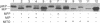Abstract
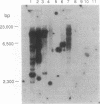
Proteolytic removal of amino-terminal octapeptides from mitochondrial intermediate proteins is a required step for a subgroup of nuclear-encoded mitochondrial precursors and is specifically catalyzed by mitochondrial intermediate peptidase (MIP). We recently reported the purification of MIP from rat liver and showed that the enzyme is a monomer of 75 kDa. We now report the sequence of a full-length rat MIP cDNA. This cDNA codes for a protein of 710 amino acids, including an amino-terminal mitochondrial leader peptide of 33 residues. The region surrounding the mature MIP amino terminus shows a cleavage site typically recognized by the general mitochondrial processing peptidase (MPP). In vitro synthesized MIP precursor is cleaved to mature MIP by purified MPP, and thus MIP is not required for its own proteolytic maturation. Comparison of the deduced MIP sequence with other sequences in the GenBank data base reveals two important similarities. The first is to a sequence encoding a putative MIP homologue in the recently reported sequence of yeast chromosome III. The putative yeast protein is predicted to be 712 amino acids long and includes a putative 23-residue mitochondrial leader peptide also with a MPP processing site. It shows 47% similarity and 24% identity to rat MIP. The second similarity is to members of a subfamily of metallopeptidases that includes rat metalloendopeptidase EC 3.4.24.15 and two bacterial proteases, oligopeptidase A and dipeptidyl carboxypeptidase. A region of greater than 50% similarity over 400 residues between MIP and these proteins is centered around the sequence motif HEXXH, typical of zinc metallopeptidases.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker K. P., Schatz G. Mitochondrial proteins essential for viability mediate protein import into yeast mitochondria. Nature. 1991 Jan 17;349(6306):205–208. doi: 10.1038/349205a0. [DOI] [PubMed] [Google Scholar]
- Conlin C. A., Miller C. G. Cloning and nucleotide sequence of opdA, the gene encoding oligopeptidase A in Salmonella typhimurium. J Bacteriol. 1992 Mar;174(5):1631–1640. doi: 10.1128/jb.174.5.1631-1640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W., Japa S., Beattie D. S. Import of the iron-sulfur protein of the cytochrome b.c1 complex into yeast mitochondria. J Biol Chem. 1990 Sep 25;265(27):16541–16547. [PubMed] [Google Scholar]
- Gavel Y., von Heijne G. Cleavage-site motifs in mitochondrial targeting peptides. Protein Eng. 1990 Oct;4(1):33–37. doi: 10.1093/protein/4.1.33. [DOI] [PubMed] [Google Scholar]
- Hamilton S., Miller C. G. Cloning and nucleotide sequence of the Salmonella typhimurium dcp gene encoding dipeptidyl carboxypeptidase. J Bacteriol. 1992 Mar;174(5):1626–1630. doi: 10.1128/jb.174.5.1626-1630.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F. U., Pfanner N., Nicholson D. W., Neupert W. Mitochondrial protein import. Biochim Biophys Acta. 1989 Jan 18;988(1):1–45. doi: 10.1016/0304-4157(89)90002-6. [DOI] [PubMed] [Google Scholar]
- Hartl F. U., Schmidt B., Wachter E., Weiss H., Neupert W. Transport into mitochondria and intramitochondrial sorting of the Fe/S protein of ubiquinol-cytochrome c reductase. Cell. 1986 Dec 26;47(6):939–951. doi: 10.1016/0092-8674(86)90809-3. [DOI] [PubMed] [Google Scholar]
- Hawlitschek G., Schneider H., Schmidt B., Tropschug M., Hartl F. U., Neupert W. Mitochondrial protein import: identification of processing peptidase and of PEP, a processing enhancing protein. Cell. 1988 Jun 3;53(5):795–806. doi: 10.1016/0092-8674(88)90096-7. [DOI] [PubMed] [Google Scholar]
- Hendrick J. P., Hodges P. E., Rosenberg L. E. Survey of amino-terminal proteolytic cleavage sites in mitochondrial precursor proteins: leader peptides cleaved by two matrix proteases share a three-amino acid motif. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4056–4060. doi: 10.1073/pnas.86.11.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E. C., Pesold-Hurt B., Suda K., Oppliger W., Schatz G. The first twelve amino acids (less than half of the pre-sequence) of an imported mitochondrial protein can direct mouse cytosolic dihydrofolate reductase into the yeast mitochondrial matrix. EMBO J. 1985 Aug;4(8):2061–2068. doi: 10.1002/j.1460-2075.1985.tb03892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaya G., Kalousek F., Fenton W. A., Rosenberg L. E. Cleavage of precursors by the mitochondrial processing peptidase requires a compatible mature protein or an intermediate octapeptide. J Cell Biol. 1991 Apr;113(1):65–76. doi: 10.1083/jcb.113.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaya G., Kalousek F., Rosenberg L. E. Amino-terminal octapeptides function as recognition signals for the mitochondrial intermediate peptidase. J Biol Chem. 1992 Apr 15;267(11):7904–7910. [PubMed] [Google Scholar]
- Jensen R. E., Yaffe M. P. Import of proteins into yeast mitochondria: the nuclear MAS2 gene encodes a component of the processing protease that is homologous to the MAS1-encoded subunit. EMBO J. 1988 Dec 1;7(12):3863–3871. doi: 10.1002/j.1460-2075.1988.tb03272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongeneel C. V., Bouvier J., Bairoch A. A unique signature identifies a family of zinc-dependent metallopeptidases. FEBS Lett. 1989 Jan 2;242(2):211–214. doi: 10.1016/0014-5793(89)80471-5. [DOI] [PubMed] [Google Scholar]
- Kalousek F., Hendrick J. P., Rosenberg L. E. Two mitochondrial matrix proteases act sequentially in the processing of mammalian matrix enzymes. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7536–7540. doi: 10.1073/pnas.85.20.7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiber J., Kalousek F., Swaroop M., Rosenberg L. E. The general mitochondrial matrix processing protease from rat liver: structural characterization of the catalytic subunit. Proc Natl Acad Sci U S A. 1990 Oct;87(20):7978–7982. doi: 10.1073/pnas.87.20.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak P., Dev I. K. Degradation of a signal peptide by protease IV and oligopeptidase A. J Bacteriol. 1988 Nov;170(11):5067–5075. doi: 10.1128/jb.170.11.5067-5075.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S. G., van der Aart Q. J., Agostoni-Carbone M. L., Aigle M., Alberghina L., Alexandraki D., Antoine G., Anwar R., Ballesta J. P., Benit P. The complete DNA sequence of yeast chromosome III. Nature. 1992 May 7;357(6373):38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- Orlowski M., Reznik S., Ayala J., Pierotti A. R. Endopeptidase 24.15 from rat testes. Isolation of the enzyme and its specificity toward synthetic and natural peptides, including enkephalin-containing peptides. Biochem J. 1989 Aug 1;261(3):951–958. doi: 10.1042/bj2610951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou W. J., Ito A., Okazaki H., Omura T. Purification and characterization of a processing protease from rat liver mitochondria. EMBO J. 1989 Sep;8(9):2605–2612. doi: 10.1002/j.1460-2075.1989.tb08400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierotti A., Dong K. W., Glucksman M. J., Orlowski M., Roberts J. L. Molecular cloning and primary structure of rat testes metalloendopeptidase EC 3.4.24.15. Biochemistry. 1990 Nov 13;29(45):10323–10329. doi: 10.1021/bi00497a006. [DOI] [PubMed] [Google Scholar]
- Pollock R. A., Hartl F. U., Cheng M. Y., Ostermann J., Horwich A., Neupert W. The processing peptidase of yeast mitochondria: the two co-operating components MPP and PEP are structurally related. EMBO J. 1988 Nov;7(11):3493–3500. doi: 10.1002/j.1460-2075.1988.tb03225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A., Behrens M., Scherer P., Pratje E., Michaelis G., Schatz G. Inner membrane protease I, an enzyme mediating intramitochondrial protein sorting in yeast. EMBO J. 1991 Feb;10(2):247–254. doi: 10.1002/j.1460-2075.1991.tb07944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider H., Arretz M., Wachter E., Neupert W. Matrix processing peptidase of mitochondria. Structure-function relationships. J Biol Chem. 1990 Jun 15;265(17):9881–9887. [PubMed] [Google Scholar]
- Sztul E. S., Chu T. W., Strauss A. W., Rosenberg L. E. Import of the malate dehydrogenase precursor by mitochondria. Cleavage within leader peptide by matrix protease leads to formation of intermediate-sized form. J Biol Chem. 1988 Aug 25;263(24):12085–12091. [PubMed] [Google Scholar]
- Sztul E. S., Hendrick J. P., Kraus J. P., Wall D., Kalousek F., Rosenberg L. E. Import of rat ornithine transcarbamylase precursor into mitochondria: two-step processing of the leader peptide. J Cell Biol. 1987 Dec;105(6 Pt 1):2631–2639. doi: 10.1083/jcb.105.6.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropschug M., Nicholson D. W., Hartl F. U., Köhler H., Pfanner N., Wachter E., Neupert W. Cyclosporin A-binding protein (cyclophilin) of Neurospora crassa. One gene codes for both the cytosolic and mitochondrial forms. J Biol Chem. 1988 Oct 5;263(28):14433–14440. [PubMed] [Google Scholar]
- Witte C., Jensen R. E., Yaffe M. P., Schatz G. MAS1, a gene essential for yeast mitochondrial assembly, encodes a subunit of the mitochondrial processing protease. EMBO J. 1988 May;7(5):1439–1447. doi: 10.1002/j.1460-2075.1988.tb02961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Jensen R. E., Yaffe M. P., Oppliger W., Schatz G. Import of proteins into yeast mitochondria: the purified matrix processing protease contains two subunits which are encoded by the nuclear MAS1 and MAS2 genes. EMBO J. 1988 Dec 1;7(12):3857–3862. doi: 10.1002/j.1460-2075.1988.tb03271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G., Steppuhn J., Herrmann R. G. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989 Apr 1;180(3):535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]