Abstract
Feed components have low water activity, making bacterial survival difficult. The mechanisms of Salmonella survival in feed and subsequent colonization of poultry are unknown. The purpose of this research was to compare the ability of Salmonella serovars and strains to survive in broiler feed and to evaluate molecular mechanisms associated with survival and colonization by measuring the expression of genes associated with colonization (hilA, invA) and survival via fatty acid synthesis (cfa, fabA, fabB, fabD). Feed was inoculated with 1 of 15 strains of Salmonella enterica consisting of 11 serovars (Typhimurium, Enteriditis, Kentucky, Seftenburg, Heidelberg, Mbandanka, Newport, Bairely, Javiana, Montevideo, and Infantis). To inoculate feed, cultures were suspended in PBS and survival was evaluated by plating samples onto XLT4 agar plates at specific time points (0 h, 4 h, 8 h, 24 h, 4 d, and 7 d). To evaluate gene expression, RNA was extracted from the samples at the specific time points (0, 4, 8, and 24 h) and gene expression measured with real-time PCR. The largest reduction in Salmonella occurred at the first and third sampling time points (4 h and 4 d) with the average reductions being 1.9 and 1.6 log cfu per g, respectively. For the remaining time points (8 h, 24 h, and 7 d), the average reduction was less than 1 log cfu per g (0.6, 0.4, and 0.6, respectively). Most strains upregulated cfa (cyclopropane fatty acid synthesis) within 8 h, which would modify the fluidity of the cell wall to aid in survival. There was a weak negative correlation between survival and virulence gene expression indicating downregulation to focus energy on other gene expression efforts such as survival-related genes. These data indicate the ability of strains to survive over time in poultry feed was strain dependent and that upregulation of cyclopropane fatty acid synthesis and downregulation of virulence genes were associated with a response to desiccation stress.
Keywords: Salmonella, poultry feed, virulence, survival, gene regulation
INTRODUCTION
Each year, 31 identified pathogens cause an estimated 9.4 million episodes of foodborne illness in the United States (Scallan et al., 2011). Among these foodborne pathogens, nontyphoidal Salmonella enterica is the leading cause of death and hospitalizations (Scallan et al., 2011). Foodborne pathogens can be acquired by food-producing animals, which may transmit zoonotic pathogens through the food chain and subsequently cause human foodborne illness (Crump et al., 2002). Poultry and poultry products are the leading source of non-Typhi serotypes of S. enterica in the United States (Braden, 2006). Poultry may be colonized with S. enterica but not cause any signs or symptoms of disease in the birds. Thus, if intestinal contents are released during processing, contamination of the carcasses may occur (Rigby et al., 1980).
The initial source of S. enterica to the birds can be transmitted from several vectors (Jarquin et al., 2009). Protein and by-product ingredients originating from animals that are used in feed have been suggested as a source of S. enterica (Williams, 1981; Davies et al., 2004). Given the conditions of the source of the main ingredients, and processing, transportation, and storage, poultry feed has a higher potential than other sources to become contaminated with S. enterica (Jones, 2011).
Currently, S. enterica serovar Kentucky is the dominant serovar isolated from poultry and poultry products in the United States (Foley et al., 2008), but this serovar rarely causes foodborne illness. Conversely, even though isolation of serovar Enteritidis from poultry products has declined, infections with this serovar have increased (CDC, 2010). Thus, it appears that survival on the farm and in other poultry-related environments including feed may not be related to the ability of S. enterica to cause disease (Foley et al., 2008). Therefore, the main objective of this study was to compare the survival capabilities of S. enterica serovars and strains in broiler feed over time in storage. A second objective was to investigate molecular mechanisms associated with survival and virulence by evaluating expression of specific genes associated with these characteristics.
MATERIALS AND METHODS
Bacteria and Culturing Conditions
In these studies a total of 11 serovars consisting of 15 strains of S. enterica were used (Table 1). All S. enterica strains were initially cultured on tryptic soy agar (Becton, Dickinson and Company, Sparks, MD) and incubated at 37°C for 24 h. After incubation, a 10-μL loop of culture was inoculated into 30 mL of tryptic soy broth (Becton, Dickinson and Company; pH 7.2) and incubated in a shaking water bath at 37°C for 15 h. From this culture, 1 mL was inoculated into tryptic soy broth and incubated in a shaking water bath at 37°C for 3 h. The culture then was centrifuged at 8,000 × g for 5 min and the supernatant discarded. The culture was washed 3 times by resuspending the pellet in PBS (Becton, Dickinson and Company), centrifuging (8,000 × g for 5 min at 25°C), and finally resuspending in PBS. Salmonella suspensions were standardized to 0.15 at 630 nm by spectrophotometry so that all serovars were used at approximately the same concentrations (7 log cfu·mL−1). A dilution series was also conducted on the suspension to precisely determine the initial S. enterica concentration.
Table 1.
A table of the Salmonella enterica serovars, the source of the strains, and references describing characteristics of the strains used in this work
| Salmonella enterica serovar | Source | Reference |
|---|---|---|
| Typhimurium DT104 | Human infection | Threlfall, 2000 |
| Typhimurium ATCC 23595 (LT2) | Laboratory strain | Swords et al., 1997 |
| Typhimurium ATCC 14028 | Laboratory strain | None |
| Enteritidis (WT) | Human infection | None |
| Enteritidis ATCC 13076 | Human infection | None |
| Kentucky | Poultry carcass | Clement et al., 2010 |
| Kentucky | Poultry carcass | Clement et al., 2010 |
| Seftenburg | Poultry farm | Rodriguez et al., 2006 |
| Heidelberg | Poultry farm | Rodriguez et al., 2006 |
| Mbandanka | Poultry carcass | Melendez et al., 2010 |
| Newport | Poultry carcass | Melendez et al., 2010 |
| Bairely | Poultry carcass | Melendez et al., 2010 |
| Javana | Poultry farm | Rodriguez et al., 2006 |
| Montevideo | Swine farm | Rodriguez et al., 2006 |
| Infantis | Poultry farm | Rodriguez et al., 2006 |
Spiking and Analysis of Feed Sample
A Chick Starter/Grower-AMP BMD feed was purchased from a local co-op (Knoxville, TN) and sieved through a screen (no. 8; 2.38-mm openings) to remove dust and small particles. The composition of the formulated starter feed is presented in Table 2. Water activity of the feed was measured using a water activity meter (Aqua Lab, Decagon Services Inc., Pullman, WA). For the survival studies, 10-µL aliquots of the S. enterica suspension prepared as described in the previous section were placed into 2 g of the feed in 5-mL tubes and mixed by agitation. The inoculated feed was stored at 25°C. At specific time points (0, 4, 8, and 24 h, and 4 and 7 d), S. enterica survival was evaluated using standard microbiological methods and a standard dilution series. We chose to use 7 d because this is the average time of storage of poultry feed on poultry farms. Briefly, the sample was suspended in 2 mL of PBS, vortexed, and a 100-µL portion of the solution was used in a dilution series that was inoculated on XLT4 (xylose lysine tergitol-4, Becton, Dickinson and Company) agar, which was incubated at 37°C for 24 h. An uninoculated sample of the poultry feed acted as the negative control. Triplicate samples were evaluated with 2 repetitions performed for each serovar.
Table 2.
Formulation and ingredient list1 of the starter/grower feed (co-op chick) used in this study
| Component | Guaranteed analysis, % |
|---|---|
| CP | 19 |
| Lysine | 0.82 |
| Methionine | 0.27 |
| Crude fat | 3.5 |
| Crude fiber | 4.5 |
| Calcium | 0.80–1.30 |
| Phosphorus | 0.7 |
| Salt | 0.25–0.75 |
| Active drug ingredient (g/t) | |
| Amprolium | 125.11 |
| Bactracin methylene disalicylate | 220.46 |
1Ingredients: grain products, plant protein products, processed grain by-products, molasses products, propionic acid, calcium carbonate, calcium phosphate, salt, choline chloride, yucca schidegera extract, Bacillus subtilis, niacin supplement, vitamin E supplement, calcium pantothenate, riboflavin supplement, vitamin A acetate, menadione dimethylpyrimidinol bisulfite, vitamin D3 supplement, biotin, vitamin B12 supplement, pyridoxine hydrochloride, folic acid, thiamine, ferrous sulfate, manganous oxide, zinc oxide, copper oxide, calcium iodate, sodium selenite, cobalt carbonate.
RNA Preparation
Total RNA was isolated from the samples as described by Gonzalez-Gil et al. (2012) with some modification. At specific time points (0, 4, 8, and 24 h) and equal volume of RNA protect bacterial reagent (Qiagen, Valencia, CA) was added to a 2-mL microfuge tube containing the Salmonella feed suspensions and allowed to stand at room temperature for 5 min. Subsequently, RNA was extracted from the samples using the RNeasy mini kit (Qiagen) as directed by the manufacturer. After extraction, the RNA samples were subjected to a DNase treatment utilizing the Qiagen DNase kit (Qiagen) as directed by the manufacturer. All samples then were quantified using spectrophotometry (Nanodrop ND-1000, ThermoScientific, Pittsburgh, PA).
Quantitative Reverse-Transcription Real-Time PCR
After purification, cDNA was synthesized from the RNA using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). All quantitative reverse-transcription real-time PCR (qRT-PCR) reactions were performed as described by Gonzalez-Gil et al. (2012) using the ABI 7100 RT-PCR system (Applied Biosystems, Carlsbad, CA). Briefly, a 20-µL total volume consisted of 10 µL of Power SYBR Green PCR Master Mix (Life Technologies Corporation, Carlsbad, CA), 300 nM of each primer, 100 ng of cDNA template, and water to volume. With the exception of hilA and 16S rRNA, primers were designed using the NCBI Primer-BLAST tool and evaluated for specificity (Table 3). All primers were synthesized by Integrated DNA Technologies (Coralville, IA). The qRT-PCR reactions were optimized to the conditions of 95°C for 15 min for the initial activation of Taq polymerase. This was followed by 35 cycles of denaturation at 94°C for 15 s, annealing at 55°C for 30 s, and amplification at 60°C for 30 s with fluorescence being measured during the extension phase. Melting curves were conducted subsequently and consisted of 95°C for 15 s, 60°C for 5 min to a final temperature of 95°C for 15 s. All reactions were performed independently and in triplicate.
Table 3.
A list of the genes, primer sequences, and references for the primers that were used to evaluate gene expression changes of Salmonella enterica strains used in this study
| Target gene1 | Sequence (5′ to 3′) | Reference |
|---|---|---|
| 16S | Forward: GCGGCCCCCTGGACAAAGAC | Gonzalez-Gil et al., 2012 |
| Reverse: TAGCTCCGGAAGCCACGCCT | ||
| hilA | Forward: ATGCCATAGCATTTTTATCC | Park et al., 2011 |
| Reverse: GATTTAATCTGTATCAGG | ||
| invA | Forward: CTGTCTGGCGGTGACGCTGG | Own design. NCBI2 reference |
| Reverse: ACGCGCCATTGCTCCACGAA | Sequence: NC_003198.1 | |
| cfa | Forward: GCTGGTGGGAATGCGAGCGT | Own design. NCBI reference |
| Reverse: CAGCACACGCATCCCCGGTT | Sequence: NC_011294.1 | |
| fabA | Forward: ACTCCCTGCGCCGAACATGC | Own design. NCBI reference |
| Reverse: CACTTCGCCCACGCCCAGAG | Sequence: NC_011294.1 | |
| fabB | Forward: CCGCGTGGTCTGAAAGCCGT | Own design. NCBI reference |
| Reverse: GGACAGTGCGCCCATCGCAT | Sequence: NC_011294.1 | |
| fabD | Forward: ACCCAGCAAGGTCCAGCGG | Own design. NCBI reference |
| Reverse: TTCGCGCCAGCGGCTTTACA | Sequence: NC_011294.1 |
116S = housekeeping gene; hilA and invA = genes involved in virulence and colonization; cfa = cyclopropane fatty acid gene; fabA, fabB, and fabD = fatty acid biosynthesis genes.
2NCBI = National Center for Biotechnology Information.
Analysis of Gene Expression
Samples were normalized using the 16S rRNA gene as an internal standard (Table 3). The relative changes (n-fold) in gene expression between samples were calculated using the 2−ΔΔC(T) method as described by Livak and Schmittgen (2001). Fold change in expression for specific target gene was determined and these data were used to generate heat maps within a Microsoft Excel 14.3.5 (Microsoft Corporation, Redmond, WA) spreadsheet using the conditional formatting and color scale functions.
Statistical Analysis
For survival and water activity experiments, each strain was sampled in duplicate with triplicate repetitions, and culturable cfu counts were analyzed via mixed ANOVA analysis (P < 0.05) to determine statistical differences between strains. Results are expressed as least squares means with SEM. For water activity measurements, each strain was sampled in triplicate for each time point and analyzed as above for the survival experiments. The software used was SAS 9.3 (SAS Institute Inc., Cary, NC).
RESULTS
The water activity of the sample of spiked feed was measured at specific times of 0, 4, 8, 24 h, and 4 and 7 d (Table 4). This was done to correlate water activity in the feed with any impact on the survival of S. enterica. Not surprisingly, there was some correlation between the water activity in the spiked feed and the survival rates of the bacteria. Water activity consistently decreased over the course of the experiments as did the counts of culturable S. enterica. However, the correlation coefficients indicated that there was no significant correlation between water activity and reduction in culturable Salmonella. This is most likely due to the large variation in reduction of Salmonella counts between each time point.
Table 4.
Measurement of water activity (aw) in the poultry feed, before being spiked with Salmonella enterica cultures, and after spiking at specific time points1
| Sample | aw |
|---|---|
| Unspiked | 0.35 ± 0.001a |
| 0 h | 0.74 ± 0.001b |
| 4 h | 0.70 ± 0.003c |
| 8 h | 0.69 ± 0.003d |
| 24 h | 0.67 ± 0.001e |
| 4 d | 0.65 ± 0.002f |
| 7 d | 0.61 ± 0.001g |
a–gMean values within a column that do not have the same superscript letter are significantly different (P < 0.05).
1Values of SEM ± from triplicates from each S. enterica strain.
The culturable S. enterica populations (log cfu·g−1) were determined at 0, 4, 8, and 24 h, and 4 and 7 d, and differences in the survival of the bacteria were found to be dependent on serovar and strain (Table 5). After 7 d, almost 3 logs (cfu per g of feed) of SalmonellaEnteriditis (WT) and Salmonella Typhimurium ATCC 23595 (LT2) were recovered from the feed samples. After 4 d of incubation at room temperature, Salmonella Typhimurium 14028 and Salmonella Montevideo could not be recovered. Both strains of Salmonella Kentucky and Salmonella Typhimurium 14028 had the most rapid decrease after 4 h with approximately 3 logs (cfu per g of feed) less than the initial inoculum recovered from the feed. Both strains of Salmonella Enteritidis, Salmonella Seftenburg, Salmonella Mbandanka, and Salmonella Infantis had the lowest decrease (approximately 1 log cfu·g−1) in recoverable bacteria after 4 h. The remaining strains decreased by approximately 2 log cfu·g−1 from the initial inoculum levels after 4 h of incubation at room temperature. Interestingly, data regarding strains of the same serovar were quite variable. The 3 Typhimurium strains had different patterns in reduction of Salmonella, whereas the Kentucky and Enteritidis strains had similar patterns when comparing data of the same serovar.
Table 5.
Changes in the counts of culturable Salmonella enterica serovars (cfu/g of feed) expressed in log recovered from artificially inoculated feed at specific time points
| Changes between time points1 | |||||
|---|---|---|---|---|---|
| Strain | 0 h to 4 h | 4 h to 8 h | 8 h to 24 h | 24 h to 4 d | 4 d to 7 d |
| Salmonella Typhimurium DT104 | 2.17 ± 0.10a | 0.38 ± 0.10bc | 0.51 ± 0.12b | 2.71 ± 0.49a | −0.58 ± 0.78abcd |
| Salmonella Typhimurium ATCC 23595 (LT2) | 1.79 ± 0.11ab | 0.03 ± 0.14bc | 0.83 ± 0.16bc | 0.73 ± 0.27ab | −0.22 ± 0.28d |
| Salmonella Typhimurium ATCC 14028 | 3.47 ± 0.80abc | −0.15 ± 0.80abc | 1.59 ± 0.29a | 1.42 ± 0.45b | NC ± 0.00d |
| Salmonella Enteritidis (WT) | 1.40 ± 0.10bc | 0.13 ± 0.05c | 0.55 ± 0.09b | 1.19 ± 0.14a | 0.42 ± 0.10cd |
| Salmonella Enteritidis ATCC 13076 | 1.03 ± 0.05c | 0.74 ± 0.28abc | 0.29 ± 0.17bcd | 1.50 ± 0.06a | 2.10 ± 0.00a |
| Salmonella Kentucky A | 3.01 ± 0.81abc | 0.36 ± 1.07abc | 0.69 ± 0.74abc | 0.75 ± 0.44ab | 0.7 ± 0.44bcd |
| Salmonella Kentucky F | 2.95 ± 0.47ab | 0.20 ± 0.64bc | 0.92 ± 0.75abc | 0.00 ± 0.94ab | 0.35 ± 0.65abcd |
| Salmonella Seftenburg | 0.97 ± 0.21abc | −0.22 ± 0.27abc | 0.38 ± 0.24bcd | 3.09 ± 0.47a | 1.05 ± 0.47abcd |
| Salmonella Heidelburg | 1.57 ± 0.35abc | 1.28 ± 0.11a | −0.38 ± 0.09d | 1.75 ± 0.54ab | 1.42 ± 0.64b |
| Salmonella Mbandanka | 1.35 ± 0.14bc | 0.59 ± 0.11b | −0.02 ± 0.08cd | 2.21 ± 0.62ab | 0.33 ± 0.72abcd |
| Salmonella Newport | 2.30 ± 0.27abc | 0.85 ± 0.24abc | 0.87 ± 0.11b | 1.15 ± 0.43ab | 1.41 ± 0.64abcd |
| Salmonella Bairely | 1.97 ± 0.20abc | 0.43 ± 0.20abc | 0.18 ± 0.21bcd | 0.94 ± 0.22ab | 2.02 ± 0.29ab |
| Salmonella Javiana | 2.09 ± 0.32abc | 0.77 ± 0.35abc | 0.44 ± 0.05b | 1.42 ± 0.60ab | 1.94 ± 0.63abcd |
| Salmonella Montevideo | 1.94 ± 0.27abc | 0.93 ± 0.49abc | 1.21 ± 0.71abcd | 2.16 ± 0.68ab | NC ± 0.00d |
| Salmonella Infantis | 0.82 ± 0.16c | 0.71 ± 0.20abc | 0.40 ± 0.11bc | 2.14 ± 0.43a | 1.75 ± 0.5abcd |
a–dMean values within a column that do not have the same superscript letter are significantly different (P < 0.05).
1Values ± SEM from triplicates with duplicate repetition samples. NC: no change between time points.
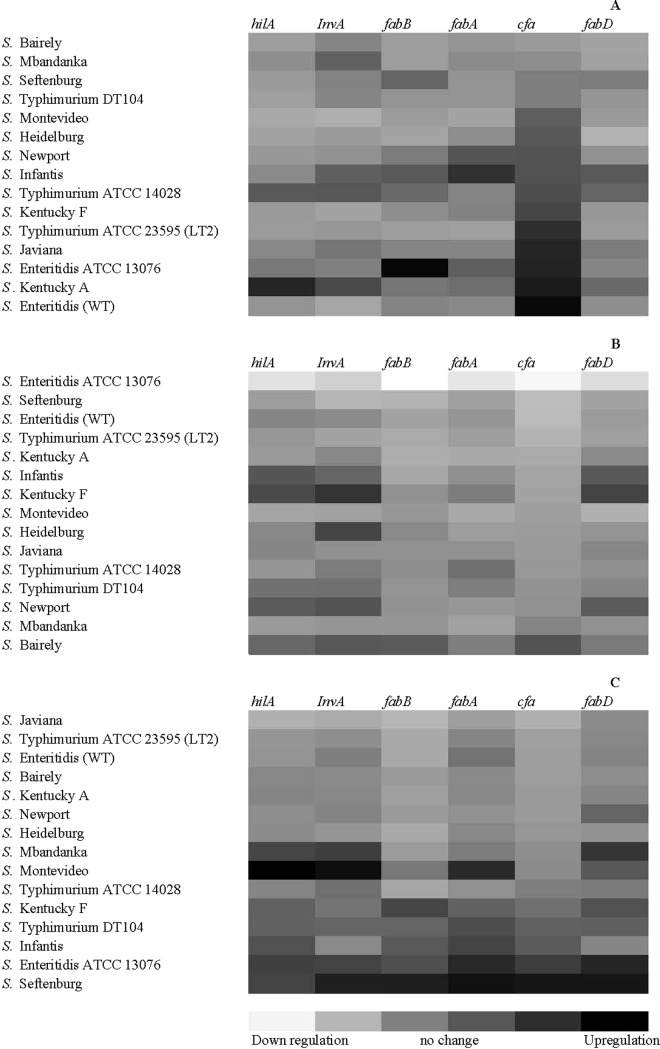
Relative fold change in gene expression for each gene was calculated and heat maps generated for the 3 time points sampled over the course of the experiment (Figure 1). These maps then were sorted from ascending to descending for each gene. In this way, it was visually apparent that the cfa gene was upregulated in most serovars after 4 h. Furthermore, it appeared that there was a correlation between regulation of the cfa gene and the fabB gene at the 8 and 24 h time points (0.93 and 0.90, respectively). There were no other apparent gene regulation and gene correlations consistent among all strains.
Figure 1.
A heat map of relative fold change in gene expression of genes involved in virulence and colonization (hilA, InvA) and fatty acid synthesis (cfa, fabB, fadD, fabA) in 15 Salmonella enterica serovars artificially inoculated into poultry feed and sampled after incubation at room temperature at 4 h (panel A), 8 h (panel B), and 24 h (panel C). Maps are sorted based on the cfa gene in ascending order of regulation for each time point. cfa = cyclopropane fatty acid gene; fabA, fabB, and fabD = fatty acid biosynthesis genes.
Correlation analysis was performed to determine if survival of the S. enterica serovars was correlated to expression of specific genes. A low positive coefficient of correlation was obtained between bacterial survival and the genes cfa, fabA, and fabB (0.23, 0.04, and 0.13, respectively). For the genes invA, fabD, and hilA, a low negative correlation (−0.24, −0.04, and −0.28) was correlated with the survival capability of the S. enterica strains tested. Although the values of correlation were numerically different, they were not statistically significant (P > 0.05).
DISCUSSION
According to Ha et al. (1998), S. enterica survival in feed can vary and is dependent on formulation. In their study, Ha et al. (1998) also found that aerobic bacterial counts recovered from feeds containing meat and bone meal were greater than those containing soybean meals. However, Pektar et al. (2011) reported that there were no differences in the abilities of S. enterica to survive in conventional versus organic feed where the conventional feed contained bone and poultry meal, which was replaced in the organic feed with alfalfa meal. Salmonella enterica contamination on individual ingredients of the feed is also an important fact to consider, because S. enterica has been isolated from feed ingredients including, grains, oilseed meal, feather and fish meal, and meat by-products (Maciorowski et al., 2004).
Survival of S. enterica in low water activity foods is well documented (Tamminga et al., 1976; Juven et al., 1984; Rowe et al., 1987; Lehmacher et al., 1995; Beuchat, 2009). Interestingly, previous studies suggest that S. enterica survival is higher in foods with water activity (aw) between 0.43 and 0.55 than foods at an aw of 0.75 (Juven et al., 1984; Pektar et al., 2011). Because water activity did not drop below 0.61 in this study, water activity may have been suboptimal for the S. enterica strains we evaluated for survival in feed.
The invA gene allows Salmonella to enter epithelial cells, playing an important role in the invasion and disease process (Galán et al., 1992). The second virulence gene evaluated in this study, hilA, regulates the expression of invasion genes in response to environmental stimuli including osmolarity, oxygen levels, and pH (Durant et al., 2000; Fluit, 2005; Chuanchuen et al., 2010; Park et al., 2011; Gonzalez-Gil et al., 2012). In the present study, there was an overall negative correlation between survival and upregulation of these 2 genes indicating that perhaps efforts for virulence were shifted away from these genes and instead focused on upregulation of stress responses (Gonzalez-Gil et al., 2012).
To survive the stress of desiccation, some bacteria increase membrane fluidity (Baysse and O’Gara, 2007). For S. enterica, membrane fluidity can be modified with an increase in de novo synthesis of unsaturated fatty acids, which occurs via the fabA-fabB pathway. Likewise, the cfa gene encodes cyclopropane fatty acid (CFA) synthase, an enzyme that cyclizes UFA to improve membrane fluidity (Kim et al., 2005). Conversely, fabD is activated to produce saturated fatty acids, which decrease membrane fluidity. Thus, the upregulation of cfa in this study at the 4-h time point was not surprising as an increase in CFA is considered to be an indicator of starvation or desiccation stress (Kieft et al., 1994).
Low water activity food products can become cross contaminated after processing by factors including poor sanitization practices, poor equipment design, and poor ingredient control, which presents a significant food safety risk (Podolak et al., 2010). Some research indicates the infectious dose of S. enterica is lower when infection occurs via a contaminated low aw food (Rowe et al., 1987; Greenwood and Hooper, 1983). The reason for this is not exactly known. However, data from this study indicate that this may not be due to upregulation of virulence-associated genes hilA and invA because our data showed a tendency for these genes to be downregulated in lower water activity. Instead, the lower infectious dose may be an adaptive tolerance response where cells that survived the low water activity are more stress resistant, making it easier for these cells to survive the subsequent stress of passage through the acidic gastrointestinal environment (Ma et al., 2009). It has also been suggested that pathogens in low water activity foods are typically metabolically inactive, and this metabolic state makes the cells less susceptible to stresses such as those encountered in the gastrointestinal environment (Barat et al., 2012).
The data indicate that differences in survival and gene expression vary by serovars of S. enterica, caution should be taken if applying the results of this study to other serovars of S. enterica that have not been evaluated. In addition, because only one type of feed and incubation temperature were used, additional experiments are necessary to understand how these variables may affect the results. In conclusion, this study demonstrated that the ability of S. enterica to survive over storage time in poultry feed was serovar and strain dependent. Furthermore, the data indicate that the upregulation of short chain fatty acid synthesis and downregulation of virulence genes may be associated with survival in the poultry feed component.
ACKNOWLEDGMENTS
This study was funded by a HATCH grant awarded to I. Hanning by the University of Tennessee Experiment Station.
REFERENCES
- Barat S., Steeb B., Maze A., Bumann D. Extensive in vivo resilience of persistent Salmonella. PLoS ONE. 2012;7:e42007. doi: 10.1371/journal.pone.0042007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysse C., O’Gara F. Role of membrane structure during stress signaling and adaptation in Pseudomonas. In: Ramos J. L., Filloux A., editors. Pseudomonas: A Model System in Biology. the Netherlands: Springer; 2007. pp. 193–224. [Google Scholar]
- Beuchat L. R., Mar. 26 Behavior of Salmonella in foods with low water activity; Presentation at IAFP Rapid response symposium “Salmonella in peanut butter products: Understanding the risk and controlling the process.”; Arlington, VA: 2009. [Google Scholar]
- Braden C. R. Salmonella enterica serotype Enteritidis and eggs: A national epidemic in the United States. Clin. Infect. Dis. 2006;43:512–517. doi: 10.1086/505973. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Atlanta, GA: US Department of Health and Human Services, CDC; 2010. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, 2009. [PubMed] [Google Scholar]
- Chuanchuen R., Ajariyakhajorn K., Koowatananukul C. Antimicrobial resistance and virulence genes in Salmonella enterica isolates from dairy cows. Foodborne Pathog. Dis. 2010;7:63–69. doi: 10.1089/fpd.2009.0341. [DOI] [PubMed] [Google Scholar]
- Clement A., Hanning I., Park S. H., Melendez S. M., Pendleton S., Woo-ming A., Scott E. E., Ricke and S. C. Processing treatments and rearing conditions effects on Salmonella, Campylobacter, and aerobic bacteria present on whole carcass chickens; American Society for Microbiology 110th General Meeting; San Diego, CA: 2010. [Google Scholar]
- Crump J. A., Griffin P. M., Angulo F. J. Bacterial contamination of animal feed and its relationship to human foodborne illness. Clin. Infect. Dis. 2002;35:859–865. doi: 10.1086/342885. [DOI] [PubMed] [Google Scholar]
- Davies P. R., Hurd H. S., Funk J. A., Fedorka-Cray P. J., Jones F. T. The role of contaminated feed in the epidemiology and control of Salmonella enterica in pork production. Foodborne Pathog. Dis. 2004;1:202–215. doi: 10.1089/fpd.2004.1.202. [DOI] [PubMed] [Google Scholar]
- Durant J. A., Corrier D. E., Ricke S. C. Short-chain volatile fatty acids modulate the expression of the hilA and invF genes of Salmonella Typhimurium. J. Food Prot. 2000;63:573–578. doi: 10.4315/0362-028x-63.5.573. [DOI] [PubMed] [Google Scholar]
- Fluit A. C. Towards more virulent and antibiotic-resistant Salmonella? FEMS Immunol. Med. Microbiol. 2005;43:1–11. doi: 10.1016/j.femsim.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Foley S. L., Lynne A. M., Nayak R. Salmonella challenges: Prevalence in swine and poultry and potential pathogenicity of such isolates. J. Anim. Sci. 2008;86:E149–E162. doi: 10.2527/jas.2007-0464. [DOI] [PubMed] [Google Scholar]
- Galán J. E., Ginocchio C., Costeas P. Molecular and functional characterization of the Salmonella invasion gene invA: Homology of invA to members of a new protein family. J. Bacteriol. 1992;174:4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gil F., Bolloch A. L., Pendleton S., Zhang N., Wallis A., Hanning I. Expression of hilA in response to mild stress in Salmonella enterica is serovar and strain dependent. J. Food Sci. 2012;77:M292–M297. doi: 10.1111/j.1750-3841.2012.02684.x. [DOI] [PubMed] [Google Scholar]
- Greenwood M. H., Hooper W. L. Chocolate bars contaminated with Salmonella napoli: An infectivity study. Br. Med. J. 1983;286:1394. doi: 10.1136/bmj.286.6375.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S. D., Maciorowsk K. G., Kwon Y. M., Jones F. T., Ricke S. C. Indigenous feed microflora and Salmonella Typhimurium marker strain survival in poultry mash diets containing varying levels of protein. Anim. Feed Sci. Technol. 1998;76:23–33. [Google Scholar]
- Jarquin R., Hanning I., Ahn S., Ricke S. C. Development of rapid detection and genetic characterization of Salmonella in poultry breeder feeds. Sensors (Basel Switzerland) 2009;9:5308–5323. doi: 10.3390/s90705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones F. T. A review of practical Salmonella control measures in animal feed. J. Appl. Poult. Res. 2011;20:102–113. [Google Scholar]
- Juven B. J., Cox J. S., Bailey J. S., Thompson J. E., Charles O. W., Shutze J. V. Survival of Salmonella in dry food and feed. J. Food Prot. 1984;47:445–448. doi: 10.4315/0362-028X-47.6.445. [DOI] [PubMed] [Google Scholar]
- Kieft T. L., Ringelberg D. B., White D. C. Changes in ester-linked phospholipid fatty acid profiles of subsurface bacteria during starvation and desiccation in a porous medium. Appl. Environ. Microbiol. 1994;60:3292–3299. doi: 10.1128/aem.60.9.3292-3299.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. H., Kim S., Kim H. G., Lee J., Lee I. S., Park Y. K. The formation of cyclopropane fatty acids in Salmonella enterica serovar Typhimurium. Microbiology. 2005;151:209–218. doi: 10.1099/mic.0.27265-0. [DOI] [PubMed] [Google Scholar]
- Lehmacher A., Bockemühl J., Aleksic S. Nationwide outbreak of human salmonellosis in Germany due to contaminated paprika and paprika-powdered potato chips. Epidemiol. Infect. 1995;115:501–511. doi: 10.1017/s0950268800058660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K., Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma L., Zhang G., Gerner-Smidt P., Mantripragada Y., Ezeoke I., Doyle M. P. Thermal inactivation of Salmonella in peanut butter. J. Food Prot. 2009;8:1596–1601. doi: 10.4315/0362-028x-72.8.1596. [DOI] [PubMed] [Google Scholar]
- Maciorowski K. G., Jones F. T., Pillai S. D., Ricke S. C. Incidence, sources, and control of foodborne Salmonellaspp. in poultry feeds. World's Poult. Sci. J. 2004;60:446–457. [Google Scholar]
- Melendez S., Hanning I., Han J., Nayak R., Clement A. R., Wooming A., Herrera P., Jones F. T., Foley S. L., Ricke S. C. Salmonella isolated from pasture poultry exhibit antimicrobial resistance and the presence of class I integrons. J. Appl. Microbiol. 2010;109:1957–1966. doi: 10.1111/j.1365-2672.2010.04825.x. [DOI] [PubMed] [Google Scholar]
- Park S. H., Jarquin R., Hanning I., Almeida G., Ricke S. C. Detection of Salmonella spp. survival and virulence in poultry feed by targeting the hilA gene. J. Appl. Microbiol. 2011;111:426–432. doi: 10.1111/j.1365-2672.2011.05054.x. [DOI] [PubMed] [Google Scholar]
- Pektar A., Alali W. Q., Harrison M. A., Beuchat L. R. Survival of Salmonella in organic and conventional broiler feed as affected by temperature and water activity. Agric. Food Anal. Bacteriol. 2011;1:175–185. [Google Scholar]
- Podolak R., Enache E., Stone W., Black D. G., Elliot P. H. Sources and risk factors for contamination, survival, persistence, and heat resistance of Salmonella in low-moisture foods. J. Food Prot. 2010;73:1919–1936. doi: 10.4315/0362-028x-73.10.1919. [DOI] [PubMed] [Google Scholar]
- Rigby C. E., Pettit J. R., Baker M. F., Bentley A. H., Salomons M., Lior H. Flock infection and transport as sources of Salmonellae in broiler chickens and carcasses. Can. J. Comp. Med. 1980;44:328–337. [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Pangloli P., Richards H. A., Mount J. R., Draughon F. A. Prevalence of Salmonella in diverse environmental farm samples. J. Food Prot. 2006;69:2576–2580. doi: 10.4315/0362-028x-69.11.2576. [DOI] [PubMed] [Google Scholar]
- Rowe B., Hutchinson D. N., Gilbert R. J., Hales B. H., Begg N. T., Dawkins H. C., Jacob M., Rae F. A., Jepson M. Salmonella ealing infections associated with consumption of infant dried milk. Lancet. 1987;2:900–903. doi: 10.1016/s0140-6736(87)91384-5. [DOI] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R. M., Angulo F. J., Tauxe R. V., Widdowson M. A., Roy S. L., Jones J. L., Griffin P. M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011;1:1–21. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swords W. E., Cannon B., Benjamin W. A virulence of LT2 strains of Salmonella Typhimurium results from a defective rpoS gene. Infect. Immun. 1997;65:2451–2453. doi: 10.1128/iai.65.6.2451-2453.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamminga S. K., Beumer R. R., Kampelmacher E. H., Van Leusden F. M. Survival of Salmonella Eastbourne and Salmonella Typhimurium in chocolate. J. Hyg. (Lond.) 1976;76:41–47. doi: 10.1017/s0022172400054929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threlfall J. Epidemic Salmonella Typhimurium DT 104—A truly international multi-resistant clone. J. Antimicrob. Chemother. 2000;46:7–10. doi: 10.1093/jac/46.1.7. [DOI] [PubMed] [Google Scholar]
- Williams J. E. Salmonellas in poultry feeds—A worldwide review. Part I: Introduction. World's Poult. Sci. J. 1981;37:6–19. [Google Scholar]