Abstract
Background
Traditional prognostic indicators of breast cancer, i.e. lymph node diffusion, tumor size, grading and estrogen receptor expression, are inadequate predictors of metastatic relapse. Thus, additional prognostic parameters appear urgently needed. Individual oncogenic determinants have largely failed in this endeavour. Only a few individual tumor growth drivers, e.g. mutated p53, Her-2, E-cadherin, Trops, did reach some prognostic/predictive power in clinical settings. As multiple factors are required to drive solid tumor progression, clusters of such determinants were expected to become stronger indicators of tumor aggressiveness and malignant progression than individual parameters. To identify such prognostic clusters, we went on to coordinately analyse molecular and histopathological determinants of tumor progression of post-menopausal breast cancers in the framework of a multi-institutional case series/case-control study.
Methods
A multi-institutional series of 217 breast cancer cases was analyzed. Twenty six cases (12 %) showed disease relapse during follow-up. Relapsed cases were matched with a set of control patients by tumor diameter, pathological stage, tumor histotype, age, hormone receptors and grading. Histopathological and molecular determinants of tumor development and aggressiveness were then analyzed in relapsed versus non-relapsed cases. Stepwise analyses and model structure fitness assessments were carried out to identify clusters of molecular alterations with differential impact on metastatic relapse.
Results
p53, Bcl-2 and cathepsin D were shown to be coordinately associated with unique levels of relative risk for disease relapse. As many Ras downstream targets, among them matrix metalloproteases, are synergistically upregulated by mutated p53, whole-exon sequence analyses were performed for TP53, Ki-RAS and Ha-RAS, and findings were correlated with clinical phenotypes. Notably, TP53 insertion/deletion mutations were only detected in relapsed cases. Correspondingly, Ha-RAS missense oncogenic mutations were only found in a subgroup of relapsing tumors.
Conclusions
We have identified clusters of specific molecular alterations that greatly improve prognostic assessment with respect to singularly-analysed indicators. The combined analysis of these multiple tumor-relapse risk factors promises to become a powerful approach to identify patients subgroups with unfavourable disease outcome.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-016-2713-3) contains supplementary material, which is available to authorized users.
Keywords: Breast cancer, Metastatic relapse, Prognostic indicators, TP53, Bcl-2, Cathepsin D, RAS
Background
Breast cancer (BC) is the most frequent malignancy in women with 800 cases out of 100,000 people, four-times as many as the second most frequent one, i.e. colorectal cancer [1]. Histopathology classification of BC according to tumor grade, stage, histotype, lymph node invasion and hormonal receptor status [2] is broadly used to draw correlations with survival. However, this classification performs poorly in predicting differential biological aggressiveness of tumors with identical grade and stage. As an example, patients with the best prognosis, i.e. bearing small size tumors, expressing estrogen receptors and without lymph node invasion, experience early tumor relapse in 10-20 % of the cases [3, 4]. Cases that relapse do not detectably differ from those that do not, as far as conventional prognostic parameters are concerned.
Determinants of tumor biological history are expected to add to traditional prognostic classification algorithms [5, 6]. Individual oncogenic determinants, e.g. p53, Her-2, E-cadherin, BRCA-1, Trops, have indeed been shown to add to prognostic and predictive procedures [5, 7–11]. However, they largely failed to outperform traditional prognostic indicators.
Tumor development depends on the accumulation of several specific genetic and epi-genetic changes [12–14]. Thus, the analysis of individual oncogenic factors is unlikely to suffice in defining the biological nature and aggressiveness of a tumor [15]. Major control pathways or clusters of drivers of cell growth, apoptosis or invasion are, on the other hand, expected to associate with tumor aggressiveness and overall malignancy much more strongly than individual factors. In this work we went on to test this model. Histopathology and oncogenically-activated determinants of tumor progression of BC were analyzed in the framework of a case-control study. The results obtained were evaluated by means of statistical analyses able to detect significant interactions of biological determinants connected with tumor relapse. This showed that correlated p53, Bcl-2 and cathepsin D specifically associate with unprecedented high levels of relative risk for local invasion and metastatic relapse. As matrix metalloproteases, which play a key role in local invasion and distant cancer spreading, were shown to be a transactivation target for mutant p53, in cooperation with oncogenic Ras, exon sequence analysis was performed for TP53 and RAS genes, and findings were coordinately analyzed with the immunohistochemistry (IHC) data and clinical phenotypes.
Methods
Breast cancer case series
A multi-institutional case series of BC patients was collected from the National Cancer Institute of Naples, together with the University of Udine, the district hospital of Venice and Rovigo, Italy. Two hundred and seventeen BC patients were analyzed (Table 1). Clinical data (age, family history, clinical stage, disease follow-up) and conventional prognostic indicators (size, pathological stage, local invasion, margin width, lymph-node invasion, histological type, necrosis, inflammatory infiltration, hormonal receptor status) were recorded [16, 17] (Table 1). Cancer grade was determined as described [18] (Table 1; Additional file 1: Table S1 and Additional file 2: Table S2). Twenty six cases (12 %) showed disease relapse during follow-up (Additional file 1: Table S1). Relapsed cases were matched with a set of control patients by tumor diameter, pathological stage, tumor histotype, age, hormone receptors and grading (Additional file 1: Table S1), and analyzed for expression of tumor progression determinants by immuno-histochemistry (IHC) and DNA sequencing, as indicated. To identify patterns of aggregation of molecular alterations associated to different classes of BC prognosis, stepwise grouping procedures were performed for model structure fitness assessment, as described.
Table 1.
Patient case series
| Age (mean ± SD) | 64.7 ± 10.9 |
|---|---|
| Type of surgery; No (%) | |
| Conservative | 111 (51.2) |
| Radical | 106 (48.8) |
| Histotype; No (%) | |
| Ductal | 186 (85.7) |
| Lobular | 20 (9.2) |
| Mixed | 11 (5.1) |
| Tumor size; No (%) | |
| T1 | 137 (63.1) |
| T2 | 76 (35.0) |
| T3 | 2 (0.9) |
| T4 | 1 (0.5) |
| T Multi | 1 (0.5) |
| Lymph node status; No (%) | |
| pN0 | 199 (91.7) |
| pN1 | 12 (5.5) |
| pN2 | 3 (1.4) |
| pN3 | 3 (1.4) |
| Grading; No (%) | |
| G1 | 27 (12.4) |
| G2 | 117 (53.9) |
| G3 | 73 (33.6) |
Histopathology
Tissue micro-arrays (TMA) of tumor samples were assembled as described [19, 20]. Briefly, whole-tumor sections of formalin-fixed paraffin-embedded (FFPE) BC samples were stained with hematoxylin-eosin, and used for guiding selection of tumor-containing areas. Three 1 mm diameter cylinders were then obtained from all tumors and transferred to recipient blocks. Filled blocks were heated for 15 min at 37 °C to induce the tumor cores to adhere to the paraffin walls. TMA sections were analysed by IHC for the expression of markers relevant to tumor development and aggressiveness (Figs. 1, 2 and 3, Table 1). Briefly, 5 μm sections of BC TMA were mounted onto Vectabond-coated slides (Vector Laboratory). Before staining, sections were heated at 56 °C and dewaxed in xylene/ethanol. Endogenous peroxidase was blocked with hydrogen peroxide in methanol. Heat-mediated ‘antigen retrieval’ was performed by treatment in pH 6 citrate buffer in a pressure cooker or microwave oven, as required for each specific target. After pre-incubation with appropriate blocking agents, e.g. species-matched normal serum, sections was incubated with the primary antibody (Additional file 3: Table S3). After washing, sections were challenged with fragment antigen-binding (Fab)2 biotinylated secondary reagents, followed by avidin-peroxidase and 3,3′-diaminobenzidine tetrachlorohydrate, activated with 0.3 % hydrogen peroxide. Nuclei were counterstained with Mayer’s hematoxylin. Appropriate normal mouse/rabbit secondary reagents or unrelated antibodies were used as negative controls. Primary antibodies directed against the chosen targets are listed in Table 2. Immunoreactivity for the various reagents was quantified on an average of 1000 cells in randomly chosen fields (40x objective). Semiquantitative scores were determined by two independent observers (M. P. and R. L.). Percent expression values of ER, PR and HER2 tended to distribute around discrete values (0, 10, 25, 50, 75 and 100 % of tumor cells) and were categorized accordingly. Percentages of Ki-67 and p53 expressing cells were analyzed without discretization, but are reported here as categorical variables for convenience (0, 1–10, 11–75, 76–100) [8]. Intensity scores varied between 0 and 3, where 0 is no reactivity, 1 is borderline detectability, 3 is the maximum observed intensity and 2 corresponds to an intermediate intensity. A combined score was obtained by multiplying percentages of positive cells by intensity. Scores were then categorized for statistical evaluation [21].
Fig. 1.
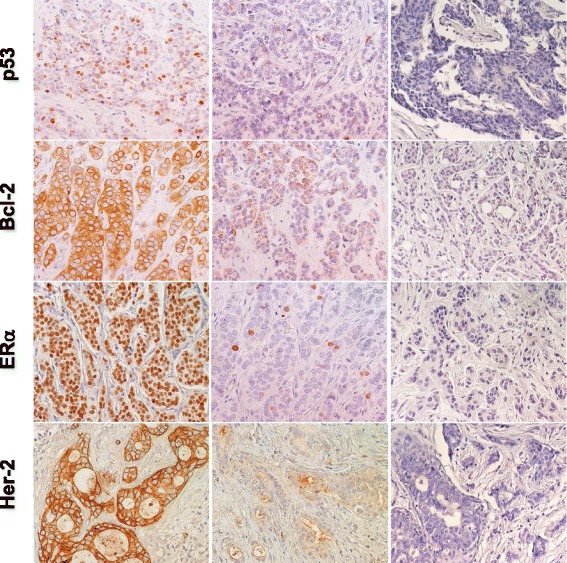
Expression of p53, Bcl-2, ERα and Her-2 in BC. Representative examples of highly expressing (left column) versus negative/low cases (middle column) are shown. Negative controls are shown on the right. Nuclei were counterstained with Mayer’s hematoxylin (in blue). Original magnification: ×400
Fig. 2.
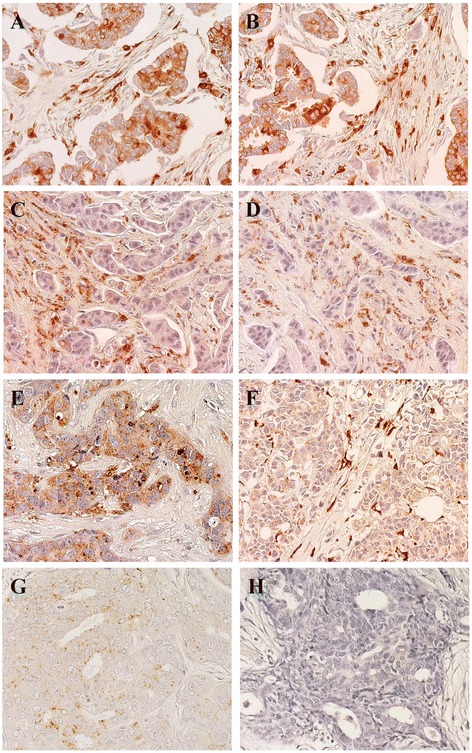
Expression of cathepsin D in BC Representative examples are shown. Positive/high (left column) versus negative/low (right column) cases for expression of cathepsin D and MMP11 are indicated. Nuclei were counterstained with Mayer’s hematoxylin (in blue). a, b Tumor cases with high cathepsin D expression both in cells and in the stroma between the tumor cells. c, d Tumor cases with low/nil cathepsin D expression in cells, but with detectable expression in the tumor stroma. e Case with high cathepsin D expression in tumor cells, but no expression in the stroma. f Case with no cathepsin D expression in tumor cells, but high expression in the stroma. g Case with low/nil cathepsin D expression in both tumor cells and stroma. h Negative control for staining. Original magnification: ×400
Fig. 3.
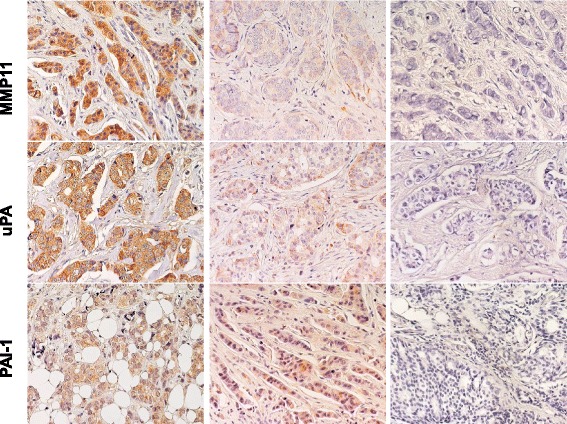
Expression of MMP11, uPA, PAI-1 in BC. Representative examples of highly expressing (left column) versus negative/low cases (middle column) are shown. Negative controls are shown on the right. Nuclei were counterstained with Mayer’s hematoxylin (in blue)
Table 2.
Antibodies utilized for immuno-histochemical detection
| Target protein | Antibody | Type | Provider |
|---|---|---|---|
| Bax | p-19a | polyclonal | Santa Cruz |
| Bcl-2 | 100/D5 | polyclonal | Novocastra/YLEM |
| Cathepsin D | cat-D | polyclonal | DAKO |
| Cyclin D1 | DCS6 | monoclonal | Novocastra/YLEM |
| Cyclin E | 13A3 | monoclonal | Novocastra/YLEM |
| ERα | MU368-UC | monoclonal | Biogenex |
| Her-2/erb-B/neu | Hercep-test | polyclonal | DAKO |
| Ki-67 | MIB-1 | monoclonal | Novocastra/YLEM |
| p16/INK4 | p16 | polyclonal | Pharmingen |
| p27/kip1 | F-8 | monoclonal | Santa Cruz |
| p53 | DO7 | monoclonal | NeoMarkers |
| PAI-1 | sc-6642 | polyclonal | Santa Cruz |
| PgR | 1A6 | polyclonal | Ventana |
| Stromelysin/MMP11 | sc-8837 | polyclonal | Santa Cruz |
| uPA | sc-6830 | polyclonal | Santa Cruz |
aantibodies were utilized as described in Methods
DNA extraction
FFPE BC sections were processed as described [22, 23]. This procedure provided with relatively crude DNA preparations, which, however, could be efficiently used as a template in ≤150 bp-long polymerase chain reaction (PCR) amplifications [24]. Briefly, four 5 μm tumor sections were deparaffinized by two extractions with either xylene or Histoclear (Carlo Erba), followed by two extractions with ethanol. Samples were then digested for 3 h at 50 °C with proteinase K (PK) 2 mg/ml, Tween 20, Tris-Cl 50 mM, EDTA 1 mM, pH 8.5, then overnight at 50 °C after PK replenishing. Samples were then incubated at 95 °C for 15 min to inactivate PK, centrifuged at top speed for 15 min at 4 °C, transferred to a fresh tube and stored at − 20 °C. DNA yields were quantified by ethidium bromide fluorescence in solution [25]. On average 30 μg DNA/sample were obtained. Size distribution of the extracted DNA [26] was profiled by ethidium bromide/agarose gel electrophoresis for sample quality assessment.
PCR amplification
After thawing, DNA samples prepared as above were incubated at 95 °C for 25 min (this step was critical for successful amplification). One μl of this crude extract was added to the amplification mix. Primers were designed using Primer3 [27, 28] (Additional file 4: Table S4). TP53 exons (from 2 to 11) were separately amplified using optimized primer sets; exons 4 and 5 were amplified as two overlapping amplicons, using non-overlapping primers, to prevent loss of mutations detection capacity in the primer-annealing region.
Reactions were performed in 30 μl total volume (15 μl KapaBlood PCR Kit B, 0.5 μl of template pretreated DNA, 20 pmol primer forward, 20 pmol primer reverse). For multiplex reactions, TP53 was amplified under the following conditions: 4 cycles (denaturation at 94 °C for 30 s; annealing at 68 °C for 45 s {-1 °C/cycle}; extension at 72 °C for 30 s), 14 cycles (denaturation at 94 °C for 30 s; annealing at 63 °C for 30 s; extension at 72 °C for 30s); 30 cycles (denaturation at 94 °C for 30 s; annealing at 60 °C for 30 s; extension at 72 °C for 30s) and a final extension at 72 °C for 10 min. PCR products were analyzed by agarose gel electrophoresis.
Sequence analysis of TP53, Ha-RAS and Ki-RAS in human tumors
In both Ha-RAS (c-Ha-RAS1) and Ki-RAS (c-Ki-RAS2) activating oncogenic mutations are found at hotspots in exon 1 and 2, at codons 12, 13 (exon 1) or 61 (exon 2). Care was taken to differentially amplify the regions of interest of functional genes versus non-expressed pseudogenes, i.e. c-Ha-RAS2 and c-Ki-RAS1. Benchmark PCR amplification of Ha- and Ki-RAS exons 1 and 2 was performed using genomic DNA and cDNA from the T24 cell line, which carries a mutated, oncogenic form of Ha-RAS with a transversion at codon 12 (from GGC to GTC). When using cDNA templates, PCR primers were designed that reside in exonic regions, for simultaneous amplification of both exon 1 and 2 of Ha-and Ki-RAS. Joint amplification of exon 1 and 2 from genomic DNA was only performed for the Ha-RAS gene (the intervening intron is only 267 bp long in the Ha-RAS gene; it is more than 12,500 bp long in the Ki-RAS gene). Additional primers were designed that included intronic regions and were therefore specific for amplification of functional genes from genomic DNA. Amplified fragments were sequenced on both strands. Insertions or deletions (indels) of the TP53 gene (Additional file 5: Table S5) were shown to carry the highest prognostic weight [29]; such mutations were identified and matched against those listed in the IARC database [29].
Statistical analysis
The independent impacts of individual risk factors on prognosis is commonly evaluated in the framework of uni-or multivariate models [8, 19, 30, 31]. Univariate analyses were performed with GraphPad Prism 6.0 (GraphPad Software Inc., La Jolla, Ca) and XLStat 2009 (Addinsoft, Paris, France). Multivariate analyses and data modeling were performed using MetaboAnalyst 2.0 [32–34] and SIMCA-P+ 11 (Umetrics, Umea, Sweden) [35] software. However, uni-or multivariate analyses do not effectively quantify interaction effects on the final outcome. To explore such interactions, a priori specified hypotheses have been used in the past as trial models, but at the risk of introducing analytical bias. To overcome these limitations, patterns of aggregation of molecular parameters affecting prognosis were modeled here through logistic regression and partial least squares discriminant analysis (PLS-DA). PLS-DA clustering was performed using relapse as a dichotomic variable. PLS-DA model validation was performed as previously described [36]. Briefly, to define the optimal number of PCs, “7-fold cross-validation” (CV) was applied [37]. Using CV, the predictive power of the model was verified through R2 (goodness of fit) and Q2 (goodness of prediction). A model with Q2 > 0.5 was considered good, Q2 > 0.9 excellent [38]. The performance of PLS-DA models was further validated by a permutation test (200 times). To help interpreting results from PLS-DA, we utilized variable importance in the projection (VIP) scores. This allowed to evaluate the parameter influence on the model and to identify the best descriptors of relapsing versus non-relapsing BC. VIP scores are weighted sums of squares of the PLS loading weights, which take into account the amount of explained Y-variation for each dimension [33]. VIP values were cumulatively calculated from all extracted PLS components, usign a threshold of 0.8 [39]. As some variables may exert effects on the whole population (global), while others can be relevant in specific subgroups only (local), procedures were utilized to identify homogeneous subgroups with respect to corresponding parameters subclasses [40–42]. Spearman’s correlation analysis was performed using MetaboAnalyst 2.0 software [32–34] and GraphPad Prism.
Results
Immunohistochemistry and correlation analysis
Histopathology and molecular biology determinations were performed as indicated [43, 44] (Figs. 1, 2 and 3; Additional file 1: Table S1 and Additional file 2: Table S2). Negative/positive correlations between histopathological and experimental parameters were assessed by Spearman’s correlation analysis (Additional file 3: Table S3). Strongest positive correlations with metastastic relapse were found for grading (rho = 0.454, p = 0.005), local relapse (rho = 0.892, p < 0.001), p53_n (rho = 0.309, p = 0.067) and uPA in the extracellular matrix (rho = 0.387, p = 0.02). Highest negative correlations (Additional file 3: Table S3) were observed between metastastic relapse and intracellular uPA (percent cytoplasmic: rho = -0.369, p = 0.027; expression intensity: rho = -0.435, p = 0.008). Of interest, p53 expression (% positive cells and intensity) was found to be associated with secreted cathepsin D (rho = 0.477, p = 0.003) and was negatively correlated with Bcl-2 (rho = -0.385, p = 0.02). p53 expression was correlated with grading (rho = 0.499, p = 0.002), cyclin E (rho = 0.335, p = 0.046), PAI-1 in the extracellular matrix (rho = 0.444, p = 0.007), Her-2 (rho = 0.368, p = 0.027) and p16 (rho = 0.514, p = 0.001) expression, first suggesting key interactions and potential synergy with other key drivers of tumor malignancy.
Prognostic parameters analysis
Internal benchmarks for additional risk determination procedures were first assessed. Biochemically-determined negativity for estrogen receptors was associated with relapse. Estrogen receptor negativity, by semi-quantitative IHC determination, was associated with twice as high relapse hazard ratio (HR) (HR = 2.0; 95 % C.I. = 0.6–7.4). An increased mitotic index (Ki-67 expression) was associated with increased risk of developing adverse events (HR = 2.6; 95 % C.I. = 0.6–11.2), consistent with previous studies [31].
Correlated analysis of the analyzed tumor determinants revealed marked increase in HR for p53, cathepsin D and Bcl-2 (Figs. 1, 2). Positivity for p53 nuclear expression was found to associate with an eleven-fold increase in relapse risk (HR = 11; 95 % C.I. = 2.5–51.8). Unprecedented increase in risk was found for cathepsin D expression (HR = 20; 95 % C.I. = 2.3–184.3). Notably, expression profiles of p53 and cathepsin D remained significantly different between cases and controls when subgrouping patients by lymph node status, supporting an independent prognostic value of these parameters. Lymph node diffusion correlated with local cancer relapse (rho = 0.405, p = 0.014), but did not with distant metastatic relapse, raising the issue that determinant of local invasion may differ from those required for metastatic diffusion. Hence, we assessed the impact of p53 and cathepsin D in lymph node-negative patients. Remarkably, tumor co-expression of p53 and cathepsin D in this patient subgroup remained associated to a sixteen-fold higher risk of experiencing relapse (HR = 16; 95 % C.I. = 1.5–171.2). Trends for association of positive lymph nodes and tumor size were found: 50 and 78 % of lymph-node-positive women were positive for p53 and cathepsin D, respectively; 63 and 74 % of women with tumors bigger than 2 cm were positive for p53 and cathepsin D, respectively.
Remarkably, the expression of Bcl-2 was associated with a markedly better prognosis, and a nine-fold reduction of risk (HR = 9.2; 95 % C.I. = 1–87.8). Bcl-2 expression was previously found to correlate with a differentiated cancer phenotype, i.e. with lower grading and lack of p53 [45]. Consistent, Bcl-2 expression was found to correlate with that of ERα and PgR, and was anti-correlated with cancer grading and with the expression of p53, Cyclin E and Her-2 (Table S3). Correspondingly, Bcl-2 expression was shown to have a beneficial influence on prognosis [46, 47], whereas loss of Bcl-2 was found in 70 % of the aggressive triple-negative BC, and was significantly associated with high proliferation, tumor progression, increased risk of death and recurrence [48]. Still, the magnitude of Bcl-2 prognostic impact observed here in metastatic versus non-metastatic BC had not been previously revealed [49], supporting a critical value of correlated evaluation of malignancy determinants (Bcl-2, p53, cathepsin D) for effective use in prognostic assessment.
To verify the strength of this unsupervised analysis, and to further build on it, we performed a supervised PLS-DA [50]. Datasets of pathological/experimental parameters were grouped using a dichotomic classification (metastatic relapse versus no relapse). This model was found to have strong goodness of fit (cumulative R2Y = 0.828) and prediction power (cumulative Q2 = 0.548) (Figs. 4, 5 and 6). PLS-DA-identified determinants clusters yielded a clear-cut discrimination between metastatic versus non metastatic tumours (Figs. 4, 5a). A PLS-DA weight plot was generated in order to identify the major discriminants between the groups analyzed (Fig. 4). Next, VIP scores were computed for each parameter. Twenty descriptors, i.e. local relapse, grading, HER-2 (membrane intensity), lymph node status, p53, p16, Bcl-2, Cyclin E, PgR, together with stromal cathepsin D, PAI-1, uPA and MMP-11 were found to markedly contribute to the classification model (VIP score ≥ 0.8) (Fig. 5b) [39]. Permutation tests were carried out in order to validate the PLS-DA model [38, 50]. The original model was found to have higher R2 and Q2 values than the permuted models, and negative Q2 values were obtained for all two permuted groups tested (Fig. 5c).
Fig. 4.

PLS-DA weight plot. Variables utilized for discrimination between the relapsing and non-relapsing groups are reported. Variables that better correlate with metastatic tumors, map in the top right quadrant; variables that correlate more with non metastatic tumors, map in the lower left quadrant. Inset: local relapse (magnified scale)
Fig. 5.

PLS-DA score plot and VIP scores. a PLS-DA score plot showing the clustering of tumor samples with (green perimeter) or without (red perimeter) metastatic relapse. Complete separation between the green versus red clusters was achieved. Clustering thresholds were applied as indicated in Methods. b Variables able to discriminate between metastatic and non metastatic tumors are presented, as ordered by VIP score. VIP scores ≥ 0.8 (above the red line) identified key variables for predicting Y responses (relapse)
Fig. 6.
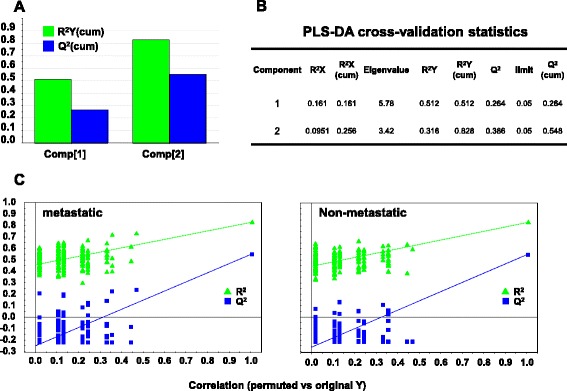
PLS-DA cross-validation and performance. a Bar plot showing the performance measures (R2Ycum and Q2cum) using different numbers of components. b R2X: portion of the variation of X explained by specified principal component; R2X (cum) Cumulative explained portion of X set variation; Eigenvalue: number of variables (K) times R2X; R2Y: portion of the Y set variation modeled by the principal component; R2Y (cum): cumulative modeled variation of Y set; Q2: overall cross-validated R2 for the specific principal component; Limit: threshold cross-validation for the specific principal component; Q2 (cum): cumulative Q2 up to the specified component, is a model predictive power according to cross validation. Unlike R2X (cum), Q2 (cum) is not additive. c Permutation tests for: metastatic (left) and non metastatic tumors (right). Permutation tests were performed by comparing R2and Q2 of the original model with R2 and Q2 of Y-class-permutated models. The correlation coefficients of original and permuted data are reported on the X axis; 200 random permutations were carried out. The values of R2 and Q2 are reported on the Y axis. The green triangles and blue squares in the upper right (ρ = 1) correspond to the values of R2 (green triangles) and Q2 (blue squares) of the original data. The low values of intercepts show that the model has high statistical significance (no over-fitting)
DNA extraction
DNA was extracted from sections of FFPE BC (Additional file 1: Table S1A). Ethidium bromide gel electrophoresis (Fig. 7) and amplification of RAS and TP53 exons benchmarked DNA as viable for downstream analyses. RAS and TP53 sequences were determined on cases and control DNA (Additional file 1: Table S1), as described (Figs. 7 and 8).
Fig. 7.

Oncogene sequence analysis. (top) Genomic DNA was extracted from BC and electrophoresed in agarose/ethidium bromide. Sample numbers are on top of each lane; mw: molecular weight markers. (mid) Exon-intron structure of the TP53 gene. (bottom) PCR amplification of the TP53 exons. EX: exon number; multiplex: simultaneous amplification of all exons with optimized primers and amplification procedure; mw: molecular weight markers
Fig. 8.
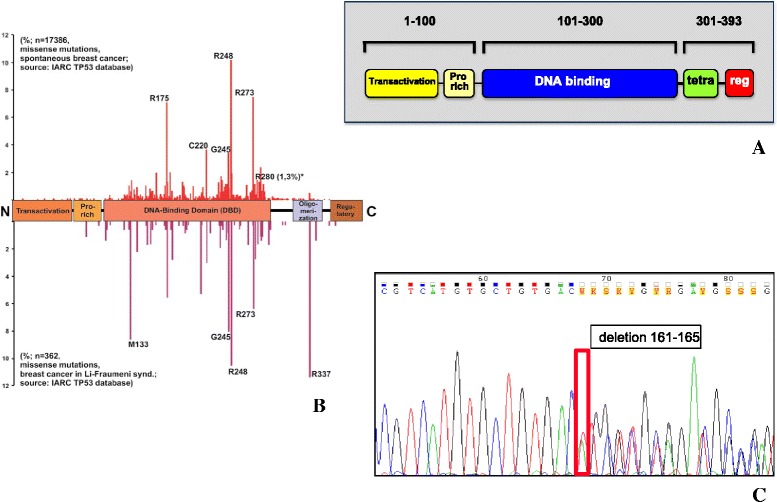
TP53 coding regions sequencing. a Block scheme of the p53 functional domains, protein sequence residues numbers are shown; Pro rich: proline-rich; tetra: oligomerization domain; reg: negative regulator of p53 function. b Prevalence of TP53 mutations in the different regions of the gene (from IARC TP53 Database, Release 17); (top) frequency of somatic mutations in BC; (bottom) frequency of germline, hereditary mutations in Li-Fraumeni syndrome. c Representative example of a DNA sequence chromatogram, containing a mutated sequence (see also Additional file 5: Table S5); the mutation site is boxed; the corresponding amino acid sequence is indicated
TP53 mutations
Case and control FFPE tumor samples, were systematically analyzed for insertions, deletions and stop codons in the coding region of the TP53 gene by PCR and sequencing of PCR amplification products (Figs. 7 and 8; Additional file 5: Table S5). Structural alterations of the TP53 gene are listed in Additional file 5: Table S5. Three indels were identified, and one stop codon, all of which led to truncation of the corresponding p53 proteins. Remarkably, all truncated p53 (8.7 % of the BC cases) were identified in relapsing cancers, three out of four cases being grade 3, i.e. those with the most malignant phenotype. These findings support models were severely damaged p53 is a strong risk factor for tumor progression in defined subgroups of BC [7, 8, 10, 31]. Notably, though, only one of these cases was a triple-negative tumor, a tumor phenotype traditionally associated with tumor aggressiveness [8], suggesting that the present molecular characterization may lead to novel subgrouping strategies of BC for risk determination. However, larger case series are needed to validate this approach.
RAS mutations
Case and control BC samples, were analyzed for mutations at codons 12, 13, 14 and 61 of the Ha-RAS and Ki-RAS genes by PCR amplification and sequencing of the first and second exon. Three cases showed a mutation at codon 12 of Ha-RAS, from GGC to GTC (Gly → Val); one case showed an additional mutation at codon 14 from GTG to ATG (Val → Met), with an overall prevalence of tumors bearing RAS mutations of 6.4 % (Additional file 1: Table S1). Of note, all mutations occurred in the metastatic and locally invasive/relapsing cases. This suggested relevance of mutated Ha-RAS in a small, distinct subset of metastatic BC. Mutations of both Ha-RAS and TP53 were identified in the same cancer, suggesting a possible cooperativity in cell transformation [51].
Discussion
Traditional prognostic indicators of BC, i.e. lymph node diffusion, tumor size, grading and estrogen receptor expression, are inadequate predictors of metastatic relapse. Therefore, identification of additional parameters versus traditional prognostic indicators is urgently needed. Several genes (oncogenes, tumor suppressor genes, transcription factors, signaling molecules, adhesion proteins, proteases) play a driving role in tumor progression [52]. Individual oncogenic determinants, e.g. p53, Her-2, E-cadherin, Trops, have been shown to possess prognostic/predictive power [7–11, 20, 53]. However, they did not outperform traditional prognostic indicators. Tumor progression is a multistep process [13, 54–58], which correlates with multiple, successive molecular modifications [13, 14]. Hence, clusters of tumor-driving traits are expected to be associated with tumor aggressiveness and overall malignancy, much more strongly than individual factors. In this work, we tested such a model in BC. Histopathological and molecular determinants of tumor progression of post-menopausal BC were analyzed, to assess impact on metastatic relapse. Aggregation of cancer determinants was expolored by modeling through discriminant analysis, logistic regression, partial least squares and partition trees. This identified upregulation of p53 and cathepsin D, together with downregulation of Bcl-2, as associated with a major increase in risk of disease relapse.
p53 is a tumor suppressor gene which is frequently mutated in cancer cells [59], and was identified as an indicator of both prognosis [8, 60–62] and response to therapy [7]. A cooperation of p53 with other drivers of tumor progression, e.g. Her-2 [8, 63] and Trop-1/Ep-CAM [10, 64] was previously shown, thus lending support our model of interaction between distinct prognostic determinants.
Bcl-2 inhibits cellular apoptosis [65]. Hower, Bcl-2 expression has a stronger impact as indicator of retained cancer differentiation, and of better disease outcome [45]. Indeed, loss of Bcl-2 was shown to have negative prognostic impact [46, 47, 49]. Bcl-2 expression was lost in 70 % of the most aggressive triple-negative BC cases, i.e. those lacking ERα, PgR and Her-2, and was significantly associated with high proliferation, tumor progression and increased risk of death and recurrence [48]. Supporting these findings, we found that Bcl-2 expression negatively correlated with cancer grading and with the expression of p53, cyclin E and Her-2. On the other hand, Bcl-2 expression was found to correlate with that of ERα and PgR, i.e. with differentiated cancer phenotypes.
Proteases, e.g. cathepsin D, uPA, MMP-11, are secreted by transformed or stromal cells of BC, and impact on tumor invasion and mestastasis [66–76]. uPA is modulated by the plasminogen activator inhibitor-1 (PAI-1), and combined assessment of uPA and PAI-1 was shown to be of value for prognostic determination [77, 78], indicating an impact of overall proteolytic balance on tumor progression. As indicated above for loss of Bcl-2, triple-negative BC were frequently associated with overexpression of cathepsin-D, and with aggressive disease course through lymph node invasion and high cancer cell proliferation/Ki-67 index [79].
As for the additional determinants we analyzed, cyclins D and E regulate the cell cycle [80], and increased levels are associated with worse prognosis and increased relapse rates in BC patients [81]. p27/kip1 and p16/INK4 are inhibitors of cyclin-dependent kinases and can prevent progression through the cell cycle [55], but can also be determinants of malignancy. High levels of the p27/kip1 cyclin inhibitor have been associated with worse prognosis and higher relapse rate in BC [82, 83]. On the other hand, deletion of p16/INK4 can be selected for in BC [84]. Consistent with an interactive predictive value, the levels of Cyclin E and of the p27 cyclin inhibitor were shown to have a higher impact when combined [82]. The mitotic index (Ki-67) is a measure of the percentage of tumor cells in active division and is a relevant prognostic indicator in BC [31]. Her-2 is a transmembrane tyrosine kinase receptor that regulates the growth of tumor cells [85]. The levels of expression of Her-2 have been shown to be independent indicators of worse prognosis, with respect to tumor relapse and overall survival in BC patients [86].
To identify interaction effects of different variables on disease outcome, expression profiles of tumor progression drivers were assessed, and results were evaluated by means of statistical analyses designed to detect significant prognostic interaction. To preempt the need for a priori specified hypotheses, patterns of aggregation of molecular parameters affecting prognosis were modeled through logistic regression and PLS-DA, using relapse as a dichotomic variable. PLS-DA score plot clustering of tumor samples with or without disease relapse, obtained separation between the two clusters. Major discriminant parameters were shown to be, HER-2, p53, p16, Cyclin E, PgR, together with stromal cathepsin D, PAI-1, uPA and MMP-11 were found to markedly contribute to the classification model; these efficiently clustered with local relapse, lymph node diffusion, tumor staging and grading. Among prognostic factors, p53 and cathepsin D stood up as major determinants of cancer relapse. Bcl-2 expression was shown to provide with unprecedented protective power versus tumor recurrence, candidating the combined assessment of these IHC parameters for use in clinical settings. Of interest, our case-control study included only one triple negative BC, indicating that a triple negative status was not a confounding variable in our study, and that p53, cathepsin D and Bcl-2 are efficient aggressiveness determinants in BC across currently categorized cancer subgroups.
Specific mutations of oncogenes and tumor suppressor genes play key roles in tumor progression. TP53 is frequently inactivated in several human tumors [87–89] and TP53 mutations help classifying and selecting patient subgroups with different biological features [8, 90], particularly in BC [8, 10, 31]. Mutations in different regions of TP53 were shown to be heterogenous in nature [91] and clinical outcome, indels having the highest impact [92]. Consistent, sequencing of the TP53 gene revealed a subgroup of BC where truncating mutations, such as indels and stop codons, were in all cases associated with cancer relapse.
The RAS genes code for small G proteins that play a critical role in signal transduction pathways downstream of growth-factor receptors. RAS mutations can affect prognosis [93–95]. Moreover, Ras downstream target genes are synergistically upregulated by mutated p53 and Ha-Ras, among them, matrix metalloproteases, which play a key role in local invasion and distant dissemination [96]. Hence, hot-spot sequence analysis was performed for Ha-and Ki-RAS, and findings were correlated with the IHC data and clinical phenotypes. The constitutive activation of the Ras proteins by point mutations, concentrated in hotspots at codons 12, 13, 61, is among the most frequently observed oncogene activation in human malignancies (75 % of adenocarcinomas of the pancreas, 40 % of adenomas and carcinomas of the colon and rectum, 25 % of carcinomas of the lung) and have been linked to worse prognosis [97]. However, although mutations in Ha-RAS and of Ki-RAS are often found in animal models of BC [98], their mutation frequency in human BC was shown to vary widely across studies. c-Ki-RAS mutations were shown to occur in 1 out of 8 BC by Yanez et al. [99]. Ha-RAS mutations were detected by Spandidos et al. [100], but not by Biunno et al. [101]. An overall low frequency of Ha-RAS mutations was found in most subsequent studies [97, 102–106]. Our findings support an incidence of mutated Ha-RAS in ≈ 5 % of BC cases. No mutations were detected in Ki-RAS. Remakably, all RAS mutations were identified in relapsed cases, suggesting impact of mutated Ha-RAS in a distinct subset of malignant BC [97, 104–106]. This finding warrants testing in a prospective clinical trial with adequate size and predictive power for relapsed cases subgroup dissection.
Conclusions
Taken together, our findings support a model of high BC aggressiveness as associated to high levels of p53 [8, 10] and cathepsin D [79], together with a downregulation of Bcl-2 [48]. An interaction between tumor-relapse risk factors may thus have a marked impact on prognosis, paving the way for using cluster molecular profiling of BC, to identify patient subgroups with distinct disease outcomes.
Acknowledgments
We thank A. Nicolucci, F. Carinci, C.A. Beltrami, C. Di Loreto, E. Bianchini, B. Murer and B. Palombo for help during the course of this work. We thank T. Soussi and C. Ishioka for discussion and support.
Funding
We gratefully acknowledge the support of the Italian Association for Cancer Research (AIRC, Italy), Italian Ministry of Development (contract n. MI01_00424 and SSI000651), Compagnia di San Paolo (Grant 2489IT), Foundation of the Cassa di Risparmio della Provincia di Chieti (Italy), Italian Ministry of University and Research (contract n. SCN_00558), and Italian Ministry of Health (RicOncol RF-EMR-2006–361866). The sponsors had no role in the design and conduct of this study, nor in the collection, analysis and interpretation of the data, or in the preparation, review or approval of the manuscript.
Availability of data and material
The data analyzed in this study are included in this published article and its supplementary information files.
Authors’ contributions
SA conceived the project; GB provided the samples from the National Naples Cancer Center, MP provided the samples and SA collected the clinical data; EG, AC and GV planned and performed the molecular analyses; VG, MDA and BS generated the sequencing data, RL and MP performed the histopathology and IHC analyses, PS and SA processed and analysed the data. All authors drafted the manuscript and approved the final version.
Competing interests
The authors declare that they have no competing interests regarding the contents of this manuscript.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This project was approved by the Italian Ministry of Health (RicOncol RF-EMR-2006–361866) and by the ethical committee of the leading institution, the National Cancer Institute of Naples (Verbale Comitato Etico n°3, 25 March 2009, Prot. CEI 1 2A/09 3 April 2009). According to national regulations no written consent was needed for this study. The presented experiments conformed to the principles set out in the World Medical Association Declaration of Helsinki [107].
Abbreviations
- CI
Confidence interval
- CV
Cross-validation
- Fab
Fragment antigen-binding
- FFPE
Formalin-fixed paraffin-embedded
- HR
Hazard ratio
- IHC
Immunohistochemistry
- PCR
Polymerase chain reaction
- PK
Proteinase K
- PLS-DA
Partial least squares discriminant analysis
- TMA
Tissue micro-array
- VIP
Variable importance in the projection
Additional files
Parameters utilized for assessing the risk of relapse. (XLSX 48 kb)
IHC analysis of consensus parameters of relapsed versus control BC cases. Table S2B. Invasion and cell cycle/apoptosis parameters of relapsed versus control BC cases. (XLSX 150 kb)
Spearman’ correlation matrix. Table S3B. Spearman’ correlation Rho coefficients. Table S3C. Spearman’ correlation P values. (XLSX 323 kb)
p53 structure and main mutation sites. Table S4B. Ha-Ras structure and main mutation sites. Table S4C. TP53 and Ha-RAS amplification primers. (XLSX 472 kb)
TP53 indels and stop codon chromatograms. (XLSX 1620 kb)
Contributor Information
Emanuela Guerra, Email: emanuela.guerra5@gmail.com.
Alessia Cimadamore, Email: alessia.cim@hotmail.it.
Pasquale Simeone, Email: simeone.pasquale@gmail.com.
Giovanna Vacca, Email: giovannavacca@gmail.com.
Rossano Lattanzio, Email: rossanolattanzio@libero.it.
Gerardo Botti, Email: g.botti@istitutotumori.na.it.
Valentina Gatta, Email: v.gatta@unich.it.
Marco D’Aurora, Email: mdaurora@unich.it.
Barbara Simionati, Email: direzione@bmr-genomics.it.
Mauro Piantelli, Email: mpiantel@unich.it.
Saverio Alberti, Email: s.alberti@unich.it.
References
- 1.Zanetti R, Buiatti E, Federico M, Micheli A, Fatti E. Cifre dei tumori in Italia. Pensiero Scientifico Editore. 1998. [Google Scholar]
- 2.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 3.Oncofem P. Carcinoma della mammella in Italia. Federazione Medica. 1992;9:11–16. [Google Scholar]
- 4.Group EBCTC. Polychemotherapy for early breast cancer: an overview of the randomised trials. Lancet. 1998;352:930–942. doi: 10.1016/S0140-6736(05)61359-1. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Melnikov AA, Levenson V, Guerra E, Simeone P, Alberti S, Deng Y. A seven-gene CpG-island methylation panel predicts breast cancer progression. BMC Cancer. 2015;15:417. doi: 10.1186/s12885-015-1412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callagy G, Cattaneo E, Daigo Y, Happerfield L, Bobrow LG, Pharoah PD, Caldas C. Molecular classification of breast carcinomas using tissue microarrays. Diagn Mol Pathol. 2003;12:27–34. doi: 10.1097/00019606-200303000-00004. [DOI] [PubMed] [Google Scholar]
- 7.Cimoli G, Malacarne D, Ponassi R, Valenti M, Alberti S, Parodi S. Meta-analysis of the role of p53 status in isogenic systems tested for sensitivity to cytotoxic antineoplastic drugs. Biochim Biophys Acta. 2004;1705:103–120. doi: 10.1016/j.bbcan.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Biganzoli E, Coradini D, Ambrogi F, Garibaldi JM, Lisboa P, Soria D, Green AR, Pedriali M, Piantelli M, Querzoli P, et al. p53 status identifies two subgroups of triple-negative breast cancers with distinct biological features. Jpn J Clin Oncol. 2011;41:172–179. doi: 10.1093/jjco/hyq227. [DOI] [PubMed] [Google Scholar]
- 9.Alberti S, Biganzoli E, Boracchi P, Ambrogi F, Querzoli P, Pedriali M, La Sorda R, Lattanzio R, Tripaldi R, Nenci I, et al. Cytoplasmic Trop-1 over-expression is associated with a favourable outcome in node-positive breast cancer patients. Jpn J Clin Oncol. 2012;42:1128–1137. doi: 10.1093/jjco/hys159. [DOI] [PubMed] [Google Scholar]
- 10.Ambrogi F, Fornili M, Alberti S, Querzoli P, Piantelli M, Pedriali M, Boracchi P, Biganzoli E, Coradini D. EpCAM Expression is an Indicator of Increased Incidence of Relapse in p53-Positive Breast Cancer. Cancer Clin Oncol. 2013;2:41–50. [Google Scholar]
- 11.Ambrogi F, Fornili M, Boracchi P, Trerotola M, Relli V, Simeone P, La Sorda R, Lattanzio R, Querzoli P, Pedriali M, et al. Trop-2 is a determinant of breast cancer survival. PLoS One. 2014;9:e96993. doi: 10.1371/journal.pone.0096993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138–141. doi: 10.1016/0168-9525(93)90209-Z. [DOI] [PubMed] [Google Scholar]
- 14.Boland CR, Sato J, Appelman HD, Bresalier RS, Feinberg AP. Microallelotyping defines the sequence and tempo of allelic losses at tumour suppressor gene loci during colorectal cancer progression. Nat Med. 1995;9:902–909. doi: 10.1038/nm0995-902. [DOI] [PubMed] [Google Scholar]
- 15.Hunter T. Oncoprotein networks. Cell. 1997;88:333–346. doi: 10.1016/S0092-8674(00)81872-3. [DOI] [PubMed] [Google Scholar]
- 16.Wold LE, Ingle JN, Pisansky TM, Johnson RE, Donohue JH. Prognostic factors for patients with carcinoma of the breast. Mayo Clin Proceedings. 1995;70:678–679. doi: 10.4065/70.7.678. [DOI] [PubMed] [Google Scholar]
- 17.Silverstein MJ, Lagios MD, Groshen S, Waisman JR, Lewinsky BS, Martino S, Gamagami P, Colburn WJ. The influence of margin width on local control of ductal carcinoma in situ of the breast. N Engl J Med. 1999;340:1455–1461. doi: 10.1056/NEJM199905133401902. [DOI] [PubMed] [Google Scholar]
- 18.Galea MH, Blamey RW, Elston CE, Ellis IO. The Nottingham Prognostic Index in primary breast cancer. Breast Cancer Res Treat. 1992;22:207–219. doi: 10.1007/BF01840834. [DOI] [PubMed] [Google Scholar]
- 19.Querzoli P, Pedriali M, Rinaldi R, Lombardi AR, Biganzoli E, Boracchi P, Ferretti S, Frasson C, Zanella C, Ghisellini S, et al. Axillary lymph node nanometastases are prognostic factors for disease-free survival and metastatic relapse in breast cancer patients. Clin Cancer Res. 2006;12:6696–6701. doi: 10.1158/1078-0432.CCR-06-0569. [DOI] [PubMed] [Google Scholar]
- 20.Querzoli P, Coradini D, Pedriali M, Boracchi P, Ambrogi F, Raimondi E, La Sorda R, Lattanzio R, Rinaldi R, Lunardi M, et al. An immunohistochemically positive E-cadherin status is not always predictive for a good prognosis in human breast cancer. Br J Cancer. 2010;103:1835–1839. doi: 10.1038/sj.bjc.6605991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lattanzio R, Marchisio M, La Sorda R, Tinari N, Falasca M, Alberti S, Miscia S, Ercolani C, Di Benedetto A, Perracchio L, et al. Overexpression of activated phospholipase Cγ1 is a risk factor for distant metastases in T1-T2, N0 breast cancer patients undergoing adjuvant chemotherapy. Int J Cancer. 2013;132:1022–1031. doi: 10.1002/ijc.27751. [DOI] [PubMed] [Google Scholar]
- 22.Castrilli G, Piantelli M, Artese L, Perfetti G, Rubini C, Fioroni M, Ranelletti FO, Piattelli A. Expression of hMSH2 and hMLH1 proteins of the human DNA mismatch repair system in ameloblastoma. J Oral Pathol Med. 2001;30:305–308. doi: 10.1034/j.1600-0714.2001.300508.x. [DOI] [PubMed] [Google Scholar]
- 23.Alberti S, Herzenberg LA. DNA methylation prevents transfection of genes for specific surface antigens. Proc Natl Acad Sci U S A. 1988;85:8391–8394. doi: 10.1073/pnas.85.22.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu B, Parsons R, Papadopoulos N, Nicolaides NC, Lynch HT, Watson P, Jass JR, Dunlop M, Wyllie A, Peltomaki P, et al. Analysis of mismatch repair genes in hereditary non-polyposis colorectal cancer patients. Nat Med. 1996;2:169–174. doi: 10.1038/nm0296-169. [DOI] [PubMed] [Google Scholar]
- 25.Bonasera V, Alberti S, Sacchetti A. Protocol for high-sensitivity/long linear-range spectrofluorimetric DNA quantification using ethidium bromide. Biotechniques. 2007;43:173–176. doi: 10.2144/000112500. [DOI] [PubMed] [Google Scholar]
- 26.Goelz SE, Hamilton SR, Vogelstein B. Purification of DNA from formaldehyde fixed and paraffin embedded human tissue. Biochem Biophys Res Commun. 1985;130:118–126. doi: 10.1016/0006-291X(85)90390-0. [DOI] [PubMed] [Google Scholar]
- 27.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 29.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 30.Biganzoli E, Boracchi P, Daidone MG, Gion M, Marubini E. Flexible modelling in survival analysis. Structuring biological complexity from the information provided by tumor markers. Int J Biol Markers. 1998;13:107–123. [PubMed] [Google Scholar]
- 31.Ambrogi F, Biganzoli E, Querzoli P, Ferretti S, Boracchi P, Alberti S, Marubini E, Nenci I. Molecular subtyping of breast cancer from traditional tumor marker profiles using parallel clustering methods. Clin Cancer Res. 2006;12:781–790. doi: 10.1158/1078-0432.CCR-05-0763. [DOI] [PubMed] [Google Scholar]
- 32.Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6:743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- 34.Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. MetaboAnalyst 2.0--a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012;40:W127–33. doi: 10.1093/nar/gks374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eriksson L, Antti H, Gottfries J, Holmes E, Johansson E, Lindgren F, Long I, Lundstedt T, Trygg J, Wold S. Using chemometrics for navigating in the large data sets of genomics, proteomics, and metabonomics (gpm) Anal Bioanal Chem. 2004;380:419–429. doi: 10.1007/s00216-004-2783-y. [DOI] [PubMed] [Google Scholar]
- 36.Vergara D, Simeone P, Del Boccio P, Toto C, Pieragostino D, Tinelli A, Acierno R, Alberti S, Salzet M, Giannelli G, et al. Comparative proteome profiling of breast tumor cell lines by gel electrophoresis and mass spectrometry reveals an epithelial mesenchymal transition associated protein signature. Mol Biosyst. 2013;9:1127–1138. doi: 10.1039/C2MB25401H. [DOI] [PubMed] [Google Scholar]
- 37.Bjerrum JT, Nielsen OH, Hao F, Tang H, Nicholson JK, Wang Y, Olsen J. Metabonomics in ulcerative colitis: diagnostics, biomarker identification, and insight into the pathophysiology. J Proteome Res. 2010;9:954–962. doi: 10.1021/pr9008223. [DOI] [PubMed] [Google Scholar]
- 38.Chan EC, Pasikanti KK, Nicholson JK. Global urinary metabolic profiling procedures using gas chromatography-mass spectrometry. Nat Protoc. 2011;6:1483–1499. doi: 10.1038/nprot.2011.375. [DOI] [PubMed] [Google Scholar]
- 39.Rudney JD, Staikov RK, Johnson JD. Potential biomarkers of human salivary function: a modified proteomic approach. Arch Oral Biol. 2009;54:91–100. doi: 10.1016/j.archoralbio.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciampi A. Computationals aspects of model choice. Heidelberg: Physica-Verlag; 1994. [Google Scholar]
- 41.Nicolucci A, Carinci F, Ciampi A. Stratifying patients at risk of diabetic complications: an integrated look at clinical, socioeconomic, and care-related factors. SID-AMD Italian Study Group for the Implementation of the St. Vincent Declaration. Diabetes Care. 1998;21:1439–1444. doi: 10.2337/diacare.21.9.1439. [DOI] [PubMed] [Google Scholar]
- 42.Breiman L, Friedman JH, Olshen RA. Classification and regression trees. Belmont: Wadsworth International Group; 1984. [Google Scholar]
- 43.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short protocols in molecular biology. 2. New York: Harvard Medical School; 1989. [Google Scholar]
- 44.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning-A laboratory manual. 2. New York: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 45.Duenas-Gonzalez A, Abad-Hernandez MM, Cruz-Hernandez JJ, Gonzalez SR. Analysis of bcl-2 in sporadic breast carcinoma. Cancer. 1997;80:2100–2108. doi: 10.1002/(SICI)1097-0142(19971201)80:11<2100::AID-CNCR9>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 46.O’Driscoll L, Linehan R, Kennedy SM, Cronin D, Purcell R, Glynn S, McDermott EW, Hill AD, O’Higgins NJ, Parkinson M, Clynes M. Lack of prognostic significance of survivin, survivin-DeltaEx3, survivin-2B, galectin-3, bag-1, bax-alpha and MRP-1 mRNAs in breast cancer. Cancer Lett. 2003;201:225–236. doi: 10.1016/S0304-3835(03)00518-4. [DOI] [PubMed] [Google Scholar]
- 47.Kroger N, Milde-Langosch K, Riethdorf S, Schmoor C, Schumacher M, Zander AR, Loning T. Prognostic and predictive effects of immunohistochemical factors in high-risk primary breast cancer patients. Clin Cancer Res. 2006;12:159–168. doi: 10.1158/1078-0432.CCR-05-1340. [DOI] [PubMed] [Google Scholar]
- 48.Abdel-Fatah TM, Perry C, Dickinson P, Ball G, Moseley P, Madhusudan S, Ellis IO, Chan SY. Bcl2 is an independent prognostic marker of triple negative breast cancer (TNBC) and predicts response to anthracycline combination (ATC) chemotherapy (CT) in adjuvant and neoadjuvant settings. Ann Oncol. 2013;24:2801–2807. doi: 10.1093/annonc/mdt277. [DOI] [PubMed] [Google Scholar]
- 49.Fitzgibbons PL, Page DL, Weaver D, Thor AD, Allred DC, Clark GM, Ruby SG, O’Malley F, Simpson JF, Connolly JL, et al. Prognostic factors in breast cancer. College of American Pathologists Consensus Statement 1999. Arch Pathol Lab Med. 2000;124:966–978. doi: 10.5858/2000-124-0966-PFIBC. [DOI] [PubMed] [Google Scholar]
- 50.Simeone P, Trerotola M, Urbanella A, Lattanzio R, Ciavardelli D, Di Giuseppe F, Eleuterio E, Sulpizio M, Eusebi V, Pession A, et al. A Unique Four-Hub Protein Cluster Associates to Glioblastoma Progression. PLoS One. 2014;9:e103030. doi: 10.1371/journal.pone.0103030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hinds PW, Finlay CA, Quartin RS, Baker SJ, Fearon ER, Vogelstein B, Levine AJ. Mutant p53 DNA clones from human colon carcinomas cooperate with ras in transforming primary rat cells: a comparison of the “hot spot” mutant phenotypes. Cell Growth Differ. 1990;1:571–580. [PubMed] [Google Scholar]
- 52.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 53.Nasr AF, Nutini M, Palombo B, Guerra E, Alberti S. Mutations ofTP53 induce loss of DNA methylation and amplification of the TROP1 gene. Oncogene. 2003;22:1668–1677. doi: 10.1038/sj.onc.1206248. [DOI] [PubMed] [Google Scholar]
- 54.Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 55.Hunter T, Pines J. Cyclins and cancer. Cell. 1991;66:1071–1074. doi: 10.1016/0092-8674(91)90028-W. [DOI] [PubMed] [Google Scholar]
- 56.Rhim JS, Yoo JH, Park JH, Thraves P, Salehi Z, Dritschilo A. Evidence for the multistep nature of in vitro human epithelial cell carcinogenesis. Cancer Res. 1990;50:5653s–5657s. [PubMed] [Google Scholar]
- 57.Chuaqui RF, Zhuang Z, Merino MJ. Molecular genetic events in the development and progression of ovarian cancer in humans. Mol Med Today. 1997;3:207–213. doi: 10.1016/S1357-4310(97)01043-5. [DOI] [PubMed] [Google Scholar]
- 58.Nagase H, Bryson S, Cordell H, Kemp CJ, Fee F, Balmain A. Distinct genetic loci control development of benign and malignant skin tumours in mice. Nat Genet. 1995;10:424–429. doi: 10.1038/ng0895-424. [DOI] [PubMed] [Google Scholar]
- 59.Davidoff AM, Humphrey PA, Iglehart JD, Marks JR. Genetic basis for p53 overexpression in human breast cancer. Proc Natl Acad Sci U S A. 1991;88:5006–5010. doi: 10.1073/pnas.88.11.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Silvestrini R, Benini E, Daidone MG, Veneroni S, Boracchi P, Cappelletti V, Di Fronzo G, Veronesi U. p53 as an independent prognostic marker in lymph node-negative breast cancer patients. J Natl Cancer Inst. 1993;85:965–970. doi: 10.1093/jnci/85.12.965. [DOI] [PubMed] [Google Scholar]
- 61.Allred DC, Clark GM, Elledge R, Fuqua SA, Brown RW, Chamness GC, Osborne CK, McGuire WL. Association of p53 protein expression with tumor cell proliferation rate and clinical outcome in node-negative breast cancer. J Natl Cancer Inst. 1993;85:200–206. doi: 10.1093/jnci/85.3.200. [DOI] [PubMed] [Google Scholar]
- 62.Joly A, Desjardins J-F, Fremond B, Desille M, Campion J-P, Malledant Y, Lebreton Y, Semana G, Edwards-Levy F, Levy M-C, Clementi B. Survival, proliferation, and functions of porcine hepatocytes encapsulated in coacted alginate beads: a step toward a reliable bioartificial liver. Transplantation. 1997;63:795–803. doi: 10.1097/00007890-199703270-00002. [DOI] [PubMed] [Google Scholar]
- 63.Geisler S, Lonning PE, Aas T, Johnsen H, Fluge O, Haugen DF, Lillehaug JR, Akslen LA, Borresen-Dale AL. Influence of TP53 gene alterations and c-erbB-2 expression on the response to treatment with doxorubicin in locally advanced breast cancer. Cancer Res. 2001;61:2505–2512. [PubMed] [Google Scholar]
- 64.Zanna P, Trerotola M, Vacca G, Bonasera V, Palombo B, Guerra E, Rossi C, Lattanzio R, Piantelli M, Alberti S. Trop-1 is a novel cell growth stimulatory molecule that marks early stages of tumor progression. Cancer. 2007;110:452–464. doi: 10.1002/cncr.22785. [DOI] [PubMed] [Google Scholar]
- 65.Berardo MD, Elledge RM, De Moor C, Clark GM, Osborne CK, Allred DC. bcl-2 and apoptosis in lymph node positive breast carcinoma. Cancer. 1998;82:1296–1302. doi: 10.1002/(SICI)1097-0142(19980401)82:7<1296::AID-CNCR12>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 66.Ahmad A, Hanby A, Dublin E, Poulson R, Smith P, Barnes D, Rubens R, Anglard P, Hart I. Stromelysin 3: an independent prognostic factor for relapse-free survival in node-positive breast cancer and demonstration of novel breast carcinoma cell expression. Am J Pathol. 1998;152:721–728. [PMC free article] [PubMed] [Google Scholar]
- 67.Ciccocioppo R, Capri MG, Alberti S. Detection of the receptor for the human urokinase-type plasminogen activator using fluoresceinated uPA. J Histochem Cytochem. 1997;45:1307–1313. doi: 10.1177/002215549704500914. [DOI] [PubMed] [Google Scholar]
- 68.Isola J, Weitz S, Visakorpi T, Holli K, Shea R, Khabbaz N, Kallioniemi OP. Cathepsin D expression detected by immunohistochemistry has independent prognostic value in axillary node-negative breast cancer. J Clin Oncol. 1993;11:36–43. doi: 10.1200/JCO.1993.11.1.36. [DOI] [PubMed] [Google Scholar]
- 69.Pyke C, Kristensen P, Ralfkiaer E, Grondahl Hansen J, Eriksen J, Blasi F, Dano K. Urokinase-type plasminogen activator is expressed in stromal cells and its receptor in cancer cells at invasive foci in human colon adenocarcinomas. Am J Pathol. 1991;138:1059–1067. [PMC free article] [PubMed] [Google Scholar]
- 70.Reilly D, Andreasen PA, Duffy MJ. Urokinase-plasminogen activator in breast cancer: assay by both catalytic and immunoassay. Blood Coagul Fibrinolysis. 1991;2:47–50. doi: 10.1097/00001721-199102000-00007. [DOI] [PubMed] [Google Scholar]
- 71.Mignatti P, Rifkin DB. Biology and biochemistry of proteinases in tumor invasion. Physiol Rev. 1993;73:161–195. doi: 10.1152/physrev.1993.73.1.161. [DOI] [PubMed] [Google Scholar]
- 72.Stephens RW, Pollanen J, Tapiovaara H, Leung KC, Sim PS, Salonen EM, Ronne E, Behrendt N, Dano K, Vaheri A. Activation of pro-urokinase and plasminogen on human sarcoma cells: a proteolytic system with surface-bound reactants. J Cell Biol. 1989;108:1987–1995. doi: 10.1083/jcb.108.5.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schultz RM, Yu H, Zhang JY. The role of urokinase and urokinase inhibitor in tumor cell metastasis. Fibrinolysis. 1992;6:23–29. doi: 10.1016/0268-9499(92)90089-Z. [DOI] [Google Scholar]
- 74.Ossowski L, Reich E. Antibodies to plasminogen activator inhibit human tumor metastasis. Cell. 1983;35:611–619. doi: 10.1016/0092-8674(83)90093-4. [DOI] [PubMed] [Google Scholar]
- 75.Hearing VJ, Law LW, Corti A, Appella E, Blasi F. Modulation of metastatic potential by cell surface urokinase of murine melanoma cells. Cancer Res. 1989;48:1270–1278. [PubMed] [Google Scholar]
- 76.Zhang YG, Du J, Tian XX, Zhong YF, Fang WG. Expression of E-cadherin, beta-catenin, cathepsin D, gelatinases and their inhibitors in invasive ductal breast carcinomas. Chin Med J (Engl) 2007;120:1597–1605. [PubMed] [Google Scholar]
- 77.Paradiso A, Volpe S, Iacobacci A, Marubini E, Verderio P, Costa A, Daidone MG, Marchetti A, Mottolese M, Amadori D, et al. Quality control for biomarker determination in oncology: the experience of the Italian Network for Quality Assessment of Tumor Biomarkers (INQAT) Int J Biol Markers. 2002;17:201–214. doi: 10.1177/172460080201700310. [DOI] [PubMed] [Google Scholar]
- 78.Sweep CG, Geurts-Moespot J, Grebenschikov N, De Witte JH, Heuvel JJ, Schmitt M, Duffy MJ, Janicke F, Kramer MD, Foekens JA, et al. External quality assessment of trans-European multicentre antigen determinations (enzyme-linked immunosorbent assay) of urokinase-type plasminogen activator (uPA) and its type 1 inhibitor (PAI-1) in human breast cancer tissue extracts. Br J Cancer. 1998;78:1434–1441. doi: 10.1038/bjc.1998.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang L, Liu Z, Chen S, Liu Y, Shao Z. A prognostic model for triple-negative breast cancer patients based on node status, cathepsin-D and Ki-67 index. PLoS One. 2013;8:e83081. doi: 10.1371/journal.pone.0083081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7:812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- 81.Pelosio P, Barbareschi M, Bonoldi E, Marchetti A, Verderio P, Caffo O, Bavilacqua P, Boracchi P, Buttita F, Barbazza R, et al. Clinical significance of cyclin D1 expression in patients with node-positive breast carcinoma treated with adjuvant therapy. Ann Oncol. 1996;7:695–703. doi: 10.1093/oxfordjournals.annonc.a010718. [DOI] [PubMed] [Google Scholar]
- 82.Porter PL, Malone KE, Heagerty PJ, Alexander GM, Gatti LA, Firpo EJ, Daling JR, Roberts JM. Expression of cell-cycle regulators p27kip1 and cyclin E, alone and in combination, correlate with survival in young breast cancer patients. Nat Med. 1997;3:222–225. doi: 10.1038/nm0297-222. [DOI] [PubMed] [Google Scholar]
- 83.Catzavelos C, Bhattacharya N, Ung YC, Wilson JA, Roncari L, Sandhu C, Shaw P, Yeger H, Morava-Protzner I, Kapusta L, et al. Decreased levels of the cell-cycle inhibitor p27kip1 protein: prognostic implications in primary breast cancer. Nat Med. 1997;3:227–230. doi: 10.1038/nm0297-227. [DOI] [PubMed] [Google Scholar]
- 84.Cairns P, Polascik TJ, Eby Y, Tokino K, Califano J, Merlo A, Mao L, Herath J, Jenkins R, Westra W, et al. Frequency of homozygous deletion at p16/CDKN2 in primary human tumors. Nat Genet. 1995;11:210–212. doi: 10.1038/ng1095-210. [DOI] [PubMed] [Google Scholar]
- 85.De Potter CR, Schelfhout AM. The neu-protein and breast cancer. Virchows Arch. 1995;426:107–115. doi: 10.1007/BF00192631. [DOI] [PubMed] [Google Scholar]
- 86.De Potter CR, Beghin C, Makar AP, Vandekerckhove D, Roels HJ. The neu-oncogene protein as a predictive factor for haematogenous metastases in breast cancer patients. Int J Cancer. 1990;45:55–58. doi: 10.1002/ijc.2910450112. [DOI] [PubMed] [Google Scholar]
- 87.Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hainaut P, Hernandez T, Robinson A, Rodriguez-Tome P, Flores T, Hollstein M, Harris CC, Montesano R. IARC Database of p53 gene mutations in human tumors and cell lines: updated compilation, revised formats and new visualisation tools. Nucleic Acids Res. 1998;26:205–213. doi: 10.1093/nar/26.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sidransky D, Tokino T, Helzlsouer K, Zehnbauer B, Rausch G, Shelton B, Prestigiacomo L, Volgestein B, Davidson N. Inherited p53 gene mutations in breast cancer. Cancer Res. 1992;52:2984–2986. [PubMed] [Google Scholar]
- 90.Network. CGA Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hartmann A, Blaszyk H, McGovern RM, Schroeder JJ, Cunningham J, De Vries EM, Kovach JS, Sommer SS. p53 gene mutations inside and outside of exons 5-8: the patterns differ in breast and other cancers. Oncogene. 1995;10:681–688. [PubMed] [Google Scholar]
- 92.Soussi T, Wiman KG. Shaping genetic alterations in human cancer: the p53 mutation paradigm. Cancer Cell. 2007;12:303–312. doi: 10.1016/j.ccr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 93.Teda M, Teratani T, Komatsu Y, Kawabe T, Shiratori Y, Omata M. Quantitative analysis of ras gene mutation in pancreatic juice for diagnosis of pancreatic adenocarcinoma. DigDisSci. 1998;43:15–20. doi: 10.1023/a:1018803532543. [DOI] [PubMed] [Google Scholar]
- 94.Lee JC, Wang ST, Lai MD, Lin YJ, Yang HB. K-ras gene mutation is a useful predictor of the survival of early stage colorectal cancers. Anticancer Res. 1996;16:3839–3844. [PubMed] [Google Scholar]
- 95.Nusko G, Sachse R, Mansmann U, Wittekind C, Hahn EG. K-RAS-2 gene mutations as predictors of metachronous colorectal adenomas. Scand J Gastroenterol. 1997;32:1035–1041. doi: 10.3109/00365529709011221. [DOI] [PubMed] [Google Scholar]
- 96.Buganim Y, Solomon H, Rais Y, Kistner D, Nachmany I, Brait M, Madar S, Goldstein I, Kalo E, Adam N, et al. p53 Regulates the Ras circuit to inhibit the expression of a cancer-related gene signature by various molecular pathways. Cancer Res. 2010;70:2274–2284. doi: 10.1158/0008-5472.CAN-09-2661. [DOI] [PubMed] [Google Scholar]
- 97.Von Lintig FC, Dreilinger AD, Varki NM, Wallace AM, Casteel DE, Boss GR. Ras activation in human breast cancer. Breast Cancer Res Treat. 2000;62:51–62. doi: 10.1023/A:1006491619920. [DOI] [PubMed] [Google Scholar]
- 98.Hokaiwado N, Asamoto M, Cho YM, Imaida K, Shirai T. Frequent c-Ha-ras gene mutations in rat mammary carcinomas induced by 2-amino-1-methyl-6-phenylimidazo [4,5-b] pyridine. Cancer Lett. 2001;163:187–190. doi: 10.1016/S0304-3835(00)00693-5. [DOI] [PubMed] [Google Scholar]
- 99.Yanez L, Groffen J, Valenzuela DM. c-K-ras mutations in human carcinomas occur preferentially in codon 12. Oncogene. 1987;1:315–318. [PubMed] [Google Scholar]
- 100.Spandidos DA. Oncogene activation in malignant transformation: a study of H-ras in human breast cancer. Anticancer Res. 1987;7:991–996. [PubMed] [Google Scholar]
- 101.Biunno I, Pozzi MR, Pierotti MA, Pilotti S, Cattoretti G, Della PG. Structure and expression of oncogenes in surgical specimens of human breast carcinomas. Br J Cancer. 1988;57:464–468. doi: 10.1038/bjc.1988.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rochlitz CF, Scott GK, Dodson JM, Liu E, Dollbaum C, Smith HS, Benz CC. Incidence of activating ras oncogene mutations associated with primary and metastatic human breast cancer. Cancer Res. 1989;49:357–360. [PubMed] [Google Scholar]
- 103.Capella G, Cronauer-Mitra S, Pienado MA, Perucho M. Frequency and spectrum of mutations at codons 12 and 13 of the c-K-ras gene in human tumors. Environ Health Perspect. 1991;93:125–131. doi: 10.1289/ehp.9193125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Miyakis S, Sourvinos G, Spandidos DA. Differential expression and mutation of the ras family genes in human breast cancer. Biochem Biophys Res Commun. 1998;251:609–612. doi: 10.1006/bbrc.1998.9527. [DOI] [PubMed] [Google Scholar]
- 105.Ward R, Hawkins N, O’Grady R, Sheehan C, O’Connor T, Impey H, Roberts N, Fuery C, Todd A. Restriction endonuclease-mediated selective polymerase chain reaction: a novel assay for the detection of K-ras mutations in clinical samples. Am J. 1998;153:373–379. doi: 10.1016/S0002-9440(10)65581-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schondorf T, Andrack A, Niederacher D, Scharl A, Becker M, Engel H, Gohring UJ. H-ras gene amplification or mutation is not common in human primary breast cancer. Oncol Rep. 1999;6:1029–1033. doi: 10.3892/or.6.5.1029. [DOI] [PubMed] [Google Scholar]
- 107.World Medical Association. http://www.wma.net/en/30publications/10policies/b3/. Accessed 25 Feb 2016.


