H3488) genome. HIS5 (fragment 2) was amplified from the S. pombae genome. The pRS426 vector was linearized by digestion with NotI (integration vector). The five fragments from the gaaC integration plasmid were assembled by recombination in yeast. The URA3 marker was used for the selection of the assembled plasmid and the transformants were selected in SCD-URA plates. The assembled plasmid was introduced to E. coli by electroporation for multiplication and digested with NotI for the isolation of the cassette (fragments 1 to 4). The integration of the gaaC cassette into the parental strain CEN.PK111-61A was done by recombination. The HIS5 marker was used for selection of the gaaC-integrated yeast and transformants were selected in SCD-HIS and confirmed by colony PCR. Activity was confirmed using the cell lysate. A colony expressing gaaC was stored as H4362.
gaaD integration
The gaaD was amplified from cDNA and restricted with BglII and ligated to the B2159 plasmid in the TPI promoter region. gaaD under TPI1 promoter was amplified and ligated to the vector pRS405 (containing LEU2 yeast marker) to form B5555 integration vector. The gaaD integration cassette with the LEU2 marker was obtained by the linearization of B5555 using AflII. The gaaD cassette was integrated into the gaaC-expressing yeast strain H4362, in the leu2-3112 locus. Transformants were selected in SCD-LEU and confirmed by colony PCR. The activity of the gaaD gene was confirmed using the cell lysate. A colony expressing gaaC and gaaD was stored as H4410.
gaaA integration
The gaaA gene was custom-synthetized as a codon-optimized ORF (Genescript). Genescript gaaA plasmid was digested with EcoRI and BamHI and ligated to B2159 plasmid in the TPI1 promoter region. The integration plasmid B5697 was obtained by ligating the TPI1/gaaA fragment to the vector pRS406 (B704), which contains the URA3 yeast marker. The gaaA integration cassette was obtained by the linearization of B5697 using BsmI. The cassette was then integrated into the ura3-52 locus of the yeast strain H4410 (gaaC + gaaD). Transformants were selected in SCD-URA medium and confirmed by colony PCR. Activity was confirmed using the cell lysate. A colony expressing gaaC, gaaD and gaaA was stored as H4425.
lgd1 integration
The integration plasmid B3013 carrying the lgd1 gene ready for integration in yeast was available from previous work [22]. This integration cassette consists of two flanking regions for CAN1, the lgd1 gene under the PGK1 promoter, and the KanMX yeast marker (resistance to G418) between two loxP sites. The plasmid was digested with SacI and KpnI and the integration cassette was transformed to the yeast strain H4425 (gaaA + gaaC + gaaD). Before plating, the yeast cells were incubated in 2 mL of SCD for 1 h. Transformants were selected in YPD + G418 and confirmed by assaying lgd1 activity of the cell lysate. The KanMX marker was then removed by transforming the selected yeast colony with plasmid pSH66, expressing cre recombinase, which removes KanMX yeast marker between the loxP sites [23]. Before plating in YPD + nourseothricin, the yeast cells were incubated in 2 mL of SCD for 1 h. After 4 days, the resulting colonies were re-plated in YPD + nourseothricin. Biomass from this plate was inoculated into 50 mL of YP + 2 % galactose and incubated overnight. The resulting culture was plated in YPD. Some isolated colonies were replated in G418 plates and SCD-URA/-LEU/-HIS medium. After 2 days, none of the colonies grew on G418, confirming that the KanMX marker had been removed. A colony expressing gaaC, gaaD, gaaA and lgd1 was stored as H4531.
gat1-gfp integration
The gat1-gfp gene was custom-synthetized based on the work of Benz [12] as a codon-optimized ORF (Genescript) The gat1-gfp fragment was digested from the Genescript plasmid and ligated to B2159 plasmid, in the TPI1 promoter region. For the integration cassette, the TPI1/gat1-gfp fragment was digested with XmaI. The vector B3531, which contains HO integration region and KanMX yeast marker, was digested with Cfr9I and ligated to the TPI1/gat1-gfp fragment. The gat1-gfp integration cassette was obtained by the digestion of the integration plasmid with XhoI and XbaI and transformed to the H4531 strain. The yeast cells were incubated in 2 mL of YPD for 2 h and then plated in YPD + G418 plates. To confirm the integration of the gat1-gfp gene, cells were incubated in SCD medium for 24 h and analysed using a fluorescence microscope. A colony showing green fluorescence (from the gfp reporter gene) was stored as H4535.
Enzymatic assays
Cell extracts, obtained as follows, were used in all the enzymatic assays. Yeast cells were grown in YPD medium and collected by centrifugation, washed with water and re-suspended in phosphate buffer 50 mM, pH 7 with addition of protease inhibitor Complete EDTA Free (Roche, Switzerland). Cells were disrupted with 0.4 mm diameter glass beads using a bead beater FastPrep (MP Biomedicals, USA) and solid residues were removed by centrifugation. Protein content was assayed by the Bradford method [24].
d-Galacturonate reductase (GAAA) and l-glyceraldehyde reductase (GAAD) activities were assayed in the forward direction by following the decrease in absorbance at 340 nm caused by the oxidation of NADPH. The reaction medium for d-galacturonate reductase contained 10 mM d-galacturonic acid (Sigma-Aldrich, Germany) and 0.2 mM NADPH (Sigma-Aldrich, Germany). The reaction medium for l-glyceraldehyde reductase contained 5 mM l-glyceraldehyde (Sigma-Aldrich, Germany) and 0.2 mM NADPH (Sigma-Aldrich, Germany). The reactions of the d-galacturonate reductase and the l-glyceraldehyde reductase were followed for 5 min.
l-Galactonate dehydratase (LGD1) and 2-keto-3-deoxy-galactonate aldolase (GAAC) activities were indirectly assayed by measuring the absorbance at 549 nm, corresponding to a chromogenic compound that forms when thiobarbituric acid combines with 2-keto-3-deoxy-galacturonate [25]. The reaction medium for l-galactonate dehydratase contained 10 mM l-galactonic acid (obtained by hydrolysis of l-galactono-1,4-lactone, Sigma-Aldrich, Germany) and proceeded for 2 h. The reaction medium for 2-keto-3-deoxy-galactonate aldolase contained 10 mM l-glyceraldehyde (Sigma-Aldrich, Germany) and 10 mM pyruvate (Sigma-Aldrich, Germany) and proceeded for 30 min. l-Galactonate dehydratase was assayed in the forward direction. 2-keto-3-deoxy-galactonate aldolase was assayed in the reverse direction as its substrate, 2-keto-3-deoxy-galacturonate, is not commercially available.
Fermentations trials
In the adaptive laboratory evolution of the transformed yeast, the medium used was YP (10 g L−1 yeast extract, 20 g L−1 peptone) pH 4,5, supplemented with 12 g L−1d-galacturonic acid. For assaying d-galacturonic acid consumption by transformed yeast in co-fermentation with d-fructose the medium used was YP pH 7 supplemented with 5 g L−1d-galacturonic acid and with 80 g L−1d-fructose. For evaluating the role of d-fructose in the d-galacturonic acid catabolism the medium used was initially YP pH 7 supplemented with 5 g L−1d-galacturonic acid and with 40 g L−1d-fructose. After 72 h, a concentrated d-fructose solution (40 % w/v) was added to result in the concentration of approximately 40 g L−1d-fructose. In the 13C tracing experiments, potassium d-[UL-13C6] galacturonate (Omicron Biochemicals, Inc., USA) was used for tracing the fate of d-galacturonic acid. The experiment was carried out with 2 g L−1 of the labeled d-galacturonic acid, 2 g L−1 of non-labeled d-galacturonic acid and 40 g L−1 of d-fructose in YP medium (neutral pH). Samples of the supernatant were taken 12 h time intervals and the biomass was collected in 96 h of fermentation. All the fermentations were carried out aerobically using 250 rpm and 30 °C. The CEN.PK 113-1A strain, which does not contain the pathway or transporter, and with similar genetic background, was used as a control in all the experiments.
Adaptive laboratory evolution
Adaptive laboratory evolution is a technique that aims for the selection of microorganisms more adapted to grow in a certain type of medium, for example, media containing toxic compounds or non-metabolizable sugars [26]. In this work, adaptive laboratory evolution was used for the selection of mutants more suited for growing with d-galacturonic acid as a carbon source. This study was carried out with the strains H4531, and H4535, both carrying the reductive fungal pathway, with the H4535 also containing the transporter gene. The cultivation was done in YP medium pH 4.5, supplemented with 12 g L−1d-galacturonic acid. A dilution of 1:50 was made in fresh medium every 7 days. The cultivations continued for 8 weeks and were analysed by HPLC, as described below.
Chemical analysis
Samples of liquid fermentation broth were taken at 12-48 h or 7-days (adaptive laboratory evolution) intervals. The concentrations of d-galacturonic acid, d-fructose and ethanol were determined by HPLC using a Fast Acid Analysis Column (100 mm × 7.8 mm, BioRad Laboratories, CA, USA) linked to an Aminex HPX-87H organic acid analysis column (300 mm × 7.8 mm, BioRad Laboratories) and 5 mM H2SO4 as eluent with the flow rate of 0.5 ml min−1. The column was maintained at 55 °C. Peaks were detected using a Waters 410 differential refractometer and a Waters 2487 dual wavelength UV (210 nm) detector.
NMR analysis
The NMR experiments were carried out either at 22 °C (culture supernatants) or at 40 °C (biomass hydrolysate) on a 600 MHz Bruker Avance III NMR spectrometer equipped with a QCI cryoprobe. The culture supernatant samples were prepared by mixing 540 µl of the supernatant with 60 µl of D2O (Aldrich, Germany). The HCl hydrolysis of the biomass as well as the 1H,13C HSQC experiments of the aliphatic area with high 13C resolution are described in detail by Jouhten and Maaheimo [27]. For 13C decoupling during the acquisition, GARP4 was used.
Authors’ contributions
AB, JK, HM and PR conceived and designed the experiments. AB, MS-G, JK, HM and PR carried out the experimental work and analyzed the data. AB, JK, HM, DM and PR wrote and revised the paper. NK, DM and PR designed the fundamental concept and participated in the coordination of the study. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank the staff of VTT for the technical support.
Competing interests
The authors declare that they have no competing interests.
Availability of data and material
The dataset supporting the conclusions of this article is included as the Additional file 1.
Ethics approval and consent for publication
Not applicable.
Funding
This research was supported by the Academy of Finland through the Sustainable energy program (Grant 271025) and also by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), a Brazilian government agency for the advancement of science and technology. Research scholarships were granted to David Mitchell, Maura Sugai-Guerios and Nadia Krieger by CNPq, and to Alessandra Biz by CNPq (PDE 207321/2015-9) and CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), a Brazilian government agency for the development of personnel in higher education. The funding body did not participate in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional file
10.1186/s12934-016-0544-1 HPLC results of the cultivations in D-galacturonic acid.
Contributor Information
Alessandra Biz, Email: ext-alessandra.biz@vtt.fi.
Maura Harumi Sugai-Guérios, Email: maurasugai@gmail.com.
Joosu Kuivanen, Email: joosu.kuivanen@vtt.fi.
Hannu Maaheimo, Email: hannu.maaheimo@vtt.fi.
Nadia Krieger, Email: nkrieger@ufpr.br.
David Alexander Mitchell, Email: davidmitchell@ufpr.br.
Peter Richard, Email: peter.richard@vtt.fi.
References
- 1.USDA. Citrus: World Markets and Trade. Foreign Agricultural Service/USDA. Office of Global Analysis. 2015 https://apps.fas.usda.gov/psdonline/circulars/citrus.pdf. Accessed 28 June 2016.
- 2.FAOSTAT. Production quantities by Country. Food and Agricultural Organization of the United Nations, Statics Division. 2013. http://faostat3.fao.org/browse/Q/QC/E. Accessed 28 June 2016.
- 3.Grohman K, Cameron R, Kim Y, Widmer W, Luzio G. Extraction and recovery of pectic fragments from citrus processing waste for co–production with ethanol. J Chem Technol Biotechnol. 2013;88:395–407. doi: 10.1002/jctb.3859. [DOI] [Google Scholar]
- 4.Edwards MC, Doran-Peterson J. Pectin-rich biomass as feedstock for fuel ethanol production. Appl Microbiol Biotechnol. 2012;95:565–575. doi: 10.1007/s00253-012-4173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lohrasbi M, Pourbafrani C, Niklasson C, Taherzadeh MJ. Process design and economic analysis of a citrus waste biorefinery with biofuels and limonene as products. Bioresour Technol. 2010;101:7382–7388. doi: 10.1016/j.biortech.2010.04.078. [DOI] [PubMed] [Google Scholar]
- 6.Grohmann K, Baldwin EA. Hydrolysis of orange peel with pectinase and cellulase enzymes. Biotechnol Lett. 1992;14:1169–1174. doi: 10.1007/BF01027023. [DOI] [Google Scholar]
- 7.Phatak L, Chang KC, Brown G. Isolation and characterization of pectin in sugar-beet pulp. J Food Sci. 2006;53:830–833. doi: 10.1111/j.1365-2621.1988.tb08964.x. [DOI] [Google Scholar]
- 8.van Maris AJ, Abbott DA, Bellissimi E, van den Brink J, Kuyper M, Luttik MA, Wisselink HW, Scheffers WA, van Dijken JP, Pronk JT. Alcoholic fermentation of carbon sources in biomass hydrolysates by Saccharomyces cerevisiae: current status. Antonie Van Leeuwenhoek J Microb. 2006;90:391–418. doi: 10.1007/s10482-006-9085-7. [DOI] [PubMed] [Google Scholar]
- 9.Huisjes EH, Luttik MA, Almering MJ, Bisschops MM, Dang DH, Kleerebezem M, Siezen R, van Maris AJ, Pronk JT. Toward pectin fermentation by Saccharomyces cerevisiae: expression of the first two steps of a bacterial pathway for d-galacturonate metabolism. J Biotechnol. 2012;162:303–310. doi: 10.1016/j.jbiotec.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Souffriau B. Transport and metabolism of d-galacturonic acid in the yeast Saccharomyces cerevisiae. Doctoral thesis. Katholieke Universiteit Leuven, Belgium, 2012.
- 11.Motter FA, Kuivanen J, Keränen H, Hilditch S, Penttilä M, Richard P. Categorisation of sugar acid dehydratases in Aspergillus niger. Fungal Genet Biol. 2014;64:67–72. doi: 10.1016/j.fgb.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Benz JP, Protzko RJ, Andrich MS, Bauer S, Dueber JP, Somerville RS. Identification and characterization of a galacturonic acid transporter from Neurospora crassa and its application for Saccharomyces cerevisiae fermentation processes. Biotechnol Biofuels. 2014;7:20. doi: 10.1186/1754-6834-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Souffriau B, Abt T, Thevelein JM. Evidence for rapid uptake of d-galacturonic acid in the yeast Saccharomyces cerevisiae by a channel-type transport system. FEBS Lett. 2012;586:2494–2499. doi: 10.1016/j.febslet.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 14.Hilditch S, Berghäll S, Kalkkinen N, Penttilä M, Richard P. The missing link in the fungal d-galacturonate pathway: identification of the l-threo-3-deoxy-hexulosonate aldolase. J Biol Chem. 2007;282:26195–26201. doi: 10.1074/jbc.M704401200. [DOI] [PubMed] [Google Scholar]
- 15.Martens-Uzunova ES, Schaap PJ. An evolutionary conserved d-galacturonic acid metabolic pathway operates across filamentous fungi capable of pectin degradation. Fungal Genet Biol. 2008;45:1449–1457. doi: 10.1016/j.fgb.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Richard P, Hilditch S. d-galacturonic acid catabolism in microorganisms and its biotechnological relevance. Appl Microbiol Biotechnol. 2009;82:597–604. doi: 10.1007/s00253-009-1870-6. [DOI] [PubMed] [Google Scholar]
- 17.Liepins J, Kuorelahti S, Penttilä M, Richard P. Enzymes for the NADPH-dependent reduction of dihydroxyacetone and d- and l-glyceraldehyde in the mould Hypocrea jecorina. FEBS J. 2006;273:4229–4235. doi: 10.1111/j.1742-4658.2006.05423.x. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritch EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Adams A, Gottschling DE, Kaiser CA, Sterns E. Methods in yeast genetics: A laboratory course manual. New York: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 20.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York: Wiley; 1994. [Google Scholar]
- 21.Gietz RD, Woods RA. Transformation of yeast by the lithium acetate/single-stranded carrier DNA/PEG method. Method Microbiol. 1998;20:53–66. doi: 10.1016/S0580-9517(08)70325-8. [DOI] [Google Scholar]
- 22.Kuorelahti S, Jouhten P, Maaheimo H, Penttilä M, Richard P. l-galactonate dehydratase is part of the fungal path for d-galacturonic acid catabolism. Mol Microbiol. 2006;61:1060–1068. doi: 10.1111/j.1365-2958.2006.05294.x. [DOI] [PubMed] [Google Scholar]
- 23.Hegemann JH, Heick SB. Delete and repeat: a comprehensive toolkit for sequential gene knockout in the budding yeast Saccharomyces cerevisiae. Methods Mol Biol. 2011;765:189–206. doi: 10.1007/978-1-61779-197-0_12. [DOI] [PubMed] [Google Scholar]
- 24.Bradford MM. Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.Buchanan CL, Connaris H, Danson MJ, Reeve CD, Hough DW. An extremely thermostable aldolase from Sulfolobus solfataricus with specificity for non-phosphorylated substrates. Biochem J. 1999;343:563–570. doi: 10.1042/bj3430563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winkler J, Reyes LH, Kao KC. Adaptive laboratory evolution for strain engineering. Methods Mol Biol. 2013;985:211–222. doi: 10.1007/978-1-62703-299-5_11. [DOI] [PubMed] [Google Scholar]
- 27.Jouhten P, Maaheimo H. Labelling analysis for 13C MFA using NMR spectroscopy. In: Nielsen LK, Blank LM, Krömer JO, editors. Metabolic flux analysis. Methods and protocols. New York: Humana Press; 2014. pp. 143–164. [Google Scholar]


