Abstract
Heterochromatin formation in fission yeast depends on RNAi machinery and histone-modifying enzymes. One of the key histone-modifying complexes is Clr4-Rik1-Cul4 methyltransferase complex (CLRC), which mediates histone H3K9 methylation, a hallmark for heterochromatin. CLRC is composed of the Clr4 histone methyltransferase, Rik1, Raf1, Raf2 and Pcu4. However, transcriptional regulation of the CLRC subunits is not well understood. In this study, we identified Set3, a core subunit of the Set3/Hos2 histone deacetylase complex (Set3C), as a contributor to the integrity and silencing of heterochromatin at centromeres, telomeres and silent mating-type locus. This novel role of Set3 relies on its PHD finger, but is independent of deacetylase activity or structural integrity of Set3C. Set3 is not located to the centromeric region. Instead, Set3 is targeted to the promoters of clr4+ and rik1+, probably through its PHD finger. Set3 promotes transcription of clr4+ and rik1+. Consistently, the protein levels of Clr4 and Rik1 were reduced in the set3Δ mutant. The heterochromatin silencing defect in the set3Δ mutant could be rescued by overexpressing of clr4+ or rik1+. Our study suggests transcriptional activation of essential heterochromatin factors underlies the tight regulation of heterochromatin integrity.
Heterochromatin represents a heavily condensed and repressive form of chromatin. It plays crucial functions in chromatin segregation, genomic transcription and epigenetic gene silencing during mitosis and meiosis, thus maintaining genomic stability. In fission yeast, constitutive heterochromatin is found mainly at telomeres, the silent mating-type locus, and centromeres1. The assembly of pericentromeric heterochromatin is tightly regulated by the interplay of RNAi machineries and enzymes that modify histones. During S phase, nascent transcripts arising from the outer repeats (divided into elements known as dh and dg) of centromeric region are transcribed by an RNA-directed RNA polymerase complex (RDRC) into double-stranded RNA (dsRNA)2,3. dsRNAs are processed into siRNAs by Dicer (Dcr1) and delivered to a Argonaute chaperone complex (ARC), whereas siRNAs are loaded onto Argonaute (Ago1)4. Ago1 is assembled with Tas3 and Chp1 into an RNA-induced transcriptional silencing complex (RITS)5. siRNAs guide RITS to target nascent transcripts by sequence complementarity, and the transcripts are sliced by Ago1 to achieve transcriptional silencing6. Transcript-bound RITS recruits RDRC to produce more dsRNA and siRNA7. The self-reinforcing loop of RNAi is strengthened by histone-modifying enzymes, mainly through the connections between RITS and Clr4-Rik1-Cul4 methyltransferase complex (CLRC). Chromatin-bound-RITS recruits CLRC to the centromeric repeats through the linker protein Stc18. Clr4, the catalytic subunit of CLRC, methylates lysine 9 of histone H3 (H3K9me)9. H3K9me is bound by Chp1, which stabilizes the association between RITS and chromatin10. H3K9me also provides binding sites for the other heterochromatin components including Swi6, Chp2, histone deacetylase complex SHREC, and Clr4 itself to result in the establishment and maintenance of heterochromatin10,11,12,13.
CLRC consists of Clr4, the β-propeller protein Rik1, the cullin protein Pcu4 (also known as Cul4), the WD-40 protein Raf1 (also known as Dos1/Clr8/Cmc1) and the Zn-finger protein Raf2 (also known as Dos2/Clr7/Cmc2)14,15,16,17,18. Not only Clr4, but all other subunits of CLRC are essential for the heterochromatin formation19. Rik1 is thought to act upstream of Clr4, and to help recruit CLRC and the RNAi machinery to chromatin15,20. Pcu4 supports the putative E3 ubiquitin ligase activity of CLRC, probably by targeting substrates essential for heterochromatin assembly17. Raf1 and Raf2 are required for the localization of Swi6, a protein crucial for organization of higher order heterochromatin16. Compared to the physical interactions between CLRC subunits and other heterochromatin components, the transcriptional regulation of CLRC subunits is less well understood.
Set3 was first characterized in budding yeast by its feature of containing PHD finger and SET domain21. The combination of both domains is a characteristic displayed by a group of trx-G proteins, including histone methyltransferase Trx and Ash122. However, no methyltransferase activity has been reported for Set3. In budding yeast, Set3, Sif2, Snt1 and Hos2 forms the functional core of a histone deacetylase complex named Set3/Hos2 Complex (Set3C), in which Hos2 harbors deacetylase activity21. The composition of core Set3C is conserved in Schizosaccharomyces pombe and Candida albicans, and closely resembles the components of metazoan NCoR/SMRT corepressor complexes23,24,25. In budding yeast, Set3C is predominantly recruited to the 5′ transcribed region of genes to reduce the histone acetylation level26. The recruitment of Set3C is possibly mediated by the recognition of H3K4me2 by the PHD finger of Set3, or by the interaction between Set3C and Ser5-phosphorylated RNA polymerase II26,27. Set3C plays both repressive and activating roles in transcription, depending on the context of the region to which it is recruited28. Although the roles of Set3 in transcriptional regulation were reported or suggested in the context of Set3C, it is unclear whether Set3 can regulate transcription independent of Set3C. Here, we show that Set3 contributes to the integrity of heterochromatin by promoting the transcription of Clr4 and Rik1, two key subunits of CLRC. Unexpectedly, this role of Set3 is independent of Set3C, indicating a novel way of Set3-mediated transcriptional regulation.
Results
Set3 contributes to the integrity of constitutive heterochromatin
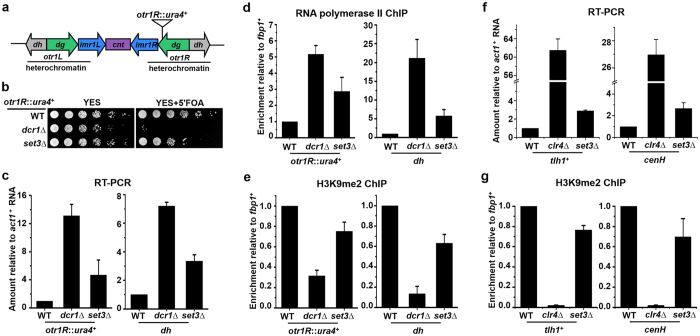
To dissect the mechanism of heterochromatin assembly, we carried out a genetic screen for mutants that displayed a defect in centromeric silencing in fission yeast. We used a parental strain in which the native ura4+ gene was deleted and a ura4+ marker gene was inserted into the outermost (otr) pericentromeric heterochromatin of chromosome 1 (otr1R::ura4+) (Fig. 1a)29. Suppression of the ura4+ marker gene by heterochromatin results in normal growth of the cells on medium containing the counterselective drug 5-fluoroorotic acid (5′-FOA), which is toxic to cells expressing ura4+. Once heterochromatin is disrupted by a mutation, the expression of ura4+ is increased and mutant cells are killed by 5′-FOA. As shown in Fig. 1b, deletion of set3+ (set3∆) in this strain derepressed the ura4+ gene, and that resulted in relatively poor growth on 5′-FOA containing medium compared to wild type (WT) cells. The phenotype of set3∆ mutant was similar to that of cells lacking an essential component of the RNAi machinery (dcr1∆), but to a much less extent. Consistently, the transcripts from the otr1R::ura4+ and endogenous pericentromeric repeat (dh), and the occupancy of RNA polymerase II (RNA Pol II) at both loci increased slightly in set3Δ cells (Fig. 1c,d). H3K9 methylation is a hallmark of constitutive heterochromatin in most eukaryotes. set3Δ cells exhibited reduced levels of H3K9 dimethylation (H3K9me2) at otr1R::ura4+ and dh, indicating impaired integrity of pericentromeric heterochromatin. Nevertheless, the drop of H3K9me2 in set3Δ cells was not dramatic as that in dcr1∆ mutant (Fig. 1e). The integrity of heterochromatin at telomeres and silent mating-type locus was also investigated. As shown in Fig. 1f, the transcription of tlh1+, a gene embedded in the subtelomeric region30, and that of cenH, an element present at the silent mating-type locus31, increased slightly in set3Δ cells. Amounts of H3K9me2 at both loci decreased in set3Δ mutant compared with WT cells (Fig. 1g). However, the defects of heterochromatin at both loci in set3Δ mutant are much weaker than those observed in a clr4Δ mutant (Fig. 1f,g). Thus, results suggest Set3 is not an essential component required for heterochromatin assembly, but is an auxiliary factor that helps maintain integrity and repressive histone modifications at constitutive heterochromatin.
Figure 1. Set3 contributes to the integrity of heterochromatin.
(a) A schematic representation of centromere 1 and the position of a inserted marker gene (otr1R::ura4). The pericentromeric heterochromatin region is indicated. (b) A fivefold serial dilution assay to examine the silencing of otr1R::ura4. Wild-type (WT) cells with silenced ura4+ grow normally on medium containing 5′-FOA, while loss of silencing kills cells on 5′-FOA. (c) RT-PCR analysis of otr1R::ura4+ and pericentromeric repeat (dh) RNA levels relative to a control act1+. The relative level in WT cells was arbitrarily designated as 1. Each column shown in (c) and below represents the mean ± s.d. from three biological repeats. (d,e) ChIP analysis of RNA Polymerase II (d) and H3K9me2 (e) at otr1R::ura4 and dh relative to fbp1+. Relative enrichment in WT cells was arbitrarily designated as 1. (f) RT-PCR analysis of tlh1+ and cenH RNA levels relative to a control act1+. (g) ChIP analysis of H3K9me2 at tlh1+ and cenH relative to fbp1+.
Role of Set3 in heterochromatin silencing depends on PHD finger, but is independent of Set3C
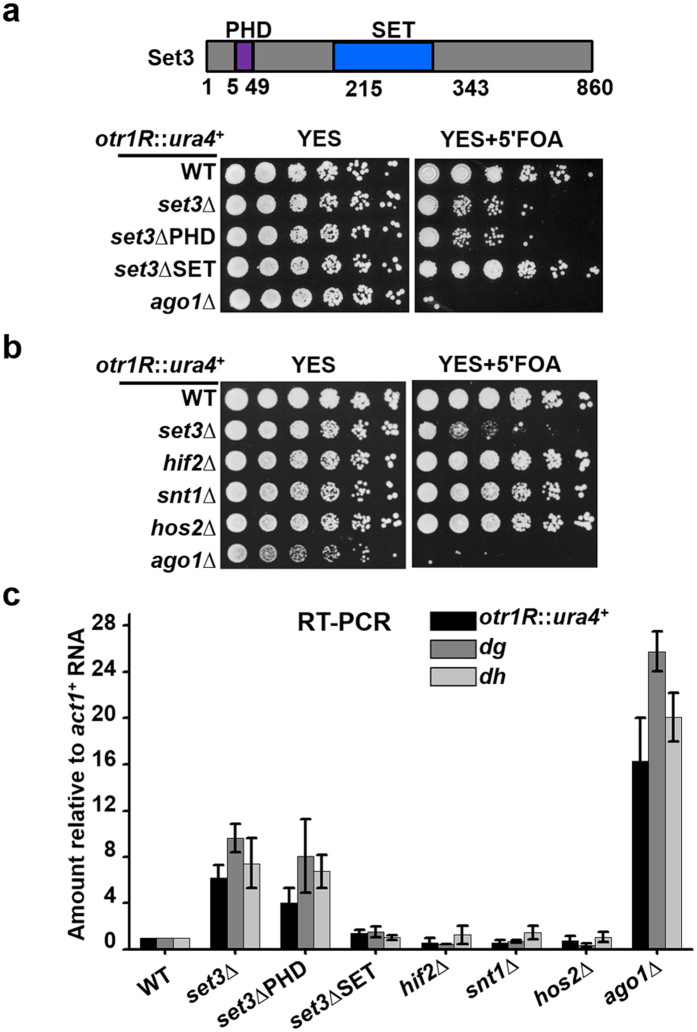
Set3 harbors a PHD finger close to its N terminus, and a SET domain around middle region (Fig. 2a). To investigate the contribution of both domains to Set3-mediated silencing, set3ΔPHD and set3ΔSET mutants were constructed in the strain carrying an otr1R::ura4+ reporter. Deletion of either PHD or SET domain does not cause the degradation of protein (Supplementary Fig. S1). The growth of set3ΔPHD mutant on the 5′-FOA containing medium was as poor as in a set3Δ deletion mutant. In contrast, the set3ΔSET mutant grew as well on 5′FOA as WT (Fig. 2a). Consistently, the transcript levels of otr1R::ura4, dg and dh increased in set3ΔPHD, but not in a set3ΔSET mutant (Fig. 2c). The results indicate that the role of Set3 in heterochromatin silencing depends on its PHD finger, but not its SET domain.
Figure 2. The role of Set3 in heterochromatin silencing relies on its PHD finger, but is independent of Set3C.
(a) A schematic representation of domains in Set3 (top). A fivefold serial dilution assay to examine the silencing of otr1R::ura4 in the mutants containing deletion of indicated domain (bottom). (b) A fivefold serial dilution assay to examine the silencing of otr1R::ura4+ in deletion mutants of Set3C subunits. (c) RT-PCR analysis of otr1R::ura4+, pericentromeric repeats (dg and dh) RNA levels relative to a control act1+. The relative level in WT cells was arbitrarily designated as 1. Each column represents the mean ± s.d. from three biological repeats.
In budding yeast, the PHD finger of Set3 recognizes H3K4me2 and is proposed to mediate the recruitment of Set3C to 5′ transcribed regions26. Therefore, we investigated whether other subunits of Set3C participate in the Set3-mediated silencing. Unexpectedly, in the strain bearing otr1R::ura4+, deletion of the catalytic subunit Hos2, or the structural subunits, including Snt1 and Hif2, did not affect the normal growth of the cells on 5′-FOA (Fig. 2b). Accordingly, transcript levels of otr1R::ura4+, dg and dh in hos2∆, snt1∆ and hif2∆ mutants were kept low as in WT cells (Fig. 2c). The results suggest that the pericentromeric region is silenced normally upon disruption of Set3C. Therefore, the role of Set3 in heterochromatin silencing is independent of enzymatic activity or the structural integrity of Set3C.
Set3 promotes the transcription of clr4 + and rik1 +
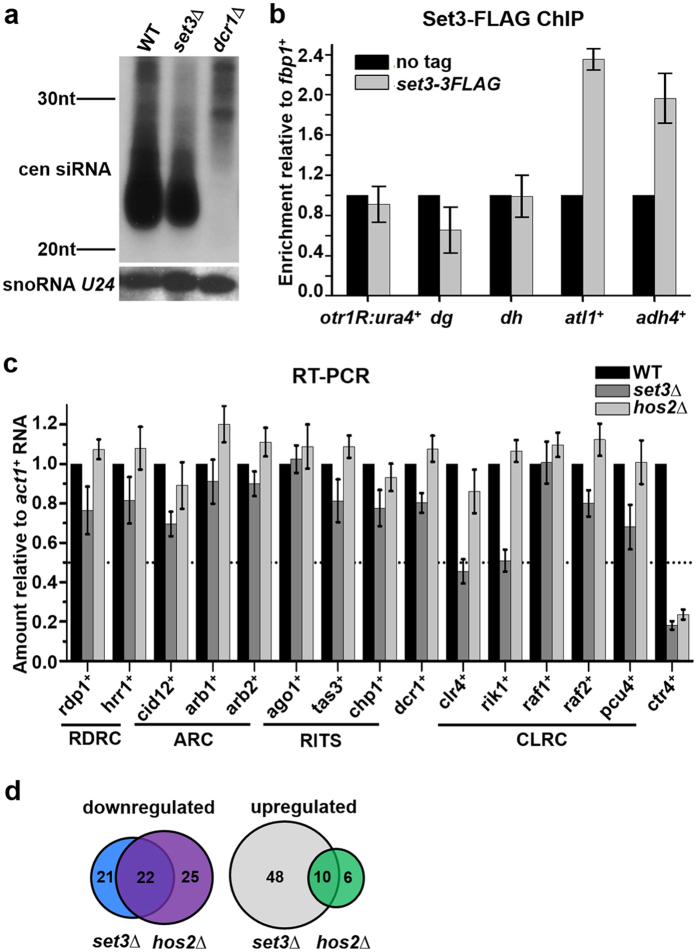
Since Set3 acts independently of Set3C in heterochromatin silencing, this novel role Set3 might have evolved in fission yeast to cope with different heterochromatin effectors not present in Saccharomyces cerevisiae, such as RNAi and CLRC. To analyze the potential role of Set3 in RNAi, we performed a Northern blotting to detect siRNAs level in a set3∆ mutant. siRNAs corresponding to the pericentromeric repeats (dg and dh) were slightly decreased in a set3∆ mutant compared to WT, while the level of non-coding snoRNA U24 was not affected (Fig. 3a). As a control, pericentromeric siRNAs were completely lost in dcr1∆ cells. The result is consistent with a weak phenotype of set3Δ mutant in heterochromatin silencing and suggests Set3 plays a non-essential role in the production of pericentromeric siRNAs.
Figure 3. Set3 promotes the transcription of clr4+ and rik1+.
(a) Northern blot analysis of centromeric siRNAs using probes against dg and dh. snoRNA U24 was detected as a loading control. (b) ChIP analysis of enrichment of Set3-3FLAG at otr1R::ura4+, dg, dh, atl1+ and adh4+ relative to fbp1+. fbp1+ is not subjected to the regulation of Set3 or Hos2 (Supplementary Fig. S3). Relative enrichment in the cells with no tag was arbitrarily designated as 1. Each column shown in (b) and below represents the mean ± s.d. from three biological repeats. (c) RT-PCR analysis of RNA levels of essential components of RNAi machinery and CLRC in set3∆ and hos2∆ mutants. The relative level to a control act1+ in WT cells was arbitrarily designated as 1. (d) Schematic representations of the genes downregulated or upregulated in the set3Δ and hos2Δ mutants, which were revealed by mRNA-seq.
Most of the RNAi components are localized at heterochromatin. To investigate whether Set3 is targeted to the pericentromeric region, we constructed a strain expressing Set3 with a C-terminal triple FLAG tag (Set3-3FLAG) and performed a ChIP assay. Note that this tag does not affect the gene silencing function of Set3 (Supplementary Fig. S2). Compared with an irrelevant euchromatic locus (fbp1+), Set-3FLAG was not enriched at the endogenous centromeric repeat (dh) and a inserted marker (otr1R::ura4+) (Fig. 3b). In contrast, substantial enrichment of Set3-3FLAG was detected at the ORFs of atl1+ and adh4+, both of which are known to be regulated by Set323. Thus, Set3 does not regulate silencing by directly binding to the centromeric chromatin.
Next, we investigated whether Set3 is involved in the transcriptional regulation of factors required for heterochromatin formation. The mRNA levels of essential components of RNAi machinery and CLRC were measured in WT and set3Δ cells. These factors include Rdp1, Hrr1 and Cid12 from RDRC; Arb1 and Arb2 from ARC; Ago1, Tas3 and Chp1 from RITS; Dcr1; Clr4, Rik1, Raf1, Dos2 and Pcu4 from CLRC. As shown in Fig. 3c, mRNA levels of clr4+ and rik1+ were reduced substantially in a set3Δ mutant. In contrast, there are no significant changes of mRNA levels in a hos2Δ mutant. Since Hos2 is the catalytic subunit of Set3C, these results suggest Set3-mediated transcription of clr4+ and rik1+ is irrelevant of the activity of Set3C. This conclusion is consistent with a Set3C-independent role of Set3 in the heterochromatin silencing.
Noting the genes differentially regulated by Set3 and Hos2, we generated expression profiles for WT, set3∆ and hos2∆ cells by mRNA-seq. 22 genes were downregulated and 10 genes were upregulated in both set3Δ and hos2Δ mutants, suggesting that these genes are subjected to regulation by Set3C (Fig. 3d, Supplementary Table S1). Among the downregulated genes, ctr4+ encodes a copper transporter complex subunit32. The positive regulation of ctr4+ by Set3 and Hos2 was further verified by RT-PCR and ChIP (Figs 3c and 4b,c), and thus ctr4+ serves as a control of Set3C-targeted gene in the following study. Meanwhile, 48 genes were upregulated and 21 genes were downregulated in a set3Δ mutant independently of Hos2 (Fig. 3d, Supplementary Table S1). This suggests a Hos2-independent role of Set3 in transcription is not rare in fission yeast.
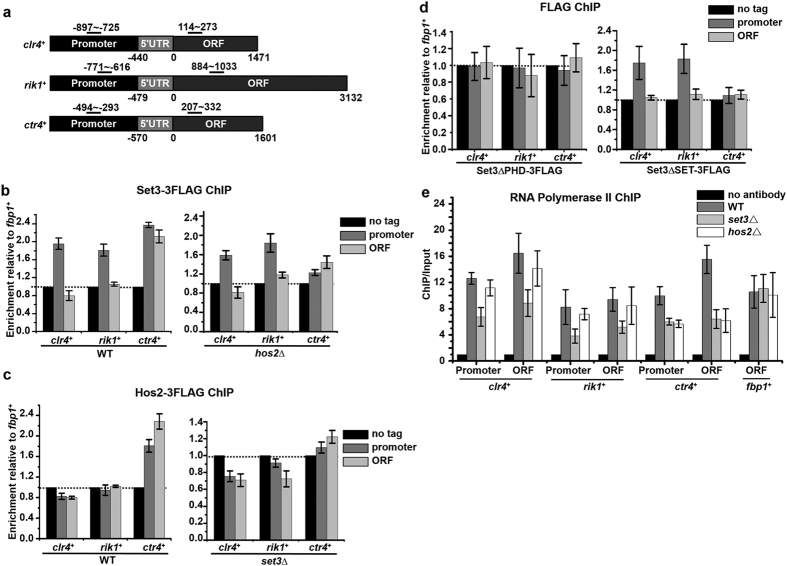
Figure 4. Set3 localizes to the promoters of clr4+ and rik1+ through its PHD finger.
(a) Schematic representations of clr4+, rik1+ and ctr4+ loci. Black bars indicate the fragments amplified in ChIP assay. (b–d) ChIP analysis of enrichments of Set3-3FLAG (b), Hos2-3FLAG (c), Set3ΔPHD-3FLAG (d) and Set3ΔSET-3FLAG (d) at promoter and ORF region of clr4+, rik1+, or ctr4+ in the indicated strains. Relative enrichment to fbp1+ in the cells with no tag was arbitrarily designated as 1. Each column shown in (b) and below represents the mean ± s.d. from three biological repeats. (e) ChIP analysis of enrichments of RNA Pol II at promoter and ORF region of clr4+, rik1+, ctr4+ or fbp1+in the indicated strains. Relative enrichment in the no antibody control was arbitrarily designated as 1.
Set3 localizes at the promoters of clr4 + and rik1 + through its PHD finger
To gain insight into the mechanism by which Set3 promotes the transcription of clr4+ and rik1+, the enrichment of Set3-3FLAG at both gene loci was investigated. As controls, the localizations of Set3 and Hos2 at ctr4+ locus was also determined (Fig. 4a). As shown in Fig. 4b, Set3 specifically localized at the promoter of clr4+ and rik1+, but not in their ORF region. There are several lines of evidence to support this localization is independent of Set3C. First, enrichment of Set3 at the promoters of clr4+ and rik1+ was not affected by the deletion of Hos2 (Fig. 4b). Second, Hos2 is not enriched at either the promoter or the ORF of either gene (Fig. 4c). Third, the conventional localization region of Set3C is not limited to the promoter region26. Consistently, Set3 and Hos2 were enriched at both the promoter and ORF region of ctr4+. The localization of Set3 at ctr4+ locus was abolished by the deletion of Hos2, and that of Hos2 was disrupted by the deletion of Set3, indicating ctr4+ is a bona fide target of Set3C (Fig. 4b,c). Therefore, our results suggest Set3 promotes the transcription of clr4+ and rik1+ by directly targeting the promoters of both genes, in a Set3C-independent manner.
The contributions of PHD finger and SET domain in the recruitment were investigated. Set3ΔPHD is not enriched at the promoters of clr4+ and rik1+, while Set3ΔSET is enriched at the promoters of both genes just like full-length Set3 (Fig. 4d). This suggests PHD finger, but not SET domain, is required for the localization of Set3 at the promoters of clr4+ and rik1+. Intriguingly, neither Set3ΔPHD nor Set3ΔSET is enriched at ctr4+ locus, suggesting PHD finger and SET domain are both required for the recruitment of Set3C to its target genes (Fig. 4d). The results strengthen the conclusion that Set3 adopts a Set3C-independent way to regulate the transcription of clr4+ and rik1+.
The role of Set3 in the transcription of clr4+ and rik1+ was further confirmed by a ChIP assay of RNA Pol II. As shown in Fig. 4e, occupancy of RNA Pol II at the promoters and coding regions of clr4+ and rik1+ was reduced substantially in set3Δ mutant, while no significant reduction was observed in hos2Δ mutant. This is consistent with the reduced mRNA levels of clr4+ and rik1+ in the set3Δ cells (Fig. 3C).
Heterochromatin silencing defect of set3Δ mutant is suppressed by overexpressing clr4 + or rik1 +
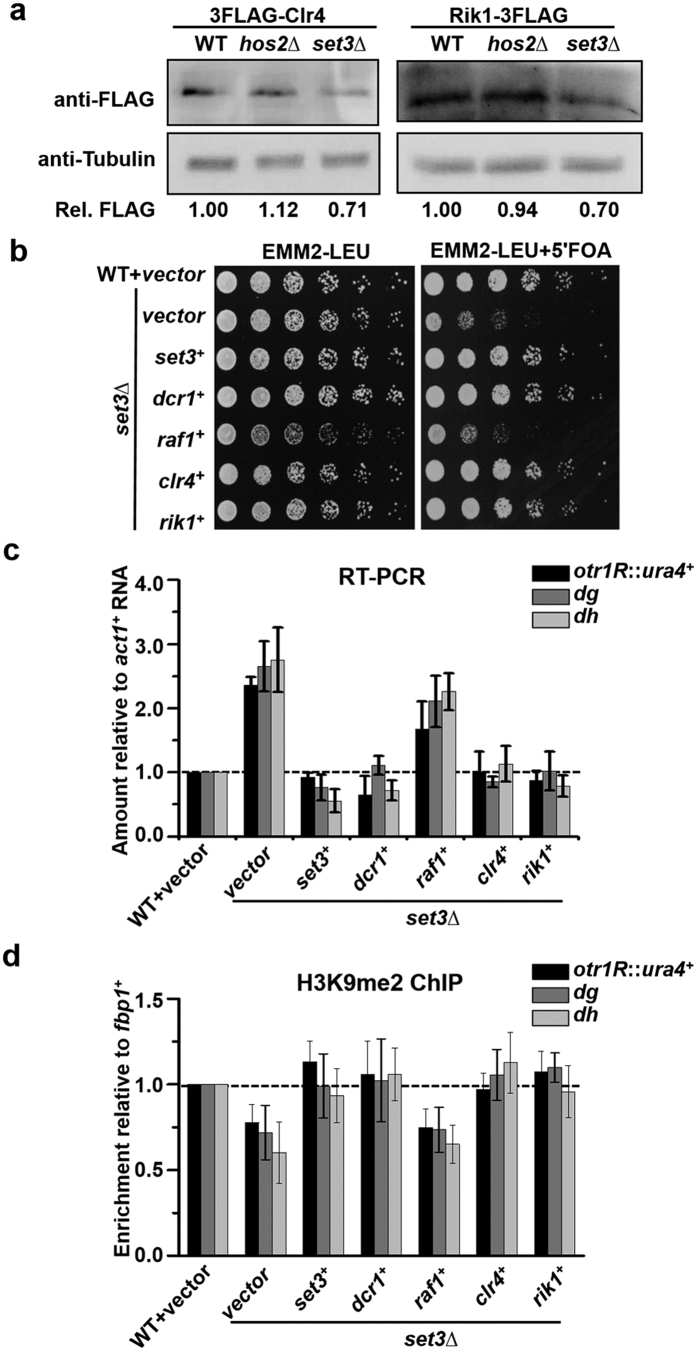
As Set3 promotes the transcription of clr4+ and rik1+, we investigated whether the protein level of both factors is regulated by Set3 accordingly. To this end, Set3 was deleted in a strain expressing FLAG-tagged Rik1 or Clr4. The tags do not affect heterochromatin silencing (Supplementary Fig. S4). The protein levels of Clr4 and Rik1 in the set3Δ mutant reduced by 30% compared to those in WT cells. As expected, deletion of Hos2 had no apparent effect on the protein levels of both factors (Fig. 5a). H3K9 methylation declines rapidly upon the transient depletion of Clr4 or Rik119,33. Thus, reduced protein levels of Clr4 and Rik1 are consistent with the decrease of H3K9me2 in set3Δ mutant (Fig. 1e,g).
Figure 5. Heterochromatin silencing defect of set3Δ mutant is suppressed by overexpressing clr4+ or rik1+.
(a) Western blotting assay to examine the level of 3FLAG-Clr4 and Rik1-3FLAG in WT, set3∆ and hos2∆ cells. Tubulin was detected as a loading control. Cropped blots are shown for clarity. Full-length blots are presented in Supplementary Fig. S5. The relative level of FLAG tagged protein (Rel. FLAG) was calculated by quantifying the relative density of 3FLAG-Clr4 or Rik1-3FLAG against tubulin. (b) Fivefold serial dilution assay to examine the silencing of otr1R::ura4+ in set3∆ cells overexpressing set3+, dcr1+, raf1+, clr4+ or rik1+. set3∆ cells were transformed with a plasmid overexpressing the indicated gene and transformants were subject to the dilution assay. (c) RT-PCR analysis of otr1R::ura4+, dg and dh RNA levels relative to a control act1+ in the transformants in (b). The relative level to a control act1+ in WT cells was arbitrarily designated as 1. Each column in (c) and below represents the mean ± s.d. from three biological repeats. (d) ChIP analysis of H3K9me2 at otr1R::ura4+, dg and dh relative to fbp1+ in the transformants in (b). Relative enrichment in WT cells was arbitrarily designated as 1.
To test whether the reduction of Clr4 and Rik1 accounts for the pericentromeric silencing defect of the set3Δ mutant, we performed a complementation assay. set3∆ mutant was transformed with a plasmid overexpressing clr4+, rik1+, raf1+, set3+, or dcr1+. The overexpression of genes was verified by RT-PCR (Supplementary Fig. S6). The silencing defect of otr1R::ura4+ in the set3∆ mutant was substantially rescued by overexpressing either clr4+ or rik1+, as shown by the improved growth on 5′FOA plate. In contrast, overexpressing raf1+, whose transcription is not affected by the deletion of Set3, could not rescue the silencing defect of the set3∆ mutant (Fig. 5b). Consistent with the good growth on 5-FOA plate, transcript levels of ura4+ and centromeric repeats (dh, dg) decreased in set3Δ cells overexpressing clr4+ or rik1+, but not in the cells overexpressing raf1+ (Fig. 5c). Dcr1 interacts with Clr4 and Rik1. Overexpressed Dcr1 promotes CLRC recruitment and H3K9 methylation at the centromeric region34. As shown in Fig. 5b, overexpressing dcr1+ was also able to rescue the growth defect of set3∆ cells on 5′-FOA, suggesting a connection between the Dcr1 and the Set3-regulated function of CLRC. Accordingly, transcript levels of ura4+ and dh/dg decreased in set3Δ mutant overexpressing dcr1+ (Fig. 5c). The integrity of pericentromeric heterochromatin was further investigated by a ChIP assay of H3K9me2. Levels of H3K9me2 at otr1R::ura4+, dg and dh in the set3Δ mutant were recovered by overexpressing clr4+, rik1+, dcr1+, but not by overexpressing raf1+ (Fig. 5d). These results suggest impaired silencing and integrity of heterochromatin in set3Δ mutant is mainly due to the reduced protein levels of Clr4/Rik1 and interfered H3K9 methylation by CLRC.
Discussion
The interplay between the components essential for the heterochromatin formation and silencing is commonly mediated by physical interactions among the various proteins. But recently, the transcriptional regulation of heterochromatin factors emerges as a novel way to regulate heterochromatin assembly. Cwf14-mediated splicing of mRNA of RNAi factors, including Ago1, Arb2, Ers1, and Dsh1, is critical for pericentromeric heterochromatin assembly35. Tls1-mediated splicing of shelterin mRNAs regulates telomeric heterochromatin formation36. However, none of CLRC subunits contain intron, thus they are not subjected to the regulation by spliceosome. In this study, we reveal a novel role of Set3 in maintaining the integrity of heterochromatin by promoting the transcription of Clr4 and Rik1, two key components of CLRC. Decreased transcription resulted in the reduction of protein levels of both factors in the set3Δ mutant. Since the expression of Clr4 and Rik1 is not totally abolished, a limited effect on the function of CLRC, including assembly, targeting and the catalytic reactions, is expected in a set3Δ mutant. Accordingly, a relative mild effect on integrity and silencing of constitutive heterochromatin is observed in the set3Δ mutant (Fig. 1c–g). A weak effect on the production of pericentromeric siRNA is also observed (Fig. 3a). As H3K9me stabilizes the interaction between RITS and chromatin10, decreased H3K9me at pericentromeric region in the set3Δ mutant might affect the proper recruitment of RITS and following siRNA production. The weak phenotype of set3Δ mutant is contrast with the dramatic defects observed in the deletions of subunit of CLRC, or deletions of other factors that directly participate in the heterochromatin assembly37,38,39. Therefore, Set3 is not an on/off regulator, but is more like a fine-tuning factor contributing to the tight regulation of heterochromatin integrity at centromeres, telomeres and silent mating-type locus. Besides the constitutive heterochromatin region, low levels of CLRC binds with euchromatin sites, including noncoding RNAs, intergenic regions and meiotic genes13. Whether the disruption of CLRC by the deletion of Set3 imposes more significant effects on the its targets in the euchromatin region is worth studying in the future.
Our study indicates that Set3 is able to promote transcription in a Set3C-independent way. This novel role of Set3 might have evolved to target new genes absent in budding yeast, such as clr4+ and rik1+. Set3C is predominantly localized to the 5′ transcribed region and promotes transcription by inhibiting the cryptic transcription inside the ORF26. In fission yeast, targeting of Set3C seems require both PHD finger and SET domain of Set3. In contrast, Set3 is specifically recruited to the promoters of clr4+ and rik1+. The recruitment is probably mediated by the interaction between PHD finger and H3K4me226, while SET domain is not necessary. Localization at the promoters suggests that Set3 contributes to the transcription initiation of clr4+ and rik1+, but underlying mechanism requires further investigation.
Methods
Yeast strains and plasmids
All the strains used in this study are listed in Supplementary Table S2. Gene deletion and tagging were performed by homologous recombination using a plasmid-based method40. Cells were grown in yeast extract medium with supplements (YES) or Edinburgh minimal medium minus leucine (EMM2-LEU) medium41. ORF of set3+, rik1+, raf2+, clr4+ or dcr1+ was cloned into pRep41 vector for overexpression42.
Fivefold serial dilution assay
Exponentially growing cells were collected and adjusted to an A600 of 1.0. Samples were diluted by fivefold for five times. 5 μl dilutions were spotted onto YES or EMM2-LEU medium supplemented with 5′FOA (003234, Fluorochem, Hadfield, UK, 1 g/Liter) as indicated. Plates were incubated for 2 or 3d at 32 °C before imaging.
RT-PCR
1 × 108 exponentially growing cells were harvested. Total RNA were extracted using the RiboPure Yeast Kit (AM1926, Life Technologies, Carlsbad, CA, USA) and reverse transcribed into cDNA by using PrimeScript RT (RR037A, Takara, Dalian, China). qPCR was performed using SYBR Premix Ex TaqII (RR820A, Takara) in a LightCycler 480 II Real-Time PCR System (Roche Applied Science, Penzberg, Upper Bavaria, Germany). Primers used are listed in Supplementary Table S3.
ChIP
3 × 108 exponentially growing cells were fixed with 1% formaldehyde for 25 min at 30 °C. After quenching by 250 mM glycine, cells were harvested and washed with Buffer 1 (50 mM HEPES (pH 7.5), 140 mM NaCl, 1 mM EDTA, 1% TritonX-100, 0.1% Na-deoxycholate). Cells were resuspended in Buffer 1 supplemented with protease inhibitors cocktail (05892970001, Roche Applied Science) and homogenized with a bead-beater (FastPrep-24, MP, California, USA) by glass beads. The cell extract was sonicated for 8 min at high frequency with a sonication system (Bioruptor UCD-300, Diagenode, Seraing, Belgium) and centrifuged. Supernatant was incubated with anti-H3K9me2 (07-441, Millipore, Massachusetts, USA), anti-RNA polymerase II 8WG16 (664906, Biolegend, San Diego, USA), or anti-FLAG (F1804, Sigma-Aldrich, St Louis, MO, USA) antibody for 4 hours. Samples were subjected to purification by using an EZ-Magna ChIP A Kit (17-408, Millipore). Eluted DNA was subjected to qPCR as described above. Primers used are listed in Supplementary Table S3.
Northern blot
Northern blot was performed as described previously39. Primers and oligos used are listed in Supplementary Table S3.
Whole cell extract
Exponentially growing yeast cells were harvested. Cells were washed and resuspended in ice-cold Buffer 1 supplemented with protease inhibitors as described above. Cells were broken in a bead-beater by glass beads. Extract was centrifuged at 4 °C. Supernatant was boiled with SDS-gel loading buffer and subjected to Western blotting by using anti-FLAG and anti-Tubulin (T6074, Sigma) antibodies. The blots were scanned and quantified by GeneGnome HR system (Syngene, Cambridge, UK).
mRNA-seq
Exponentially growing yeast cells were harvested and total RNA was extracted by using RiboPure Yeast Kit. Enrichment of mRNA, fragmentation, reverse transcription, library construction, Hi-seq and data analysis were performed by Genergy Biotechnology (Shanghai, China).
Additional Information
How to cite this article: Yu, Y. et al. Set3 contributes to heterochromatin integrity by promoting transcription of subunits of Clr4-Rik1-Cul4 histone methyltransferase complex in fission yeast. Sci. Rep. 6, 31752; doi: 10.1038/srep31752 (2016).
Supplementary Material
Acknowledgments
We thank Robin Allshire (The University of Edinburgh, UK) and Shiv Grewal (National Institutes of Health, USA) for providing valuable strains, Rolf Sternglanz (Stony Brook University, USA) for his valuable comments on the manuscript. This work is supported by the National Natural Science Foundation of China (91129730 and 21132004 to HL, 31571287 to YY), the program 863 (2014AA021301 and 2013AA102803B to HL), independent research funding of Fudan University (20520133231 to YY), and the Open Fund of State Key Laboratory of Genetic Engineering.
Footnotes
Author Contributions Y.Y. and H.Z. designed and performed experiments. X.D. performed the Northern blot and assisted ChIP assay. W.W. assisted the construction of strains and ChIP assay. Y.Y. and H.L. supervised the project. Y.Y. and H.Z. wrote the manuscript. All co-authors critically reviewed the manuscript.
References
- Grewal S. I. & Jia S. Heterochromatin revisited. Nat Rev Genet. 8, 35–46 (2007). [DOI] [PubMed] [Google Scholar]
- Colmenares S. U., Buker S. M., Buhler M., Dlakic M. & Moazed D. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell. 27, 449–461 (2007). [DOI] [PubMed] [Google Scholar]
- Chen E. S. et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 451, 734–737 (2008). [DOI] [PubMed] [Google Scholar]
- Holoch D. & Moazed D. Small-RNA loading licenses Argonaute for assembly into a transcriptional silencing complex. Nat Struct Mol Biol. 22, 328–335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A. et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 303, 672–676 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine D. V. et al. Argonaute slicing is required for heterochromatic silencing and spreading. Science. 313, 1134–1137 (2006). [DOI] [PubMed] [Google Scholar]
- Motamedi M. R. et al. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 119, 789–802 (2004). [DOI] [PubMed] [Google Scholar]
- Bayne E. H. et al. Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell. 140, 666–677 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J., Rice J. C., Strahl B. D., Allis C. D. & Grewal S. I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 292, 110–113 (2001). [DOI] [PubMed] [Google Scholar]
- Schalch T. et al. High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin. Mol Cell. 34, 36–46 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M., Iida T., Urano T. & Nakayama J. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 23, 3825–3835 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T. et al. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 128, 491–504 (2007). [DOI] [PubMed] [Google Scholar]
- Zhang K., Mosch K., Fischle W. & Grewal S. I. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 15, 381–388 (2008). [DOI] [PubMed] [Google Scholar]
- Hong E. J., Villen J., Gerace E. L., Gygi S. P. & Moazed D. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol. 2, 106–111 (2005). [DOI] [PubMed] [Google Scholar]
- Jia S., Kobayashi R. & Grewal S. I. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol. 7, 1007–1013 (2005). [DOI] [PubMed] [Google Scholar]
- Li F. et al. Two novel proteins, dos1 and dos2, interact with rik1 to regulate heterochromatic RNA interference and histone modification. Curr Biol. 15, 1448–1457 (2005). [DOI] [PubMed] [Google Scholar]
- Horn P. J., Bastie J. N. & Peterson C. L. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev. 19, 1705–1714 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G. et al. The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe. Genetics. 171, 1583–1595 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanker S. et al. Continuous requirement for the Clr4 complex but not RNAi for centromeric heterochromatin assembly in fission yeast harboring a disrupted RITS complex. PLoS Genet 6, e1001174 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace E. L., Halic M. & Moazed D. The methyltransferase activity of Clr4Suv39h triggers RNAi independently of histone H3K9 methylation. Mol Cell. 39, 360–372 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijnappel W. W. et al. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 15, 2991–3004 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon S. C., Zhang X., Trievel R. C. & Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 6, 227 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentas S. et al. The SET domain protein, Set3p, promotes the reliable execution of cytokinesis in Schizosaccharomyces pombe. PLoS One 7, e31224 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D. et al. A histone deacetylase adjusts transcription kinetics at coding sequences during Candida albicans morphogenesis. PLoS Genet. 8, e1003118 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R. et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol. 9, 1401–1412 (2007). [DOI] [PubMed] [Google Scholar]
- Kim T. & Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 137, 259–272 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind C. K. et al. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell. 39, 234–246 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Xu Z., Clauder-Munster S., Steinmetz L. M. & Buratowski S. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 150, 1158–1169 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire R. C., Nimmo E. R., Ekwall K., Javerzat J. P. & Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9, 218–233 (1995). [DOI] [PubMed] [Google Scholar]
- Hansen K. R., Ibarra P. T. & Thon G. Evolutionary-conserved telomere-linked helicase genes of fission yeast are repressed by silencing factors, RNAi components and the telomere-binding protein Taz1. Nucleic Acids Res. 34, 78–88 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall I. M. et al. Establishment and maintenance of a heterochromatin domain. Science. 297, 2232–2237 (2002). [DOI] [PubMed] [Google Scholar]
- Labbe S., Pena M. M., Fernandes A. R. & Thiele D. J. A copper-sensing transcription factor regulates iron uptake genes in Schizosaccharomyces pombe. J Biol Chem. 274, 36252–36260 (1999). [DOI] [PubMed] [Google Scholar]
- Partridge J. F. et al. Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Mol Cell. 26, 593–602 (2007). [DOI] [PubMed] [Google Scholar]
- Yu R., Jih G., Iglesias N. & Moazed D. Determinants of heterochromatic siRNA biogenesis and function. Mol Cell. 53, 262–276 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallgren S. P. et al. The proper splicing of RNAi factors is critical for pericentric heterochromatin assembly in fission yeast. PLoS Genet. 10, e1004334 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. et al. Tls1 regulates splicing of shelterin components to control telomeric heterochromatin assembly and telomere length. Nucleic Acids Res. 42, 11419–11432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi A. et al. Heterochromatin protein 1 homologue Swi6 acts in concert with Ers1 to regulate RNAi-directed heterochromatin assembly. Proc Natl Acad Sci USA 109, 6159–6164 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K., Hayashi A., Nakayama J. & Murakami Y. A novel RNAi protein, Dsh1, assembles RNAi machinery on chromatin to amplify heterochromatic siRNA. Genes Dev. 26, 1811–1824 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X. et al. Sgf73, a subunit of SAGA complex, is required for the assembly of RITS complex in fission yeast. Sci Rep. 5, 14707 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan J. et al. High-throughput knockout screen in fission yeast. Nat Protoc. 1, 2457–2464 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A. & Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823 (1991). [DOI] [PubMed] [Google Scholar]
- Basi G., Schmid E. & Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 123, 131–136 (1993). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.