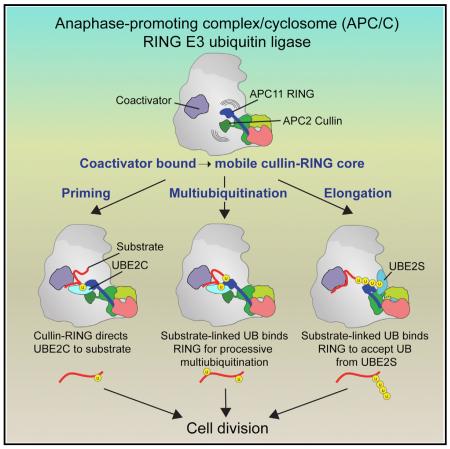
SUMMARY
Protein ubiquitination involves E1, E2, and E3 trienzyme cascades. E2 and RING E3 enzymes often collaborate to first prime a substrate with a single ubiquitin (UB) and then achieve different forms of polyubiquitination: multiubiquitination of several sites and elongation of linkage-specific UB chains. Here, cryo-EM and biochemistry show that the human E3 anaphase-promoting complex/cyclosome (APC/C) and its two partner E2s, UBE2C (aka UBCH10) and UBE2S, adopt specialized catalytic architectures for these two distinct forms of polyubiquitination. The APC/C RING constrains UBE2C proximal to a substrate and simultaneously binds a substrate-linked UB to drive processive multiubiquitination. Alternatively, during UB chain elongation, the RING does not bind UBE2S but rather lures an evolving substrate-linked UB to UBE2S positioned through a cullin interaction to generate a Lys11-linked chain. Our findings define mechanisms of APC/C regulation, and establish principles by which specialized E3–E2–substrate-UB architectures control different forms of polyubiquitination.
Graphical abstract
INTRODUCTION
Posttranslational modification by multiple ubiquitins (UBs) or UB chains is a predominant eukaryotic mechanism regulating protein half-life, location, interactions, or other functions. After an E1 enzyme links UB to the catalytic Cys of an E2 enzyme (~30 in humans), the thioester-bonded E2~UB intermediate is employed by an E3 enzyme (~ 600 in humans) (Deshaies and Joazeiro, 2009). Most E3s display a domain that recruits a substrate's degron motif and a hallmark RING domain thought to bind a cognate E2~UB intermediate that determines acceptor Lys properties (Metzger et al., 2014; Streich and Lima, 2014). Some E2s promiscuously modify lysines irrespective of context, while others generate linkage-specific UB chains (Christensen et al., 2007; Mattiroli and Sixma, 2014). Our current understanding is based on a limited number of landmark structures showing how RING domains align E2~UB active sites for nucleophilic attack, how a RING E3–E2 complex can target a preferred acceptor Lys, and how one RING forms homologous complexes with different E2~UB intermediates (Branigan et al., 2015; Dou et al., 2012; McGinty et al., 2014; Plechanovová et al., 2012; Pruneda et al., 2012; Reverter and Lima, 2005; Scott et al., 2014). However, structural models for dynamic polyubiquitination of substrates remain elusive.
Visualizing substrate polyubiquitination is challenging because proteins are modified on assorted sites, and UB chains are often elongated in a linkage-specific manner where each distal UB progressively added to a growing chain is successively presented to the catalytic center to accept another UB. The multiple ubiquitination sites are essentially moving targets for a catalytic RING–E2~UB assembly. Furthermore, E3 RING and degron-binding domains are often flexibly tethered, raising the question of how catalytic encounter could be achieved (Berndsen and Wolberger, 2014; Streich and Lima, 2014). Here, we addressed how mobile RING E3–E2 assemblies and UB-linked substrates are positioned for modification of multiple substrate lysines and evolving UBs by the essential human E3, the 1.2 MDa multisubunit anaphase-promoting complex/cyclosome (APC/C) (King et al., 1995; Sudakin et al., 1995).
APC/C catalyzes polyubiquitination of key cell cycle regulators to control the metaphase-to-anaphase transition, exit from mitosis, and maintenance of G1 (Primorac and Musacchio, 2013; Wieser and Pines, 2015). A seemingly simplistic code for substrate binding, with degrons such as KEN- or D-boxes recruited via a coactivator (CDC20 or CDH1) and APC10, is complemented by assorted catalytic mechanisms to achieve the variety and timing of polyubiquitination required to distinctly regulate vastly diverse substrates. Polyubiquitination is catalyzed with two E2s (Rodrigo-Brenni and Morgan, 2007), in humans UBE2C and UBE2S, in multiple steps (Figure 1A) (Aristarkhov et al., 1996; Garnett et al., 2009; Williamson et al., 2009; Wu et al., 2010; Yu et al., 1996). The priming reaction, where a substrate receives a single UB from UBE2C, was explained by prior structures that revealed how KEN- and D-box substrates bind APC/CCDH1 and how APC/C's cullin–RING catalytic core recruits and positions UBE2C adjacent to the preferred target lysine in a substrate (Brown et al., 2015; Buschhorn et al., 2011; Chao et al., 2012; da Fonseca et al., 2011; Tian et al., 2012). Furthermore, an atomic structure showed APC/C is blocked by the inhibitor EMI1 (Chang et al., 2015). However, the structures did not explain APC/C-catalyzed polyubiquitination.
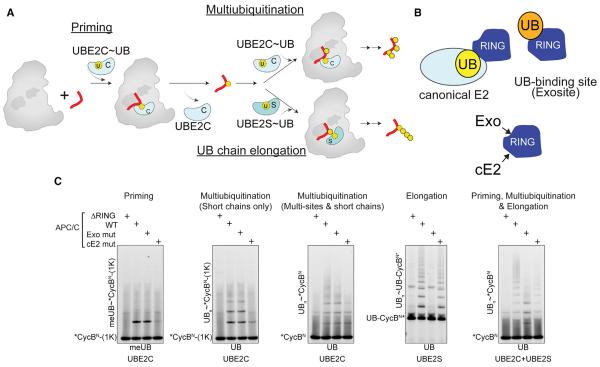
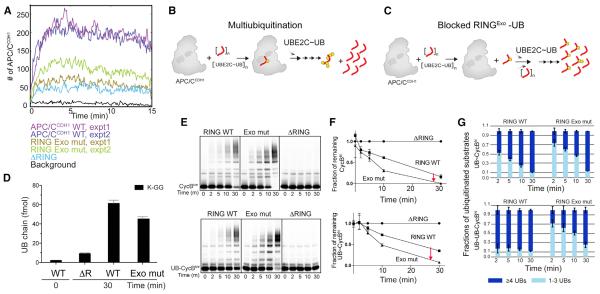
Figure 1. APC/C and Two E2 Partners Use Distinct Mechanisms for Polyubiquitination.
(A) Distinct mechanisms of priming, multiubiquitination and UB chain elongation by APC/C, UBE2C, and UBE2S.
(B) Distinct APC11 RING surfaces involved in polyubiquitination: canonical E2 binding site - cE2; UB-binding exosite - Exo.
(C) APC11 RING mutants define distinctive priming, multiubiquitination, and UB chain elongation by APC/CCDH1 with UBE2C and/or UBE2S, using WT or methyl UB (meUB), during a single encounter of the indicated version of CycBN (1K = K67 only; UB- = UB-fusion).
After the priming reaction, APC/C catalyzes two forms of substrate polyubiquitination: multiubiquitination and chain elongation. During multiubiquitination, UBE2C adds more UBs either individually or as short chains with various linkages, while UBE2S catalyzes Lys11-linked chain elongation from a substrate-linked UB (Williamson et al., 2009; Wu et al., 2010; Yu et al., 1996). Ultimately, different forms of polyubiquitination– i.e., multiple monoUBs, multiple Lys11-linked chains, and branched chains–direct APC/C substrate degradation with different efficiencies (Dimova et al., 2012; Grice et al., 2015; Lu et al., 2015b; Meyer and Rape, 2014). Furthermore, processively polyubiquitinated substrates are degraded earlier in the cell cycle, whereas degradation occurs later for substrates receiving fewer UBs in a single encounter with APC/C. These latter substrates are subject to deubiquitination and competition with other substrates delaying their acquisition of a degradation signal (Kamenz et al., 2015; Lu et al., 2015a; Lu et al., 2014; Lu et al., 2015b; Rape et al., 2006). This is controlled in part by feed-forward processive affinity amplification (PAA), whereby relative to unmodified substrates, UB-linked substrates display higher affinity for APC/C, and are preferentially subjected to further rapid multiubiquitination by UBE2C followed by slower UB chain elongation by UBE2S (Lu et al., 2015c).
How does a single E3 produce such polyubiquitination variety? Although UBE2C is activated by the canonical E2-binding site on APC11's RING domain (Brown et al., 2014), mechanisms underlying PAA remain unknown (Lu et al., 2015c). And while UBE2S requires a distinct UB-binding site on the RING, hereafter referred to as “exosite” for simplicity, UBE2S lacks a standard E2 RING-binding signature sequence and instead its residues required for activity do not correspond to known RING E3 functions (Figure 1B) (Brown et al., 2014; Kelly et al., 2014; Williamson et al., 2009; Wu et al., 2010). Structural mechanisms explaining APC/C's massive enhancement of UBE2S's efficiency for generating Lys11-linked di-UB linkages remain elusive (Brown et al., 2014; Kelly et al., 2014).
Here, we describe structural studies that relied on protein engineering and crosslinking to overcome transient interactions to visualize APC/C complexes with their E2s representing different forms of polyubiquitination. The data reveal how a malleable E3 synergizes with different E2s and evolving ubiquitinated substrates to adopt distinct catalytic architectures that define assorted products of polyubiquitination reactions.
RESULTS
Distinct RING Roles in Priming, Multiubiquitination, and UB Chain Elongation
Using our recombinant APC/CCDH1 system (Weissmann et al., 2016), we discovered mechanistic differences between priming, multiubiquitination, and UB chain elongation in reactions with two different RING mutants. The “RINGcE2” mutant impairs canonical E2~UB activation, while “RINGexo” bears Ala replacements for residues essential for recruiting the acceptor UB for chain elongation by UBE2S (Figure 1B) (Brown et al., 2014; Dou et al., 2012; Plechanovová et al., 2012; Pruneda et al., 2012). In reactions with either WT or methylated UB (meUB) that cannot form chains, we assayed modification of three variants of the canonical APC/C substrate cyclin B N terminus (CycBN) during a single encounter with the different versions of APC/C (Figure 1C). Consistent with canonical RING activation of UBE2C, priming (monitored with a single Lys version of *CycBN and meUB) and multiubiquitination (monitored with single Lys and WT versions of *CycBN and WT UB) were impaired by the RINGcE2 mutant. Also as expected, UBE2S-mediated UB chain elongation on a UB-fused CycBN* substrate that bypasses the need for priming was eliminated by the APC/C RINGexo mutant. Unexpectedly, however, this mutant also subdued formation of high molecular weight conjugates by UBE2C, thereby uncovering that multiubiquitination by UBE2C is mechanistically distinct.
Anchoring the RINGexo Site, UBE2C, and UBE2S for Structural Studies of Polyubiquitination
Structural studies of E3–E2–substrate complexes have depended on artificial reinforcement because the interactions are fleeting (Brown et al., 2015; McGinty et al., 2014; Reverter and Lima, 2005; Scott et al., 2014). To visualize APC/C–E2–substrate architectures underlying polyubiquitination we used protein engineering to strengthen interactions with the RING exosite, and then crosslinking to stabilize UBE2C and UBE2S positioned for multiubiquitination and UB chain elongation, respectively.
First, a UB variant (UBv) with substantially increased affinity (1.6 μM KD) for APC11's RING was selected by phage display (Figure 2A) (Ernst et al., 2013). After determining the crystal structure of the APC11 RING–UBv complex, NMR and enzyme assays demonstrated that the corresponding surfaces mediate APC11 RING domain binding to UBv and to UB and vice-versa, titrating free UBv impedes multiubiquitination and UB chain elongation by WT APC/C in a manner paralleling the APC/C RINGexo mutant, and the UBv is specific for APC/C and does not affect activities of a related RING or an unrelated UB-binding E3 (Figures 2A–2E, S1, Table S1). Furthermore, biological relevance was highlighted using a Xenopus egg extract system: the RINGexo-binding UBv, but not the negative control mutant, substantially slowed APC/C-dependent cyclin B degradation (Figure 2F). Although we cannot be certain that APC11-UBv and APC11-UB interactions are identical (a combination of mutations collaterally displace Lys11 so the UBv structure does not permit UBE2S-dependent chain elongation), the similarity between UBv and UB (0.37Å RMSD), and the numerous experiments indicating that to a first approximation UBv binds the same surface as a substrate-linked UB suggested that UBv would be a useful tool for anchoring the dynamic APC/CCDH1 RING exosite.
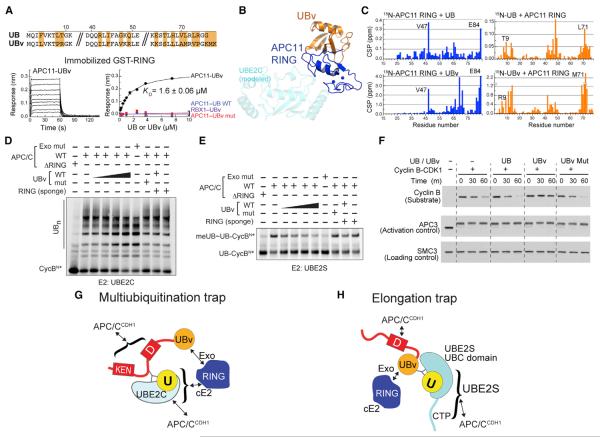
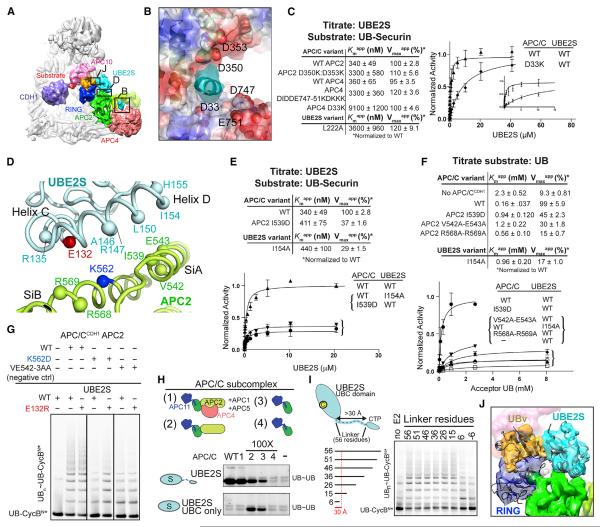
Figure 2. Anchoring the RINGexo Site, UBE2C, and UBE2S for Structural Studies of Polyubiquitination.
(A) Phage display selected UB variant (UBv) binds APC11 RING with high affinity and selectivity, measured by BLI.
(B) Crystal structure UBv (orange)–APC11 (navy) confirms binding to exosite, opposite canonical E2 site (modeled, cyan).
(C) UB and UBv bind similar APC11 RING surface and vice-versa, based on NMR chemical shift perturbations (CSPs).
(D) Similar effects on UBE2C-dependent multiubiquitination for blocking RING exosite with either UBv or APC11 mutations. Sponge = excess APC11 RING sequestering UBv.
(E) Similar effects on UBE2S-dependent UB chain elongation for blocking RING exosite with either UBv or APC11 mutations.
(F) UBv inhibits APC/C-dependent degradation of cyclin B in mitotic X. laevis egg extracts, examined by indicated westerns.
(G) Scheme of 3-way crosslinked complex used to trap APC/CCDH1–UBE2C architecture representing multiubiquitination.
(H) Scheme of 3-way crosslinked complex used to trap APC/CCDH1–UBE2S architecture representing UB chain elongation.
As a prelude to structural studies, we performed crosslinking based on the notion that connecting several weak interactors would enable avidly capturing multiple sites in ubiquitination complexes. The 3-headed sulfhydryl-reactive crosslinker TMEA simultaneously joined a C-terminal Cys on UB representing the donor, the active site Cys on either UBE2C or UBE2S, and optimized sites to represent multiubiquitination or UB chain elongation in surrogates for UB-linked substrates where UBv was fused to fragments derived from the substrate Hsl1 (Figures 2G, 2H, and S2). Avid binding of the respective substrate and/or E2 portions of the crosslinked products was confirmed by the multiubiquitination trap inhibiting overall UBE2C activity, substrate-binding, and UBE2C activation, and the UB chain elongation trap inhibiting UBE2S activation at substantially lower concentrations than individual components (Figure S2).
Cryo-EM Reconstructions of APC/C–E2 Complexes Poised for Polyubiquitination
Each trap was purified with APC/CCDH1, and cryo-EM was used to determine 3D reconstructions of the complex representing multiubiquitination with UBE2C at overall resolution of 6.4Å and that representing UB chain elongation with UBE2S to 6Å. The catalytic core, RING-UBv and E2 portions of maps displayed local resolutions of ~6–10 Å, apparently limited by conformational heterogeneity consistent with the dynamic mechanisms of polyubiquitination. Initial models constructed by docking atomic structures were improved by molecular dynamics flexible fitting (Figure S3, Movies S1 and S2). The donor UB is not visible even at low contour in either complex, consistent with local variability of the “closed” E2~UB conjugate in solution (Pruneda et al., 2012; Wickliffe et al., 2011). Overall, the EM data, together with structure-guided biochemical analyses, reveal molecular mechanisms of multiubiquitination and UB chain elongation.
Structural “Snapshots” of APC/C–E2 Architectures Representing Steps in Polyubiquitination
To visualize steps in polyubiquitination, we compared the new EM reconstructions with prior data to provide “snapshots” along the process. EM maps were aligned representing substrate-bound APC/CCDH1 alone (Figure 3A) (Chang et al., 2014) and with UBE2C representing the priming reaction (Figure 3B) (Brown et al., 2015), alongside the maps representing multiubiquitination (Figure 3C) and UB chain elongation (Figure 3D). Side-by-side comparisons show the remarkable ways the dynamic APC2–APC11 cullin-RING catalytic core, evolving ubiqui tinated substrates and E2s synergize to adopt distinct catalytic architectures specifying each reaction, illustrated beneath the EM reconstructions in Figure 3.
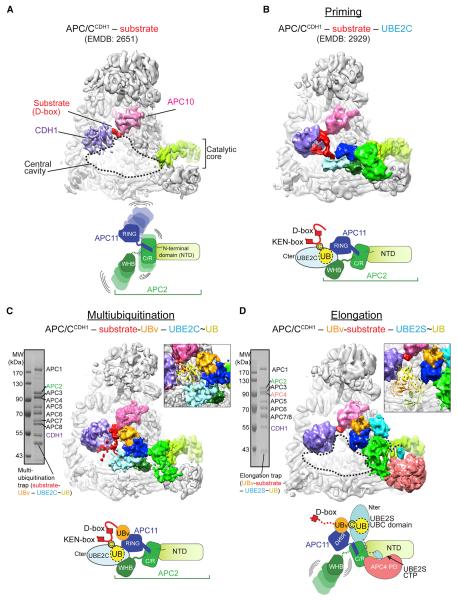
Figure 3. “Snapshots” of Distinct APC/C–E2 Architectures for Polyubiquitination.
(A) Prior APC/CCDH1–substrate complex (Chang et al., 2014). CDH1-purple, APC10-pink, APC2 NTD-light green, substrate-red.
(B) Prior structural data for complex representing priming (Brown et al., 2015), with UBE2C in light blue, APC2–APC11 intermolecular cullin–RING (C/R) domain green, and APC2 WHB domain in forest.
(C) Cryo-EM reconstruction representing multiubiquitination. UBv - orange. Inset, one UB (1UBQ, yellow) is shown fitting between substrate and active site.
(D) Cryo-EM reconstruction representing UB chain elongation. UBE2S - teal. Inset, distance between substrate binding and active sites accommodates polyUB, shown by tetraUB (2XEW, cartoon).
Prior structural data showed how a substrate degron is anchored on one side of APC/C, with the D-box corecruited by CDH1 and APC10 (Figure 3A). However, without interacting partners APC/C's cullin-RING catalytic core is mobile and not well resolved (Chang et al., 2014). How the mobile cullin-RING delivers the UBE2C~intermediate to substrate was revealed in a cryo-EM reconstruction representing “priming,” which showed UBE2C's active site juxtaposed with the preferred target site, stabilized by crosslinking (Figure 3B) (Brown et al., 2015). UBE2C~UB is positioned by both the APC11 RING and APC2 WHB domains, which emanate from the intermolecular APC2–APC11 cullin-RING interaction domain (C/R) (Figure 3B). UBE2C~UB is activated by binding the canonical E2 site on the RING, although primary recruitment occurs through APC2's WHB domain engaging the backside of UBE2C (Brown et al., 2015).
The cryo-EM reconstruction representing multiubiquitination provides insights into subsequent additional ubiquitination of a UB-linked substrate by APC/CCDH1–UBE2C (Figure 3C). UBE2C is placed in the same manner for multiubiquitination as for priming (Figure 3B, C). This positioning of UBE2C, through co-binding the APC11 RING and the APC2 WHB domains, condenses the search radius for multiple fluctuating lysines emanating from a tethered substrate. Reinspection of this geometry to understand multiubiquitination shows the active site of UBE2C located ~20 Å from the D-box. Importantly, this distance would allow numerous substrate lysines to access UBE2C and is large enough to accommodate a substrate-linked UB but small enough to constrain the catalytic geometry for generation of short chains (Figure 3C inset). UBv as a proxy for substrate-linked UB is bound to the RING exosite, providing a model for how this interaction would increase affinity and enhance processivity.
Finally, the new cryo-EM data reveal a completely different catalytic arrangement underlying APC/C-UBE2S-mediated UB chain elongation (Figure 3D). The “substrate” is recruited via a multisite mechanism, with the D-box binding CDH1 and APC10 on one side of APC/C, and the linked UBv localized by interactions with the RING exosite. This could place a homologous UB's K11 adjacent to UBE2S's active site. In agreement with prior mutational data (Brown et al., 2014; Kelly et al., 2014; Williamson et al., 2009), UBE2S is not recruited to the RING (Figure 3D). Furthermore, whereas RING domains typically bind the N-terminal portion of an E2 catalytic UBC domain, an APC2/APC4 groove binds UBE2S's unique C-terminal peptide (CTP), and APC2 interacts with the C-terminal portion of UBE2S's UBC domain (Figure 3D). The density for the N-terminal portion of the UBE2S UBC domain disappears at higher resolution, presumably due to the combination of a lack of direct contacts and greater relative motion furthest from the APC/CCDH1-binding site (Figure S3B). Another difference from the priming and multiubiquitination catalytic architectures is that in the complex with UBE2S, APC2's WHB domain is not visible (Figure 3D), consistent with dispensability for UB chain extension (Brown et al., 2015).
Different positions of the E2s in the complexes representing multiubiquitination and UB chain elongation would explain differences in products of these distinct polyubiquitination reactions. During multiubiquitination, UBE2C is placed in the central cavity, with its active site facing inside APC/C and proximal to substrate. This confines the space available for a modified substrate and would account for the limited number of UBs typically linked to substrates in reactions with UBE2C. By contrast, UBE2S is placed at the edge of APC/C (Figure 3D). This open architecture would allow growth of a polyUB chain either entirely outside APC/C altogether, or inside the central cavity (Figure 3D, inset). Further experiments examining how multiubiquitination is amplified by a substrate-linked UB binding the RING exosite and how UB chain elongation is determined are described in separate sections below.
RING Exosite Binding to Substrate-Linked UB Influences Processivity of Multiubiquitination
The structure representing multiubiquitination suggested that a substrate-linked UB binding to the RING exosite could provide a secondary interaction (Figure 3C). This may underlie findings from a recent single-molecule study, which showed that ubiquitinated substrates have a relatively higher propensity to bind APC/C, which drives their further multiubiquitination (Lu et al., 2015c). We used the single molecule assay to confirm the role for the RING exosite. Indeed, during multiubiquitination reactions with UBE2C where the UB linkages to substrate are evolving, the RING exosite mutant showed relatively decreased binding as probed by the number of APC/CCDH1 complexes detected in a field-of-view with saturating substrate (Securin) molecules, ~2 × 105, on the surface of a chip (Figure 4A).
Figure 4. Substrate-Linked UB Binding to RING Exosite Contributes to Processive Affinity Amplification for Multiubiquitination.
(A) Single-molecule time traces for binding to evolving ubiquitinated immobilized Securin molecules during multiubiquitination by UBE2C and APC/CCDH1 or indicated RING mutants.
(B) In processive multiubiquitination by APC/C and UBE2C, multiple UBs are added to substrate in a single binding event.
(C) Model if blocking substrate-linked UB binding to the RING exosite reduces processivity and shifts to distributive mode of multiubiquitination. A larger fraction of substrate would be modified, but with fewer UBs.
(D) RING exosite contributes to quantity of UB chains formed during CycBN* multiubiquitination by APC/CCDH1 and UBE2C, measured by AQUA mass spectrometry.
(E) Role of UB-binding RING exosite on processivity, monitored by formation of high molecular weight conjugates and fraction of substrate modified during multiple turnover UBE2C-catalyzed multiubiquitination of CycBN* (top) or UB-CycBN* (bottom).
(F) Role of RING exosite on fraction of substrate modified over time, in assays as in (E), quantifying depletion of unmodified CycBN* (top) or UB-CycBN* (bottom). Error bars: SEM, N = 3.
(G) Role of RING exosite on extent of substrate modification during multiubiquitination with UBE2C. Ubiquitinated products generated as in (E) were divided into 2 categories, with ≥ 4 UBs (navy) or 1–3 UBs (blue) as resolved by SDS-PAGE to examine extent of generation of highly multiubiquitinated products. Error bars: SD, N = 3.
This led to several predictions regarding mechanisms of multiubiquitination (Figures 4B and 4C). Enhanced lifetime of a ubiquitinated substrate on APC/C would increase processivity, thereby increasing the extent of modification while concomitantly decreasing catalytic turnover of substrate. Thus, mutating the RING exosite would be predicted to both decrease the extent of modification, and to correspondingly increase the fraction of substrate modified during the reaction. The notable exception would be during a single substrate encounter with APC/CCDH1, if a particular substrate-linked UB could not access the RING, then blocking the exosite would not impact the extent of ubiquitination. CycBN* could be a suitable substrate for these experiments, because the EM data predict that only one of its 16 potential ubiquitination sites (Lys51) could not access UBE2C, and a short UB chain linked to all the sites could access the RING exosite. We confirmed the predictions for ubiquitination site usage using a proteolytic strategy that isolates most sites and subsequent semiquantitative mass spectrometry analysis of the resultant peptides (Figures S4A–S4C). Mutating the RING exosite only subtly perturbed the site usage.
The predicted roles of the RING exosite on the extent of multiubiquitination were tested with several assays. First, we quantified UB chain formation using UB-AQUA, which showed a ~20 percent reduction in UB chains formed upon mutating the RING exosite without apparent discrimination toward a specific chain linkage (Figures 4D, S4B, and S3C). Second, we assayed various forms of multiubiquitination for a suite of CycBN* mutants with different numbers and locations of lysines. All acquired fewer UBs with the APC/CCDH1 RINGexo mutant, except the one mutant predicted to have limited potential for PAA due to few target lysines: the RINGexo mutant does not substantially affect modification of the single Lys substrate in a single encounter with APC/C (Figures 1C, 4E, S4D–S4J).
The role of the RING exosite on the fraction of substrate turned over was monitored during multibiquitination time-courses. As predicted (Figure 4C), free substrate was more rapidly depleted in reactions with the RING exosite mutant (Figures 4E–4G, S4B, S4D–S4J). Quantifying the effects showed that a higher proportion of the CycBN* was modified by at least one UB in reactions with the RING exosite mutant (Figures 4F and 4G). However, a higher proportion of the ubiquitinated CycBN* substrate received four or more UBs from UBE2C with wild-type APC/CCDH1. The effects were magnified for the UB-fused CycBN* substrate, further implicating a role for substrate-linked UB binding to the RING exosite (Figures 4E–4G). Taken together, the results suggested that UB-binding to the APC11 RING exosite supports processive affinity amplification during multiubiquitination.
Unique UBE2S Tethering
UB chain extension is achieved by an entirely distinct mechanism. Our EM data reveal how APC/C uses a unique E3 architecture that (1) anchors UBE2S via a tether, (2) positions the active site, and (3) delivers the acceptor UB to UBE2S. Although the structural data agree with prior scanning mutagenesis data for the acceptor UB, UBE2S, and APC11's RING domain (Brown et al., 2014; Kelly et al., 2014; Wickliffe et al., 2011), we performed mutational studies of APC/C to both confirm the structure and define how the novel APC/C catalytic architecture establishes UB chain elongation.
We identified key APC/C residues recruiting UBE2S based on interactions with the basic/hydrophobic tip of UBE2S's unique, flexible 66-residue C-terminal peptide (CTP) that is both necessary and sufficient for binding to APC/C (Williamson et al., 2009; Wu et al., 2010). The EM reconstruction shows this nestled in a complementary acidic and hydrophobic groove at the interface between APC2's N-terminal domain (NTD), C/R domain, and APC4's β-propeller (Figures 5A and 5B). Although the homologous sequence from the inhibitor EMI1 was assigned to comparable density in a prior EM structure, individual side-chains have yet to be resolved for this region for either UBE2S or EMI1, and roles of the APC2/APC4 groove have not been tested (Chang et al., 2015). Our molecular model shows the APC2/APC4 groove including APC2's Asp350 and Asp353, and APC4's Asp33 and the loop spanning from Asp747 through Glu751, and their mutation specifically impairs APC/CCDH1-UBE2S-catalyed UB chain elongation without affecting multiubiquitination with UBE2C (Figure 5B, S5A–S5E). These APC2/APC4 groove mutations and the corresponding mutation from UBE2S's CTP (L222A) caused parallel defects, ~10- to 30-fold increases in apparent Km (Kmapp) with little effect on apparent Vmax (Vmaxapp) in reactions monitoring polyubiquitination of a UB-fused substrate while titrating UBE2S (Figure 5C). Thus, the interaction between UBE2S's CTP and the APC2/APC4 groove plays the predominant role in recruiting UBE2S, but other elements are crucial to activate UBE2S-mediated UB chain elongation.
Figure 5. Distinctive Multisite Interactions Establish Unique Catalytic Architecture Specifying UB Chain Elongation by APC/C and UBE2S.
(A) Cryo-EM reconstruction of APC/C-CDH1 complex with UBE2S representing UB chain elongation as in Figure 3D, indicating regions with close-ups in panels (B), (D), and (J).
(B) Model for APC2/APC4 groove interactions with UBE2S CTP, based on docking APC/C structure (Chang et al., 2015) in cryo-EM reconstruction. APC2/APC4 groove is shown as a surface colored by electrostatic potential, with selected side-chains lining the groove in spheres. EM density for UBE2S CTP - cyan.
(C) Role of APC2/APC4 groove in recruiting UBE2S CTP, as determined from kinetic parameters for the indicated mutants during polyubiquitination of a UBSecurin substrate while titrating UBE2S. SEM, n ≥ 3.
(D) Placement of UBE2S C- and D-helices (cyan) by APC2 Si helices modeled based on (Chang et al., 2015) (green).
(E) Role of APC2 placement of UBE2S UBC domain in substrate polyubiquitination, from kinetic parameters for mutants during polyubiquitination of a UB-Securin substrate while titrating UBE2S. SEM, n ≥ 3.
(F) Role of APC2 placement of UBE2S UBC domain in activating UB chain synthesis, from kinetic parameters for mutants upon titrating acceptor UB during APC/CCDH1-UBE2S-mediated di-UB synthesis. SEM, n ≥ 3.
(G) APC2 placement of UBE2S UBC domain tested by charge-swap rescue assaying UBE2S E132R restoring UB chain elongation specifically to APC/CCDH1 with APC2 K562D mutant.
(H) Importance of placing UBE2S's UBC domain, or recruiting the CTP, determined from minimal APC/C subcomplexes (schematics on top) required to stimulate di-UB synthesis by WT UBE2S or isolated UBC domain lacking the CTP (bottom). Reactions with APC/C WT and subcomplexes 1–3 with WT UBE2S are controls based on (Brown et al., 2014).
(I) Importance of placing both UBE2S's UBC domain and CTP for APC/C activation of UB chain elongation. UBE2S deletion mutants with progressively shorter linkers between the two domains were assayed for APC/CCDH1-dependent polyubiquitination of UB-CycBN*. Reactions with WT UBE2S and linker deletions to 26 are controls based on (Brown et al., 2014).
(J) Model for acceptor UB (orange, UBv as proxy) in active site of UBE2S (cyan).
Unique E2 and Acceptor Placement Promote Chain Elongation
UB chain elongation requires juxtaposition of an E2 active site with an acceptor UB. The EM density showed an unprecedented cullin-RING mechanism, whereby the cullin, not the RING, positions UBE2S. This provides a rationale for deleterious effects of mutating UBE2S's “C” and “D” helices (Brown et al., 2014; Kelly et al., 2014), which straddle a pair of APC2 C/R domain helices that we term SiA and SiB, for UBE2S-interacting A and B-helices (Figure 5A and 5D, Movie S2). UBE2S binding to APC2's SiA and SiB-helices orients the active site toward the APC/C central cavity and APC11's tethered RING domain, and places the machinery catalyzing UB chain elongation at the extreme edge of APC/C's central cavity. This cullin function of APC2 may be specialized for APC/C, as the corresponding region in canonical cullins is not known to play a catalytic role.
Several assays validated that APC2 placement of UBE2S's UBC domain is important to activate UB chain elongation. Parallel effects are observed for mutations in the APC2 Si-helices and the corresponding interacting C- and D-helices in UBE2S. Monitoring APC/CCDH1-dependent chain elongation on a UB-primed substrate while titrating UBE2S showed that mutations disrupting the APC2–UBE2S UBC domain interface decreased Vmaxapp, without substantially impacting the Kmapp value for UBE2S (Figures 5E and S5F). In assays monitoring fluorescent UB transfer from UBE2S while titrating free UB as acceptor, the mutations caused increased Kmapp for the acceptor UB (3.5- to 7.5-fold) and decreased Vmaxapp (2- to 6-fold) (Figures 5F and S5G). Some mutations almost eliminated UB chain formation. Further support for APC2 placement of UBE2S's catalytic UBC domain comes from a compensatory charge-swap experiment, as defective UB chain elongation caused by the deleterious APC2 SiB-helix K562D mutation was specifically rescued by the structurally complementary E132R mutation from UBE2S's helix C (Figure 5G).
Subunit and domain deletion mutagenesis experiments confirmed that an APC2–APC11 subcomplex containing the Si helices and RING domain is minimally required to activate di-UB chain synthesis by UBE2S's isolated catalytic domain, albeit at 100-fold higher E3 concentrations due to lack of CTP-recruitment (Figures 5H and S5H). In agreement with the structural data, robust CTP-dependent activation required preserving the APC2/APC4 groove (Figure 5H). Importantly, even in the minimized systems, structure-based point mutations in APC2's Si helices thwarted activation of UBE2S-mediated di-UB synthesis (Figures S5I–S5K). NMR experiments further confirmed the distinctive interaction between APC/C's C/R and UBE2S's UBC domains: extreme line-broadening for resonances corresponding to 15N-UBE2S's folded UBC domain occurred upon adding an APC2–APC11 subcomplex. This depended on intact Si helices but not the RING (Figures S6A–S6E).
To test if UB chain elongation involves APC/CCDH1 simultaneously engaging UBE2S's UBC and CTP domains as in the structure, we assayed a series of mutants with progressively shorter linkers connecting the domains. Indeed, only UBE2S variants with linkers that could span the 30 Å distance separating the CTP and UBC domains retained full activity (Figure 5I). Overall, the EM data suggest that it is essential to place UBE2S adjacent to the acceptor UB delivered by the RING exosite, as visualized by modeling UB in place of UBv in the EM reconstruction (Figure 5J).
UBE2S-Specific Assembly Elements Cannot Support UBE2C
Despite differences in catalytic architecture and the specific domains mediating interactions, there are some common principles underlying multiubiquitination and UB chain elongation (Figure 6A). First, APC/C recruits each E2 via auxiliary interactions: APC2's WHB binds UBE2C's backside while the APC2/APC4 groove recruits UBE2S's CTP. Second, the cullin-RING catalytic core positions both E2s proximal to their distinctive acceptors, albeit by APC2's WHB and APC11's RING domains co-positioning UBE2C and by APC2's Si-helices guiding UBE2S. To test if each E2 depends on its own interactions with APC/C, we wished to assay effects of transplanting their unique elements. Although we were unable to relocalize UBE2S to the site occupied by UBE2C, we were able to test if UBE2S-specific features could support APC/CCDH1 activity with UBE2C as follows.
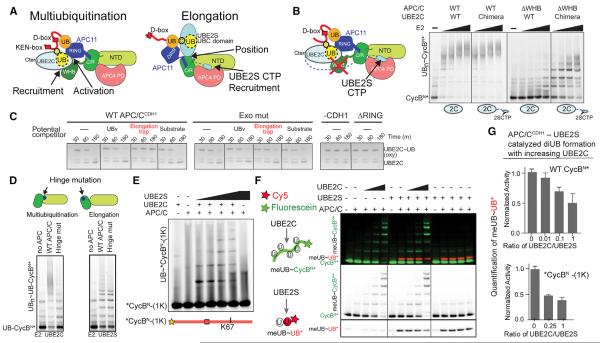
Figure 6. Functional Specialization of Each Polyubiquitination Architecture.
(A) Distinct APC/C mechanisms recruiting, positioning, and/or activating UBE2C or UBE2S for multiubiquitination and UB chain elongation, respectively.
(B) UBE2S CTP is a poor substitute for APC2 WHB in supporting UBE2C-dependent substrate multiubiquitination, as shown from assays with WT UBE2C or a chimera with appended UBE2S's CTP, and WT APC/CCDH1 or a deletion mutant lacking APC2 WHB domain.
(C) Shackling the RING away from the multiubiquitination architecture by the elongation trap inhibits APC/C activation of intrinsic UBE2C activity, monitored by inhibition of APC/CCDH1-stimulated hydrolysis of oxyester-linked UBE2C~UB.
(D) Specific APC2 cullin conformation is required for multiubiquitination. Top – schematic of distinctive APC2 NTD-C/R domain orientations showing burial or exposure of hinge. Distinct defects with hinge mutant (APC2 I501D, I502D) for multiubiquitination or UB chain elongation of UB-CycBN*.
(E) UBE2C and UBE2S build a UB chain on *CycBN-(1K) during the substrate's single encounter with APC/CCDH1.
(F) Competition between APC/CCDH1 activities with UBE2C and UBE2S probed simultaneously with 2 colors. MeUB can only be donor and not acceptor. Only fluorescein-CycBN* (green) accepts meUB from UBE2C. Only Cy5-UB* (red) with blocked C terminus accepts meUB from UBE2S.
(G) Bar graphs showing reduced APC/CCDH1-UBE2S-catalyzed meUB~UB* formation in (F) and S6G in the presence of UBE2C activity. SD, n ≥ 2.
First, we asked if UBE2S's CTP could substitute for APC2's WHB in recruiting UBE2C. We assayed a chimeric E2 harboring UBE2S's CTP grafted onto UBE2C (Chang et al., 2015). Control reactions showed that appending UBE2S's CTP does not hinder multiubiquitination with wild-type APC/CCDH1 (Figure 6B), and deleting the APC2 WHB domain does not impact UBE2S-catalyzed chain synthesis (Brown et al., 2015). However, >30-fold more of the UBE2C-UBE2SCTP chimera was required to multiubiquitinate CycBN* with the APC/CCDH1 mutant lacking APC2's WHB domain (Figure 6B). Thus, UBE2C requires its distinctive APC/C binding mechanism.
Next, we asked if specific RING positioning is important using the elongation trap to shackle the RING away from the UBE2C-specific architecture (Figures 2H and 3D). The elongation trap blocked APC/CCDH1-dependent hydrolysis of an oxyester-linked UBE2C~UB complex, which requires RING-mediated activation independent of substrate (Figure 6C). Inhibition depended on redirecting the RING, because mutating the exosite to prevent UBv tethering of the RING restored activation in the presence of the elongation trap, and neither UBv nor substrate alone impaired activity.
Finally, we noted that the two catalytic architectures display different relative orientations for APC2's NTD and C/R domains. The helix at the hinge comprising residues 500–506 is largely buried in the EM reconstructions with UBE2C, but is partially exposed in complex with UBE2S. Accordingly, there is little effect of mutating hinge-helix isoleucines (501 and 502) to aspartates on UBE2S-dependent polyubiquitination of a UB-fused substrate, whereas this substantially impairs multiubiquitination with UBE2C (Figure 6D). Thus, the distinctive cullin conformation is required for multiubiquitination.
Weak RING Interactions May Limit Competition between Multiubiquitination and Chain Elongation
Previous studies raised the question if multiubiquitination and UB chain elongation can occur simultaneously (Williamson et al., 2009; Wu et al., 2010). Indeed, priming, multiubiquitination, and UB chain elongation can occur independently (Figure 1C), and both E2s can function during a substrate's single encounter with APC/CCDH1 (Wang and Kirschner, 2013), including on a substrate with a single Lys (Figure 6E). Nonetheless, there has been no evidence of synergy. Instead, single molecule kinetic experiments indicated that UBE2S can act after UBE2C in a second gradual phase of polyubiquitination (Lu et al., 2015c).
To further explore the extent to which UBE2C and UBE2S could compete or catalyze their respective reactions simultaneously, we monitored activities of both E2s independently of each other but in the same tube using a donor meUB that cannot serve as acceptor, Cy5-labeled C-terminally blocked UB that only accepts meUB from UBE2S, and fluorescein-CycBN* or the single Lys version that only accept meUB from UBE2C. Increasing UBE2C to concentrations ~2-fold above Km slightly but reproducibly reduced di-UB synthesis by UBE2S (Figure 6F, 6G, S6F, and S6G), although no further inhibition was observed by adding more UBE2C. Such minor inhibition could be explained by competition between a UB-modified substrate and UBE2S acceptor UB for the RING exosite. Or RING positions for the two reactions could be mutually exclusive as observed in the EM reconstructions for the UBv-trapped structures (Figures 3C and 3D). Alternatively, it is also possible that WT UB binds the RING differently and could allow simultaneous engagement of UBE2C at the cE2 site and UBE2S's acceptor UB at the exosite. Irrespective of the mechanism, competition would be limited if RING interactions with different partners were fleeting. Indeed, UB-binding to the RING exosite is weak (Figure 2A) (Brown et al., 2014) and NMR chemical shift perturbation experiments did not detect interaction between 100 μM 15N-labeled APC11 RING domain and 400 μM UBE2C or a disulfide-linked proxy for a UBE2C~UB intermediate (Figures S6H–S6J). Overall, it seems that the RING interactions visualized by trapping occur transiently in the context of multi-site binding during APC/CCDH1-mediated polyubiquitination (Figure 3).
DISCUSSION
A major challenge in understanding RING E3 catalyzed polyubiquitination has been to explain how dynamically tethered substrates and mobile RING-E2 assemblies collide. We were able to address this through cryo-EM and biochemical analyses, which revealed distinct APC/C–E2–UB-linked substrate architectures specialized for distinct geometric challenges of multiubiquitination and UB chain elongation.
Multiubiquitination is specified by multisite interactions between APC/C's cullin-RING core, UBE2C, and a tethered UB-linked substrate that (1) secure UBE2C~UB to condense the search volume for catalytic encounter with the fluctuating substrate; (2) localize the active site proximal to substrate yet in a position that also accommodates a substrate-linked UB for further modification; and (3) additionally bind a primed substrate's linked UB to increase lifetime on APC/C and enhance processivity (Figure 7A). Constraining the proximity of substrate and UBE2C may contribute to preferential addition of individual UBs or short chains.
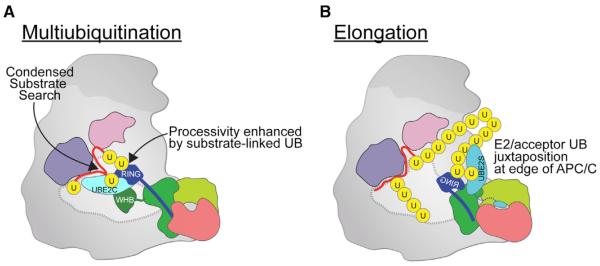
Figure 7. Specialized APC/C–E2 Architectures for Multiubiquitination and UB Chain Elongation.
(A) Processive multiubiquitination occurs by APC/C's APC2 cullin (green)-APC11 RING (blue) positioning UBE2C proximal to substrate (red), reducing the search volume for catalytic encounter while substrate-linked UB (yellow) binds the RING exosite to increase the evolving ubiquitinated substrate's lifetime on APC/C and enhance processivity. Each UB transfer cycle is accompanied by catalytic core dynamics releasing the used UBE2C for replacement by a charged UBE2C~UB to donate the next UB for ligation.
(B) Specialized architecture for UB chain elongation. APC2/APC4 recruits UBE2S's CTP, APC2 (cullin) places UBE2S's catalytic UBC domain, and APC11's RING guides the acceptor UB's Lys11 to the active site. Location of UBE2S at the edge of APC/C would accommodate growth of long UB chains.
Topological requirements for linkage-specific UB chain elongation are satisfied by specialized placement of UBE2S, which (1) employs an extraordinary cullin-RING mechanism–where the cullin binds E2 and RING binds the substrate-linked UB–to juxtapose UBE2S's active site and the acceptor UB and (2) localizes the catalytic center at the edge of APC/C, spatially allowing growth of long UB chains (Figure 7B). Because elements of the catalytic complex are flexible relative to each other, catalytic engagement of APC2–APC11, UBE2S, and the acceptor UB may occur dynamically, as in the various orientations observed for 3D classes in negative stain EM (Movie S2). Although future studies will be required to determine if and how UB binding to the RING and UBE2S binding to APC2 synergize for catalysis, a rationale for APC/C tracking substrate-linked UBs for chain elongation (Kelly et al., 2014) comes from the RING–UBv structure: interactions with a homologous acceptor UB would include the β1/β2-loop harboring Lys11, which after linkage to another UB would disengage from the catalytic assembly, thereby promoting further chain elongation.
Each architecture is optimal for its own form of polyubiquitination but suboptimal for the other activity. For example, the constrained assembly for multiubiquitination would hinder growth of long UB chains due to limited space between a substrate degron and E2 active site (Figure 7A). Meanwhile, the placement of UBE2S would be suboptimal for priming or multiubiquitination because the long distance between a tethered substrate's D-box and the active site would limit opportunities for catalytic encounter with substrate lysines (Figure 7B).
It seems that the RING acting as a hub depends on individual interactions being weak, with multiple contacts converging to avidly support polyubiquitination architectures. While RING-UBE2C binding is undetectable in isolation, multisite interactions promote catalytic encounter (Figure 3C and S6H–S6J), after which the used UBE2C must dissociate from APC2–APC11 to be replaced by a charged UBE2C~UB for another UB to be ligated (Brown et al., 2015). On the opposite side of the RING, evolving substrate-linked UBs apparently transiently sample the exosite either to promote PAA during UBE2C-catalyzed multiubiquitination or for chain elongation by UBE2S without jamming the system. Although future studies and new tools will be required determine the precise structure of UB bound to APC11's RING during these reactions, UB binding may be “fuzzy” due to dynamic hydrophobic interactions, or may involve specific contacts dynamically presenting UB from various formats akin to UB chain binding to the proteasome. We speculate that APC/C, like the proteasome (Shi et al., 2016), has multiple weak UB binding site(s) awaiting discovery as either promotiong PAA, UB chain elongation, or regulation.
APC/C's diverse catalytic architectures could comprise a combinatorial system for decorating substrates with various UB conjugate topologies. Although the rules determining whether a ubiquitinated substrate is preferentially modified by UBE2C and/or UBE2S remain to be determined, we envision that numerous input signals, for example D- and KEN-box affinities for CDH1, relative substrate lysine positions, and propensity for further polyubiquitination by UBE2C or UBE2S establish a “mix-and-match” system for differentially modifying substrates to regulate their proteasomal degradation (Dimova et al., 2012; Grice et al., 2015; Kirkpatrick et al., 2006; Lu et al., 2015b; Meyer and Rape, 2014).
The different catalytic architectures are likely to be distinctly regulated. Some APC/C inhibitors, perhaps Mitotic Checkpoint Complex, could inhibit activity with UBE2C but still allow binding of UBE2S (Herzog et al., 2009; Kelly et al., 2014). Also, APC/C's assembly with the E2s is differentially regulated, for example by phosphorylation and protein interactions (Craney et al., 2016). Furthermore, UBE2C and UBE2S themselves undergo APC/C-dependent ubiquitin-mediated proteolysis through poorly characterized mechanisms (Garnett et al., 2009; Rape and Kirschner, 2004; Williamson et al., 2009; Wu et al., 2010). It is tempting to speculate that the two architectures enable the E2s to regulate each other. It seems likely that future studies will reveal how, when, and where the two catalytic architectures contribute to distinct APC/C functions and cell division.
Principles derived from the APC/C–E2 structures may apply to polyubiquitination by many RING E3s (Figure 7). Indeed, key features have parallels in other systems, including RING E3 collaboration with multiple E2s, multisite E3–E2 interactions, polyubiquitinating E2s forming an active conformation even without RING-binding, and UB-binding exosites in other E3s or E2s (Figure S7) (Metzger et al., 2014; Streich and Lima, 2014). APC/C presents fascinating re-use of a UB-binding exosite for different functions in distinct polyubiquitination architectures (Figure 7). The stage is now set for future studies aimed at understanding how other multidomain RING E3s dynamically respond in their own specialized ways to the distinctive features of multiple assorted E2~UB and ubiquitinated substrate partners to establish the enormous conjugate variety associated with cellular polyubiquitination.
EXPERIMENTAL PROCEDURES
Protein Purification
Proteins described are human, except sequences derived from Hsl1, the tight-binding APC/CCDH1 substrate from S. cerevisiae. Baculoviruses expressing APC/C and subcomplexes were generated using biGBac (Weissmann et al., 2016). APC/C, subcomplexes, and UBA1 were expressed insect cells, and all other proteins in bacteria, and purified as described (Brown et al., 2015; Brown et al., 2014). The multiubiquitination and elongation traps were generated in a manner similar to that described for the priming trap (Brown et al., 2015) but with modifications. For multiubiquitination, the “substrate” was a fusion between a KEN- and D-box containing fragment of Hsl1 and C-terminal UBv, with a single Cys in place of Hsl1 K788, and the E2 was a catalytic Cys only version of UBE2C (C102A) that is active in multiubiquitination. For elongation, the “substrate” was a fusion between UBv at the N terminus and a D-box containing fragment of Hsl1 with a single Cys in place of UBv K11, and the E2 was a catalytic Cys only version of UBE2S (C118F, linker 15, see Figures S2G–S2L). 3-way crosslinking and trap purification were performed largely as described (Brown et al., 2015), except using the trifunctional sulfhydryl cross-linker TMEA (Pierce). For generation of samples for cryo-EM, APC/CCDH1 was first affinity purified based on tags on APC/C, the traps were added, and complexes were enriched by anti-Flag purification based on tags on the traps.
Structural Studies
NMR, X-ray crystallography, and cryo-EM were performed largely as described previously (Brown et al., 2015).
Assays
Ubiquitination assays were performed largely as described (Brown et al., 2014), with some differences. APC/C, CDH1, substrate, E1, and E2 were mixed on ice prior to initiating reactions, which were performed at room temperature in buffer used for purification (20 mM HEPES pH 8.0, 200 mM NaCl, 1 mM DTT) supplemented with 5 mM MgCl2, 5 mM ATP, and 0.25 mg/mL BSA. For kinetic analyses, product bands were quantitated based on a fluorescein label on *UB, UB-CycBN*, or UB-Securin* (* denotes location of fluorescein, N- or C terminus) using a Typhoon FLA 9500 PhosphorImager. For APC/C-dependent substrate ubiquitination reactions, APC/C-independent products were subtracted as background. Details of assays in each figure are in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Cryo-EM shows mechanisms of polyubiquitination by cell-cycle regulator APC/C
Distinct cullin-RING-E2 architectures for multiubiquitination and chain elongation
RING activates UBE2C and binds substrate-linked ubiquitin to amplify processivity
RING delivers ubiquitin for K11-linked chain elongation by UBE2S placed by cullin
ACKNOWLEDGMENTS
We thank J. Bollinger, J. Rogers, J.R. Michael, D.W. Miller, K. Kavdia, J. Peng, S. Frase. Funding: Jane Coffin Childs, Leukemia & Lymphoma Society (N.G.B.); Edward R. & Anne G. Lefler Center (A.O.); CIHR and Mitacs Elevate (W.Z.); FWF–Hertha Firnberg Program (R.Q.); Damon Runyon, Lallage Feazel Wall Fund (Y.L.); Japan Society for the Promotion of Science (M.Y.); CIHR MOP#111149 and 136956 (S.S.S.); NIH R01GM073960 (B.K.), R01GM026875 (M.W.K.), R01AG011085 (J.W.H.), R37GM065930 and P30CA021765 (B.A.S.), P41GM103403 (NECAT); Boehringer Ingelheim, the Austrian Science Fund (SFB-F34 and Wittgenstein award), the Austrian Research Promotion Agency (Headquarter grants FFG-832936 and FFG-852936, Laura Bassi Centre for Optimized Structural Studies grant FFG-840283), and the European Community (FP7/2007–2013, grant 241548, MitoSys) (J.-M.P.); DFG Sonderforschungsbereich 860 (H.S.); ALSAC, HHMI (B.A.S.). NECAT and APS - NIH P41GM103403, DOE DE-AC02-06CH11357. J.W.H. consults for Millennium: the Takeda Oncology Company and Biogen.
Footnotes
ACCESSION NUMBERS The accession numbers reported in this paper include: cryo-EM reconstructions representing multiubiquitination and elongation, EMDB: EMD-3432 and EMD-3433, respectively; crystal structure of APC11-UBv, PDB: 5JG6; and NMR assignments related to the APC11-UBv interaction, BMRB: 26783, 26784, and 26785.
SUPPLEMENTAL INFORMATION Supplemental Information includes Supplemental Experimental Procedures, seven figures, one table, and two movies and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2016.05.037.
AUTHOR CONTRIBUTIONS N.G.B., R.V., E.R.W., A.O., W.Z., Y.L., J.-M.P., H.S., B.A.S. designed research supervised by N.G.B., S.S.S., B.K., M.W.K., J.W.H., B.A.S., J.M.P., B.A.S.; N.G.B., R.V., E.R.W., F.W., A.O., K.-P.W., W.Z., S.Y., J.S.H., R.Q., I.F.D., Y.L., P.D., M.R.B., C.R.R.G., D.J.M., D.H., D.Y., M.A.J., M.Y., P.Y.M., and G.P. performed research and/or contributed new reagents; N.G.B., R.V., E.R.W., A.O., W.Z., Y.L., J.S.H., C.R.R.G., J.-M.P., H.S., and B.A.S. analyzed data; and N.G.B., R.V., E.R.W., K.-P.W., J.W.H. and B.A.S. wrote the paper.
REFERENCES
- Aristarkhov A, Eytan E, Moghe A, Admon A, Hershko A, Ruderman JV. E2-C, a cyclin-selective ubiquitin carrier protein required for the destruction of mitotic cyclins. Proc. Natl. Acad. Sci. USA. 1996;93:4294–4299. doi: 10.1073/pnas.93.9.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 2014;21:301–307. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- Branigan E, Plechanovová A, Jaffray EG, Naismith JH, Hay RT. Structural basis for the RING-catalyzed synthesis of K63-linked ubiquitin chains. Nat. Struct. Mol. Biol. 2015;22:597–602. doi: 10.1038/nsmb.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NG, Watson ER, Weissmann F, Jarvis MA, VanderLinden R, Grace CR, Frye JJ, Qiao R, Dube P, Petzold G, et al. Mechanism of polyubiquitination by human anaphase-promoting complex: RING repurposing for ubiquitin chain assembly. Mol. Cell. 2014;56:246–260. doi: 10.1016/j.molcel.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NG, VanderLinden R, Watson ER, Qiao R, Grace CR, Yamaguchi M, Weissmann F, Frye JJ, Dube P, Ei Cho S, et al. RING E3 mechanism for ubiquitin ligation to a disordered substrate visualized for human anaphase-promoting complex. Proc. Natl. Acad. Sci. USA. 2015;112:5272–5279. doi: 10.1073/pnas.1504161112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschhorn BA, Petzold G, Galova M, Dube P, Kraft C, Herzog F, Stark H, Peters JM. Substrate binding on the APC/C occurs between the coactivator Cdh1 and the processivity factor Doc1. Nat. Struct. Mol. Biol. 2011;18:6–13. doi: 10.1038/nsmb.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Molecular architecture and mechanism of the anaphase-promoting complex. Nature. 2014;513:388–393. doi: 10.1038/nature13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature. 2015;522:450–454. doi: 10.1038/nature14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao WC, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- Craney A, Kelly A, Jia L, Fedrigo I, Yu H, Rape M. Control of APC/C-dependent ubiquitin chain elongation by reversible phosphorylation. Proc. Natl. Acad. Sci. USA. 2016;113:1540–1545. doi: 10.1073/pnas.1522423113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Fonseca PC, Kong EH, Zhang Z, Schreiber A, Williams MA, Morris EP, Barford D. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470:274–278. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Dimova NV, Hathaway NA, Lee BH, Kirkpatrick DS, Berkowitz ML, Gygi SP, Finley D, King RW. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nat. Cell Biol. 2012;14:168–176. doi: 10.1038/ncb2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat. Struct. Mol. Biol. 2012;19:876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst A, Avvakumov G, Tong J, Fan Y, Zhao Y, Alberts P, Persaud A, Walker JR, Neculai AM, Neculai D, et al. A strategy for modulation of enzymes in the ubiquitin system. Science. 2013;339:590–595. doi: 10.1126/science.1230161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Mansfeld J, Godwin C, Matsusaka T, Wu J, Russell P, Pines J, Venkitaraman AR. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat. Cell Biol. 2009;11:1363–1369. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice GL, Lobb IT, Weekes MP, Gygi SP, Antrobus R, Nathan JA. The Proteasome Distinguishes between Heterotypic and Homotypic Lysine-11-Linked Polyubiquitin Chains. Cell Rep. 2015;12:545–553. doi: 10.1016/j.celrep.2015.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog F, Primorac I, Dube P, Lenart P, Sander B, Mechtler K, Stark H, Peters JM. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323:1477–1481. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenz J, Mihaljev T, Kubis A, Legewie S, Hauf S. Robust Ordering of C Events by Adaptive Thresholds and Competing Degradation Pathways. Mol. Cell. 2015;60:446–459. doi: 10.1016/j.molcel.2015.09.022. [DOI] [PubMed] [Google Scholar]
- Kelly A, Wickliffe KE, Song L, Fedrigo I, Rape M. Ubiquitin chain elongation requires E3-dependent tracking of the emerging conjugate. Mol. Cell. 2014;56:232–245. doi: 10.1016/j.molcel.2014.09.010. [DOI] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- Lu D, Hsiao JY, Davey NE, Van Voorhis VA, Foster SA, Tang C, Morgan DO. Multiple mechanisms determine the order of APC/C substrate degradation in mitosis. J. Cell Biol. 2014;207:23–39. doi: 10.1083/jcb.201402041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Girard JR, Li W, Mizrak A, Morgan DO. Quantitative framework for ordered degradation of APC/C substrates. BMC Biol. 2015a;13:96. doi: 10.1186/s12915-015-0205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Lee BH, King RW, Finley D, Kirschner MW. Substrate degradation by the proteasome: a single-molecule kinetic analysis. Science. 2015b;348:1250834. doi: 10.1126/science.1250834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Wang W, Kirschner MW. Specificity of the anaphase-promoting complex: a single-molecule study. Science. 2015c;348:1248737. doi: 10.1126/science.1248737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiroli F, Sixma TK. Lysine-targeting specificity in ubiquitin and ubiquitin-like modification pathways. Nat. Struct. Mol. Biol. 2014;21:308–316. doi: 10.1038/nsmb.2792. [DOI] [PubMed] [Google Scholar]
- McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514:591–596. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger MB, Pruneda JN, Klevit RE, Weissman AM. RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta. 2014;1843:47–60. doi: 10.1016/j.bbamcr.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HJ, Rape M. Enhanced protein degradation by branched ubiquitin chains. Cell. 2014;157:910–921. doi: 10.1016/j.cell.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plechanovová A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primorac I, Musacchio A. Panta rhei: the APC/C at steady state. J. Cell Biol. 2013;201:177–189. doi: 10.1083/jcb.201301130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda JN, Littlefield PJ, Soss SE, Nordquist KA, Chazin WJ, Brzovic PS, Klevit RE. Structure of an E3:E2~Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol. Cell. 2012;47:933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432:588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Scott DC, Sviderskiy VO, Monda JK, Lydeard JR, Cho SE, Harper JW, Schulman BA. Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell. 2014;157:1671–1684. doi: 10.1016/j.cell.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Chen X, Elsasser S, Stocks BB, Tian G, Lee BH, Shi Y, Zhang N, de Poot SA, Tuebing F, et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science. 2016;351:831. doi: 10.1126/science.aad9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streich FC, Jr., Lima CD. Structural and functional insights to ubiquitin-like protein conjugation. Annu. Rev. Biophys. 2014;43:357–379. doi: 10.1146/annurev-biophys-051013-022958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca FC, Ruderman JV, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol. Biol. Cell. 1995;6:185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Li B, Warrington R, Tomchick DR, Yu H, Luo X. Structural analysis of human Cdc20 supports multisite degron recognition by APC/C. Proc. Natl. Acad. Sci. USA. 2012;109:18419–18424. doi: 10.1073/pnas.1213438109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Kirschner MW. Emi1 preferentially inhibits ubiquitin chain elongation by the anaphase-promoting complex. Nat. Cell Biol. 2013;15:797–806. doi: 10.1038/ncb2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann F, Petzold G, VanderLinden R, Huis in 't Veld PJ, Brown NG, Lampert F, Westermann S, Stark H, Schulman BA, Peters JM. biGBac enables rapid gene assembly for the expression of large multi-subunit protein complexes. Proc. Natl. Acad. Sci. USA. 2016;113:E2564–E2569. doi: 10.1073/pnas.1604935113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieser S, Pines J. The biochemistry of mitosis. Cold Spring Harb. Perspect. Biol. 2015;7:a015776. doi: 10.1101/cshperspect.a015776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc. Natl. Acad. Sci. USA. 2009;106:18213–18218. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Merbl Y, Huo Y, Gallop JL, Tzur A, Kirschner MW. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc. Natl. Acad. Sci. USA. 2010;107:1355–1360. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, King RW, Peters JM, Kirschner MW. Identification of a novel ubiquitin-conjugating enzyme involved in mitotic cyclin degradation. Curr. Biol. 1996;6:455–466. doi: 10.1016/s0960-9822(02)00513-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.