Abstract
A growing body of evidence has shown that alcohol alters the activity of the innate immune system and that changes in innate immune system activity can influence alcohol-related behaviors (Cui et al., 2014; Vetreno & Crews, 2014). Here we show that the Toll innate immune signaling pathway modulates the level of alcohol resistance in Drosophila. In humans, a low level of response to alcohol is correlated with increased risk of developing an alcohol use disorder (Schuckit, 1994). The Toll signaling pathway was originally discovered in, and has been extensively studied in Drosophila. The Toll pathway is a major regulator of innate immunity in Drosophila, and mammalian Toll-like receptor signaling has been implicated in alcohol responses. Here, we use Drosophila-specific genetic tools to test eight genes in the Toll signaling pathway for effects on the level of response to ethanol. We show that increasing the activity of the pathway increases ethanol resistance while decreasing pathway activity reduces ethanol resistance. Furthermore, we show that gene products known to be outputs of innate immune signaling are rapidly induced following ethanol exposure. The interaction between the Toll signaling pathway and ethanol is rooted in the natural history of Drosophila melanogaster.
Keywords: Alcohol Resistance, Alcohol Sensitivity, Innate Immunity, Neuroimmune, Addiction, Alcoholism, NF-κB, Tolerance, Mutations, RNAi, Ethanol
Introduction
Alcohol use is pervasive in our society, and alcohol abuse has been estimated to cost the United States economy $223.5 billion per year (Bouchery et al., 2011). A survey of more than 36,000 American adults found that 29.1% of respondents met the criteria for DSM-5 diagnosis of an alcohol use disorder at some time in their life, and 13.9% had met the criteria in the past 12 months (Grant et al., 2015). A 25 year longitudinal study in humans found that baseline resistance to alcohol was the strongest predictor of future alcoholism (Schuckit, 1994; Schuckit & Smith, 2011). Alcoholism has a strong genetic component, and while no single alcoholism gene has been identified, large networks of genes with small individual effects sum to generate predisposition for addiction (Enoch, 2013).
A growing body of evidence shows that chronic alcohol consumption changes the expression of conserved gene networks in the human brain, members of which have been demonstrated to regulate alcohol behaviors in model systems (Ponomarev et al., 2012; Zhou et al., 2011; Liu et al., 2007; Liu et al., 2006; Iwamoto et al., 2004; Flatscher-Bader et al., 2005; Liu et al., 2004; Sokolov et al., 2003; Mayfield et al., 2002; Lewohl et al., 2000; Farris et al., 2014). One such network contains a set of genes that control the innate immune system. Studies in fruit flies, rodents, and humans have all shown that innate immune system genes increase expression after alcohol exposure (Crews et al., 2013; Liu et al., 2006; Kong et al., 2010; Zou & Crews, 2014). Recent rodent work has shown that numerous innate immune system pathways affect alcohol consumption, including chemokines, interleukins, peroxisome proliferator-activated receptors (PPARs), and Toll-like receptor pathways (Robinson et al., 2014). Furthermore, it has been reported that Toll-like receptor signaling can modulate neural activity. In brain slice preparations from the central amygdala of mice, treatment with lipopolysaccharide, the activator of Toll-like receptor 4 (TLR4), can directly modulate GABAergic signaling, and ethanol and lipopolysaccharide treatment can have additive effects on GABAergic signaling (Bajo et al., 2014).
The innate immune system is a branch of the immune system that invokes a rapid, preprogrammed, and generalized response to pathogens. Whereas the adaptive immune system recognizes, responds to, and remembers essentially any foreign antigen, the innate immune system is hardwired to respond to the antigens stereotypical of pathogens. Innate immune system responses include inflammation to seal off a site of infection, recruitment of immune cells, and production of antimicrobial peptides (Turvey & Broide, 2010). In Drosophila, there are two major branches of the innate immune system: the Toll pathway and the immune deficiency (IMD) pathway (Buchon et al., 2014).
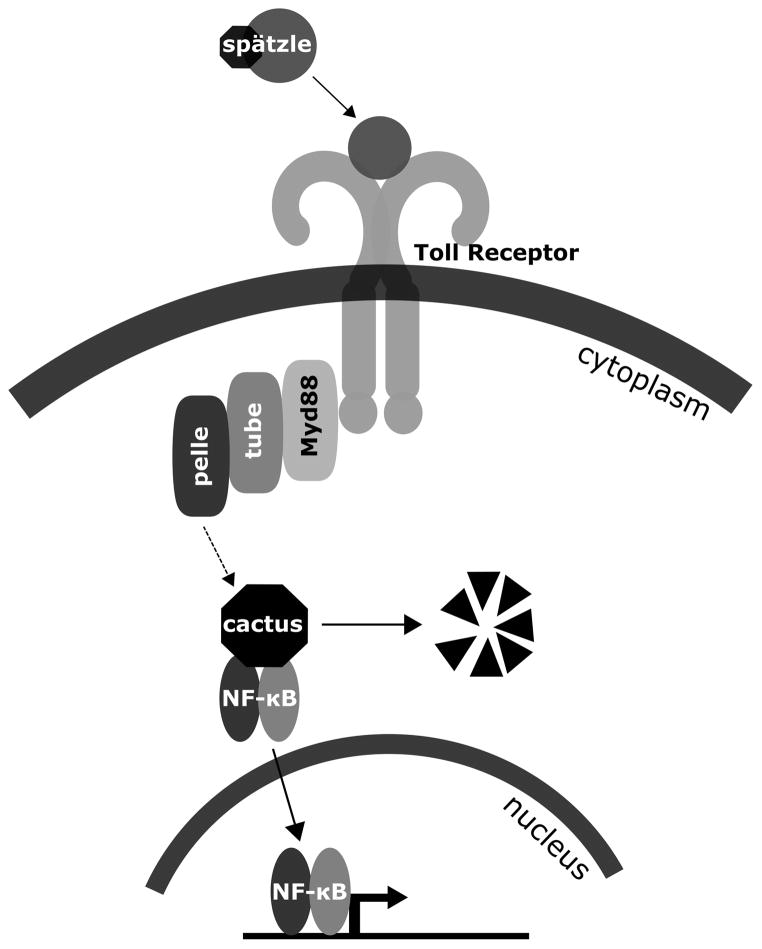
The Toll pathway was initially described by the Nüsslein-Volhard lab for its role in the establishment of the dorsal-ventral axis during embryonic development of Drosophila melanogaster. In larvae and adult flies however, the same pathway is reused to regulate the innate immune system (Lemaitre et al., 1996). The Toll pathway is conserved across metazoans, from sponges to humans (Song et al., 2012), and the Drosophila Toll pathway is related to the mammalian Toll-like receptor Myd88-dependent pathway. As depicted in Fig. 1, in Drosophila the Toll ligand is a protein called Spätzle, which circulates in the hemolymph as an inactive precursor. Upon fungal or Gram-positive bacterial infection Spätzle is cleaved, binds to Toll, and activates the pathway. Myd88, Tube, and Pelle are adaptor proteins that associate with Toll. After activation, Pelle phosphorylates the NF-κB inhibitor Cactus, which causes Cactus degradation and allows NF-κB homologs Dorsal and Dif to enter the nucleus and activate transcription of target genes (reviewed in Imler, 2014).
Figure 1.
Schematic diagram of the interacting proteins in the Toll signaling pathway. An inactive Spätzle precursor is cleaved after infection and binds to the Toll receptor. Upon Toll pathway activation, Myd88, Tube, and Pelle associate with Toll, and Cactus is phosphorylated. Phosphorylation of Cactus leads to degradation of Cactus at the proteasome, which releases the inhibition of NF-κB proteins. Once released, NF-κB transcription factor family members (Dif, Dorsal) enter the nucleus and can regulate target genes.
The other main branch of the Drosophila innate immune system is the IMD pathway, which responds to Gram-negative bacterial infection. There is evidence for cross-talk between the Toll and IMD signaling pathways: simultaneous stimulation of the Toll and IMD pathways has an additive effect on expression of some antimicrobial peptide genes (Tanji et al., 2007) and infection with some pathogens leads to activation of both pathways (Mansfield et al., 2003; Hashimoto et al., 2009; Luce-Fedrow et al., 2008; Lau et al., 2003). Relish is the Drosophila NF-κB that is primarily associated with and activated by the IMD pathway (Buchon et al., 2014). All three NF-κB proteins (Dif, Dorsal, and Relish) can form heterodimers with one another, providing a means for integration of information from the Toll and IMD pathways (Tanji et al., 2010).
In recent years, an intriguing connection between ethanol consumption and the innate immune pathway has become apparent in Drosophila. In experiments detailed in Milan et al. (2012) and Kacsoh et al. (2013) it was found that fruit flies use alcohol to help fight infection by parasitic wasps. There are several species of endoparasitoid wasps of the genus Leptopilina that inject their eggs into fruit fly larvae. The wasp offspring then develop within and feed on the fruit fly larva, eventually killing them during pupariation. The Toll innate immune pathway is a regulator of the anti-parasite immune response in Drosophila (Paddibhatla et al., 2010; Schlenke et al., 2007; Sorrentino et al., 2004; Small et al., 2012). Under standard culture conditions fly survival is quite low after infection by these wasps. However, when standard fly food is replaced with food containing 6% ethanol, wasp survival decreases and fly survival increases. When given a choice between standard food and ethanol-containing food, a greater portion of infected larvae chose ethanol food than did uninfected larvae (Milan et al., 2012). A subsequent study showed that, after seeing a female wasp, female Drosophila were more likely to lay eggs on ethanol-containing food. This reaction was sex-specific and did not occur in response to a male wasp (Kacsoh et al., 2013). Indeed, flies respond to a parasitic infection in a way that suggests that the fly innate immune system modulates ethanol-related behaviors. Here we ask whether genetically manipulating the Toll pathway affects resistance to ethanol.
Materials and Methods
Stocks and Fly Husbandry
All flies were maintained on a standard cornmeal/molasses/agar medium under 12:12 light:dark conditions. In order to collect age-matched flies, a bottle with eclosing flies is cleared of adult flies and three days later the adult females are harvested and allowed to age for three more days, yielding a group of flies that are 3–6 days old. The full genotype of stocks used in this study and additional information about alleles or transgenes can be found in Table S2. Canton S was used as the wild-type control where appropriate. Heterozygous animals were produced by crossing stocks to our wild-type Canton S stock before testing. All transgenes were tested as heterozygotes.
Stocks were from the Bloomington Drosophila Stock Center (NIH P40OD018537) or the Tübingen Drosophila Stock Collection (provided by Dr. David Stein at the University of Texas at Austin). The UAS-V5-Dif, UAS-V5-RelN, and UAS-Toll10B-FLAG stocks were provided by Dr. Y. Tony Ip (University of Massachusetts Medical School). Animals carrying the Actin-GeneSwitch transgene were derived from the stock BSC# 9431. This stock carries other mutations that did not interest us. A stock bearing only the w1118 allele and the Actin-GeneSwitch transgene was generated by genetic crossing. Each of the UAS overexpression transgenes and the Actin-GeneSwitch line were backcrossed to our wild type Canton S stock seven times to remove any second-site mutations and to ensure they are in the same genetic background. RNAi lines acquired from the TRiP consortium (Transgenic RNAi Project) were used to knock down expression of eight Toll pathway genes. The TRiP stocks have the same genetic background and all carry their respective RNAi construct at the same attP insertion site (except for the cactus RNAi line). A stock with no RNAi construct inserted at the attP insertion site was used as a control for RNAi experiments (BSC# 36303). Dif1 was acquired in a background that contained Dipt and Drs reporter constructs, but was separated from these transgenes by crossing.
Activation of GeneSwitch Transgenes
The GeneSwitch system has been described in Osterwalder et al. (2001) and Roman et al. (2001), and makes use of a fusion of Gal4 and progesterone receptor domains to generate a transcription factor that activates UAS transgenes in the presence of RU-486 (a.k.a. mifepristone, Cayman Chemical, Ann Arbor, MI, USA). To generate drug-laced food, a stock solution of 25 mM RU-486 in 80% ethanol was added to molten fly food to produce food with a final concentration 200 μM RU-486, alongside control food that was melted and mixed with carrier (modified from McGuire et al., 2004). The RU-486 food was distributed to fly vials and allowed to cool and dry for at least one hour. Flies were kept on drug- or carrier-containing food for three days. Because RU-486 has poor solubility in water, the stock solution used 80% ethanol as the solvent, and therefore both the RU-486 and carrier-fed flies were housed on food that initially contained 0.64% ethanol v/v.
Ethanol Resistance Assay
Experiments were performed in the inebriator as described in Krishnan et al. 2012 and Cowmeadow et al. 2005. All experiments are performed with age-matched female flies being sedated with ethanol for the first time. Groups of 10 flies are placed in plastic vials and exposed to a stream of concentrated ethanol vapor until all flies are sedated (typically 15-18 minutes). Then the ethanol-saturated air stream is replaced with a humidified air stream and recovery from sedation is recorded. Flies are considered recovered when they regain postural control. n=4–6 vials for each group. All experiments were performed between 11:00 and 16:00 (zeitgeber time 3–8).
Determination of Ethanol Concentration in flies
The Enzymatic Ethanol Assay Kit (Diagnostic Chemicals Ltd. Oxford, CT) was used to measure internal ethanol concentration of flies. Flies were sedated in the inebriator, and then collected at the designated time. 10 flies were ground with a plastic pestle in 500μl of 50mM TRIS pH 7.5, vortexed, centrifuged for 2 minutes, and the supernatant was transferred to a fresh tube. 6μl of this solution was incubated in 370μl of reagent solution for 10 minutes at 37°C. 340nm absorbance was then recorded with a NanoDrop ND-1000 (Thermo Scientific, Wilmington, DE) and plotted against a standard curve. The ethanol concentration in fly hemolymph was then calculated assuming a 0.85μl volume per fly (Cowmeadow et al., 2005).
Statistical Analysis
Behavioral recovery data was entered into GraphPad Prism 6 for graphing and statistical analysis. Statistical significance was determined using the log rank test. Error bars represent standard error of the mean in all figures. Ethanol absorption and metabolism data were analyzed in Prism 6 using linear regression to compare the slope and intercepts of the data sets, and using multiple Student t-tests and the Holm-Sidak method to correct for multiple comparisons.
RNA Isolation and Sequencing
RNA was extracted from the heads of 3–5 day old adult female flies that were either treated with ethanol or left untreated as controls. Approximately 180 fly heads were used per group. The ethanol treated group was exposed to an ethanol-saturated air stream until all flies were sedated (15 minutes), followed by a 30 minute ethanol-free air stream. For the untreated control group, flies were exposed to an ethanol-free air stream for the entire 45 minutes. At the end of the treatment (30 minutes post-sedation), both groups of flies were transferred to a 50 ml conical tube and snap frozen in liquid nitrogen. Heads were snapped off from the body by briefly vortexing the tubes. The frozen heads were sorted from the bodies using a series of cooled metal mesh sieves. Total RNA was isolated from the heads using the guanidinium thiocyanate single-step method (Ausubel, 1994). After isolation, RNA was treated with DNAse I (Life Technologies, Grand Island, NY) and purified by acid-phenol/chloroform extraction and ethanol precipitation. RNA concentration and quality was assessed using a 2100 Bioanalyzer RNA 6000 Pico Chip (Agilent Technologies, Inc., Santa Clara, CA). RIN values for the control and ethanol-treated samples were 6.80 and 6.40, respectively.
Poly(A)+ RNA was prepared from an aliquot of each total RNA sample with magnetic oligo-(dT) beads (Dynabeads® Oligo (dT), Life Technologies, Grand Island, NY). cDNA synthesis and Illumina library construction were performed with the TruSeq RNA Library Preparation Kit using standard Illumina protocols and sequenced to at least 20M reads in an Illumina HiSeq 2000 (Illumina, Inc., San Diego, CA) using paired-end chemistry and 100-bp cycles. Raw sequences have been deposited in the public functional genomics data repository from NCBI: Gene Expression Omnibus (GEO). Data can be found on the GEO website (http://www.ncbi.nlm.nih.gov/geo/) using accession number GSE77792. All essential sample annotation and experimental design information including sample data relationships have been included in the repository according to the Minimum Information About a Microarray Experiment (MIAME) guidelines (Brazma et al., 2001).
RNA-Seq Data Processing and Statistical Analysis
RNA-seq reads were aligned and mapped to the Drosophila reference genome (BDGP Release 5) using SOAPaligner/SOAP2, allowing no more than 5 bp mismatches. Low-quality reads (containing adapters or high content of unknown bases) were filtered out. Expression levels for genes were calculated using RPKM (reads per kilobase transcriptome per million mapped reads) method (Mortazavi et al., 2008) using the CLC Genomics Workbench (CLC bio, Boston, MA). Differential expression analysis was conducted using Cluster 3.0, and Java TreeView software (Eisen et al., 1998; Saldanha, 2004) and expressed as log2 Ratios (EtOH/Ctrl). We used FDR ≤ 0.0001 and an absolute value of log2 Ratio ≥ 1 as the threshold to judge the significance of expression difference. Gene ontology analysis was conducted using the Database for Annotation, Visualization and Integrated Discovery (DAVID) web-accessible tool, version 6.7 (Huang et al., 2009b; Huang et al., 2009a). For gene ontology annotation search and clustering, significant gene categories for each cluster were identified using default High Classification Stringency parameters (Kappa Similarity Term Overlap: 3; Similarity Threshold: 0.85; Initial Group Membership: 3; Final Group Membership: 3; Multiple Linkage Threshold: 0.5) and official gene symbols as input. The statistical significance of over-representation of antimicrobial peptide genes was determined using a binomial test in GraphPad Prism. The observed frequency of eight antimicrobial peptide genes in the 137 differentially expressed genes was compared to the 124 Humoral Immune Response genes (GO:0006959) in the Drosophila melanogaster genome (17,717 genes in FlyBase release 6.06).
Results
To test whether the Toll pathway modulates ethanol-induced behaviors in Drosophila melanogaster, we tested the effect of mutations, RNAi knockdown, and overexpression of Toll pathway genes on the rate of recovery from ethanol sedation. In this study, experimental and control animals were simultaneously exposed to vaporized ethanol until sedated, then switched to a humidified air stream to recover and the rate of recovery was measured (Krishnan et al., 2012; Cowmeadow et al., 2005). If the experimental group recovered from sedation faster than the control group, the experimental group is said to be more resistant to ethanol. Conversely, flies that recovered significantly slower than the control are said to be less resistant (or more sensitive). There is day-to-day variability in resistance to ethanol, for this reason all of the direct comparisons that we make are sedated in tandem on the same day. As per convention, in the text, gene names are italicized while the encoded protein is capitalized and typeset in roman.
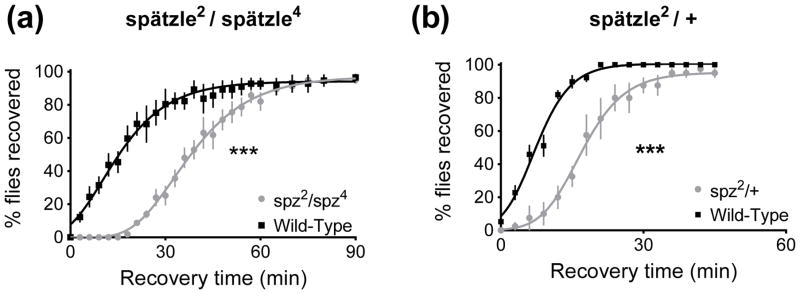
Spätzle is the Drosophila ligand of Toll, and Spätzle binding to Toll activates the pathway (Lemaitre et al., 1996; Weber et al., 2003). Transheterozygotes are often used when studying maternal effect lethal mutations. This is necessary because the stocks are maintained over a balancer chromosome, and it is thought that the chromosome of interest accumulates secondary lethal mutations, making homozygotes unobtainable without first backcrossing the mutation. Thus, animals lacking functional spätzle were obtained as a heteroallelic combination of two null alleles, spz2 and spz4 (Lemaitre et al. 1996).These spätzle null animals were less resistant to ethanol than wild-type controls, as can be seen by their slower recovery rate (p<0.0001, Fig. 2A). Additionally, animals heterozygous for a null allele of spätzle (spz2) and a wild-type chromosome were less resistant to ethanol than wild-type controls (p<0.0001, Fig. 2B). If the spätzle mutant is less ethanol resistant because of reduced signaling down the Toll pathway, then one would expect that reducing signaling at subsequent steps in the pathway would also reduce ethanol resistance. We tested this idea with a variety of genetic tools.
Figure 2.
spätzle mutants show reduced resistance to ethanol in a recovery from ethanol sedation assay. Age-matched females are placed in vials and exposed to ethanol vapor until sedated, and then animals are allowed to recover in a humidified air stream. 0 minutes denotes the beginning of the recovery. A) spätzle-null transheterozygotes recover from sedation more slowly than Canton S wild-type control animals (***, p<0.0001). B) Heterozygotes carrying a null allele of spätzle and a wild type chromosome recover from sedation more slowly than Canton S wild-type control animals (***, p<0.0001).
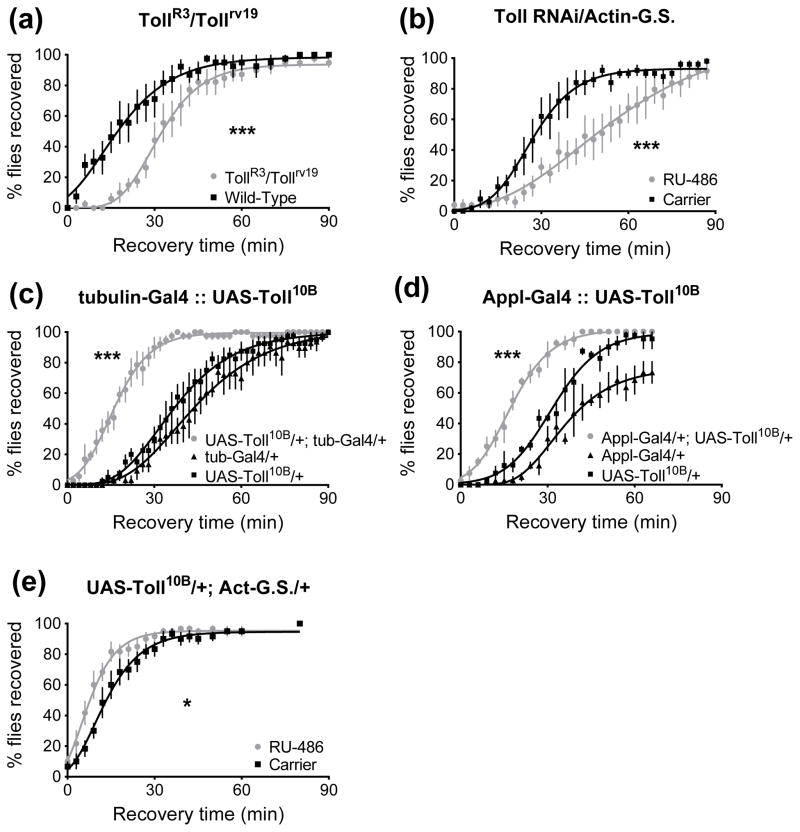
We manipulated Toll receptor activity with mutant alleles, a gene-specific RNAi knockdown transgene, and a Toll overexpression transgene (Fig. 3). Toll mutant animals were generated by combining two loss-of-function alleles of Toll (TollR3 and Tollrv19) to yield a transheterozygous animal. When compared to wild-type, these Toll mutant animals were less resistant than controls in a recovery from sedation assay (p<0.0001, Fig. 3A). Other researchers have reported similar findings for Toll mutants: animals with a transposon inserted near the Toll locus were less resistant than wild-type controls in an ethanol sedation assay (Morozova et al., 2007; Morozova et al., 2011). To confirm the involvement of Toll, we suppressed Toll expression with an RNAi transgene. For this experiment we used the Actin-GeneSwitch driver. An important advantage of the GeneSwitch system is that it permits adult-specific expression, which avoids disruption of normal development. In this system, a fusion protein between the Gal4 transcription factor and progesterone receptor activates UAS transgenes only in the presence of the RU-486 inducer (Osterwalder et al., 2001; Roman et al., 2001). The GeneSwitch system allowed us to perform experiments in which all flies have the same genotype, and the transgene of interest is expressed only in adults fed an inducer for three days prior to the experiment. Feeding RU-486 to animals carrying the Actin-GeneSwitch transgene but not a UAS responder transgene has no effect on ethanol resistance at the dose used here (Fig. S1 C & D). In concert with the Toll mutant analysis, animals in which Toll expression was suppressed with an Actin-GeneSwitch driven RNAi transgene also showed reduced ethanol resistance (p<0.0001, Fig. 3B).
Figure 3.
A Toll loss-of-function mutation or a Toll knockdown decreases resistance, while expression of a constitutively active Toll increases resistance in a recovery from ethanol sedation assay. 0 minutes denotes the beginning of the recovery period. A) Transheterozygous TollR3/Tollrv19 loss-of-function animals recover more slowly than wild-type Canton S (***, p<0.0001). B) RNAi knockdown of Toll via a UAS transgene driven by Actin-GeneSwitch, in which the GeneSwitch inducer was provided for three days prior to testing. Inducer-fed animals (RU-486) recovered more slowly than carrier-fed controls (***, p<0.0001). C) Overexpression of the constitutively active Toll10B allele using the ubiquitous tubulin-Gal4 driver increased resistance (***, p<0.0001 vs. either parental control). D) Overexpression of Toll10B in neurons using the Appl-Gal4 driver increased resistance (***, p<0.0001 vs. either parental control). E) Inducible overexpression of Toll10B only in adults increased resistance (*, p=0.03).
Increased Toll activity produced the opposite response. To increase Toll signaling activity we used a transgene that expresses a constitutively active Toll variant (Toll10B). Expression of the UAS-Toll10B transgene (Hu et al., 2004) enhanced ethanol resistance in our sedation assay. Driving the UAS-Toll10B construct ubiquitously with tubulin-Gal4 increased behavioral ethanol resistance (p<0.0001 vs. either parental control, Fig. 3C). Additionally, overexpressing Toll10B in neurons with the Appl-Gal4 driver increased ethanol resistance (p<0.0001 vs. either parental control, Fig. 3D). Appl-Gal4 is a neuron-specific, pan-neural driver (Torroja et al., 1999; Scholz et al., 2005; Fang et al., 2013). In addition, the Actin-GeneSwitch driver was used to overexpress UAS-Toll10B. When these animals were fed RU-486 inducer for three days, resistance increased compared to carrier-fed controls (p=0.03, Fig. 3E). Increasing or reducing Toll activity transgenically does not affect ethanol absorption or metabolism (Fig. S1 A and B). In all Gal4 overexpression experiments, we also individually examine the parental Gal4 driver line (black triangles in plots) and the parental UAS-responder line (black square in plots) to verify that changes in behavior are caused by expression of the responder transgene and are not the consequence of mutational insertion of a single transgene into the genome nor are they caused by off-target effects of the Gal4 transcription factor. The parental lines were crossed to wild-type Canton S so that the parental controls carry only a single copy of the transgene, as is the case in the experimental group.
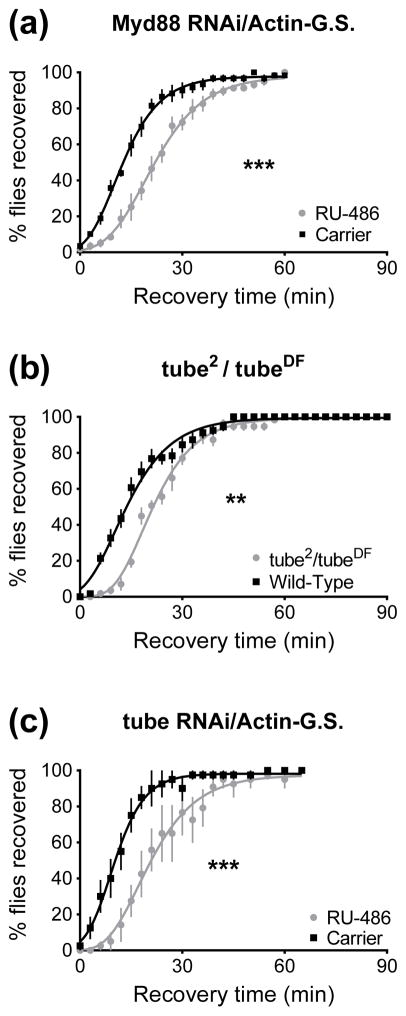
Myd88 is an adapter protein that interacts with Toll and is required for the immune response to infection (Tauszig-Delamasure et al., 2001; Marek & Kagan, 2012). Only transgenic RNAi knockdown of Myd88 was tested in this study. When Myd88 was knocked down for three days with the Actin-GeneSwitch driver, resistance to ethanol was reduced (p<0.0001, Fig. 4A). Tube is part of the adapter complex that assembles along with Toll, Myd88, and Pelle during Toll activation and is required for Toll signaling (Letsou et al., 1991; Moncrieffe et al., 2008). We generated tube null animals by a transheterozygous cross; animals carrying the tube2 null allele were mated with animals carrying a chromosomal deficiency (deletion) that removes the tube locus, producing tube2/tubeDF null animals (Hecht & Anderson, 1993). These mutant animals are less resistant to ethanol than the wild-type control (p=0.0013, Fig. 4B). Furthermore, a reduction in resistance was also produced when tube was knocked down in an adult-specific manner using an Actin-GeneSwitch driven RNAi transgene (p<0.0001, Fig. 4C).
Figure 4.
Loss of Myd88 or tube reduces resistance in a recovery from ethanol sedation assay. 0 minutes denotes the beginning of the recovery period. A) When an RNAi transgene is expressed in the adult to knock down Myd88 expression, resistance to ethanol is reduced compared to carrier-fed controls (***, p<0.0001). B) tube null transheterozygotes recover from sedation more slowly than wild type flies (**, p=0.0013). C) Knockdown of tube in the adult reduces resistance (***, p<0.0001).
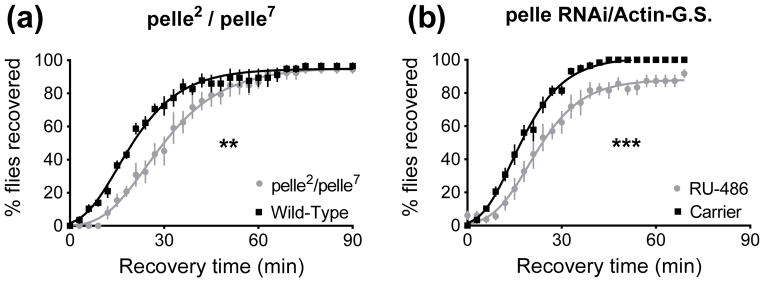
Pelle is a kinase that assembles with Toll, Myd88, and Tube after Toll pathway activation, and there is indirect evidence that Pelle is the kinase that phosphorylates Cactus (Huang et al., 2010; Towb et al., 2001). The phosphorylation of Cactus causes its degradation, which frees the Dif and Dorsal transcription factors that were sequestered outside of the nucleus by Cactus. Blocking the destabilization of Cactus reduces downstream nuclear signaling by Dif and Dorsal (Fig. 1). When two pelle loss-of-function alleles (pelle2 and pelle7) were combined to generate a pelle mutant transheterozygote (Hecht & Anderson, 1993; Anderson & Nüsslein-Volhard, 1984), ethanol resistance was reduced (p=0.0058, Fig. 5A). We also examined the effect of pelle knockdown using the inducible GeneSwitch system to drive ubiquitous expression of a pelle RNAi transgene. Similar to the pelle mutant, a three day pelle RNAi knockdown reduced resistance (p=0.0008, Fig. 5B).
Figure 5.
Loss of pelle via mutation or knockdown reduces resistance in a recovery from ethanol sedation assay. 0 minutes denotes the beginning of the recovery period. A) pelle loss-of-function transheterozygotes recover from sedation more slowly than wild type flies (**, p=0.0058). B) Knockdown of pelle in the adult reduces resistance (***, p=0.0008).
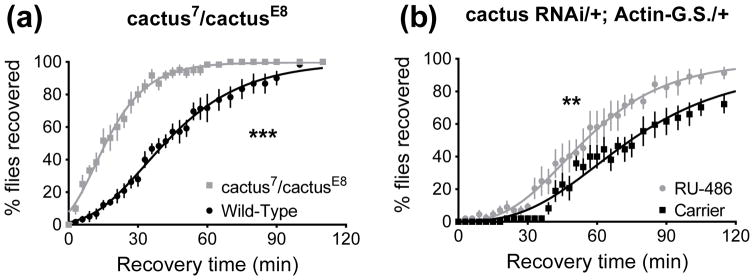
When the Toll pathway is inactive, the NF-κB inhibitor Cactus sequesters Dif and Dorsal transcription factors in the cytoplasm. After stimulation of the Toll pathway, Cactus is degraded and the NF-κB proteins Dif and Dorsal are able to enter the nucleus and activate target genes (Fig. 1, Geisler et al., 1992; Roth et al., 1991). Thus, reduction of cactus expression via mutation or knockdown should mimic a stimulated pathway where NF-κB proteins are allowed to enter the nucleus. We show that cactus mutant animals (transheterozygous for two separate loss-of function alleles, cactus7 and cactusE8) were more resistant to ethanol (p<0.0001, Fig. 6A) than the paired control, and that adult-specific RNAi knockdown of cactus expression increases resistance (p=0.0091, Fig. 6B). These findings align with the phenotypes seen when Toll pathway activity was stimulated with the UAS-Toll10B transgene (Fig. 3C–E).
Figure 6.
Loss of cactus increases resistance in a recovery from ethanol sedation assay. 0 minutes denotes the beginning of the recovery period. A) Transheterozygous cactus loss-of-function animals (cactus7/cactusE8) are more resistant to ethanol than wild type (***, p<0.0001). B) RNAi knockdown of cactus increases resistance to ethanol (**, p=0.0091). In A and B all animals recovered.
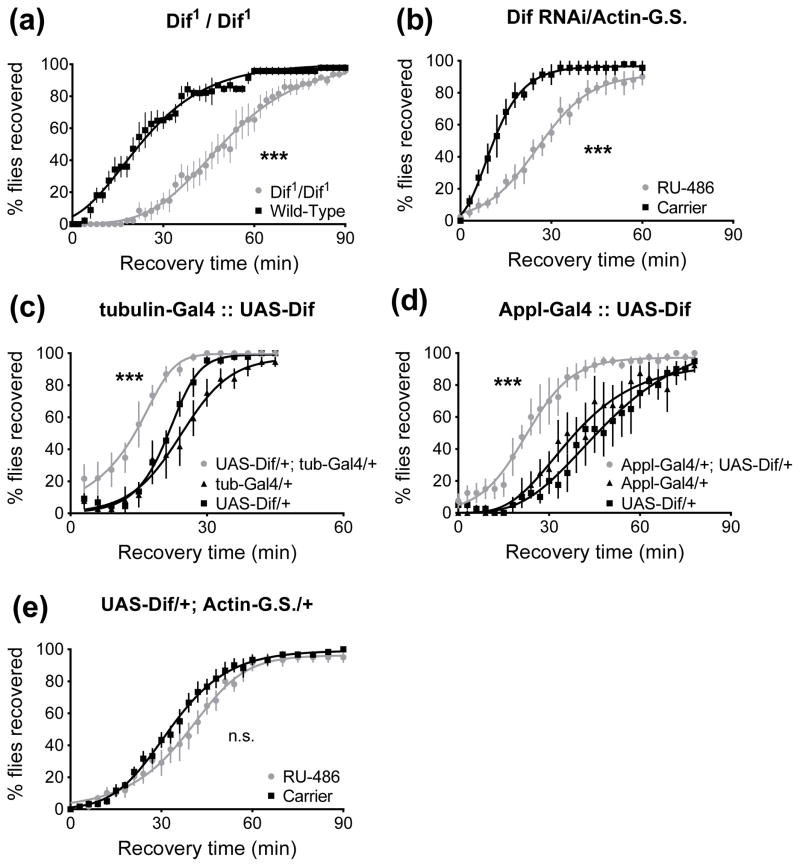
Dif is one of the three NF-κB family members in Drosophila, and upon activation of the Toll pathway functions in the induction of target genes including antimicrobial peptides (Petersen et al., 1995). Dif mutant animals (Dif1/Dif1) are less ethanol resistant than wild-type animals (p<0.0001, Fig. 7A), and RNAi knockdown of Dif expression in the adult decreases resistance (p<0.0001, Fig. 7B). Next we used overexpression of a Dif transgene to mimic active Toll pathway signaling. We observed that overexpression of Dif (UAS-Dif, Yagi & Ip, 2005) leads to increased resistance when driven by the ubiquitous tubulin-Gal4 driver (p<0.0001 vs. either parental control, Fig. 7C). Resistance is also increased when the neural Appl-Gal4 driver is used to drive expression of the UAS-Dif transgene (p<0.0001 vs. UAS-Dif alone, p=0.0005 v Appl-Gal4 alone, Fig. 7D). However, three day overexpression of Dif using the Actin-GeneSwitch driver had no effect on ethanol resistance (p=0.4895, Fig. 7E).
Figure 7.
Reduction of Dif reduces resistance, and overexpression of Dif can increase resistance as measured in a recovery from ethanol sedation assay. 0 minutes denotes the beginning of the recovery period. A) Dif mutant animals show reduced resistance (***, p<0.0001), as seen by their slower recovery from sedation. B) Three day knockdown of Dif using the inducible GeneSwitch system leads to decreased resistance (***, p<0.0001). C) Overexpression of a Dif transgene with the ubiquitous tubulin-Gal4 driver increases resistance (***, p<0.0001 vs. either parental). D) Overexpression of Dif in neurons increases ethanol resistance (***, p<0.0001 vs. UAS-Dif alone, p=0.0005 v Appl-Gal4 alone). E) Three day overexpression of Dif using the GeneSwitch system had no effect on alcohol resistance (p=0.4895).
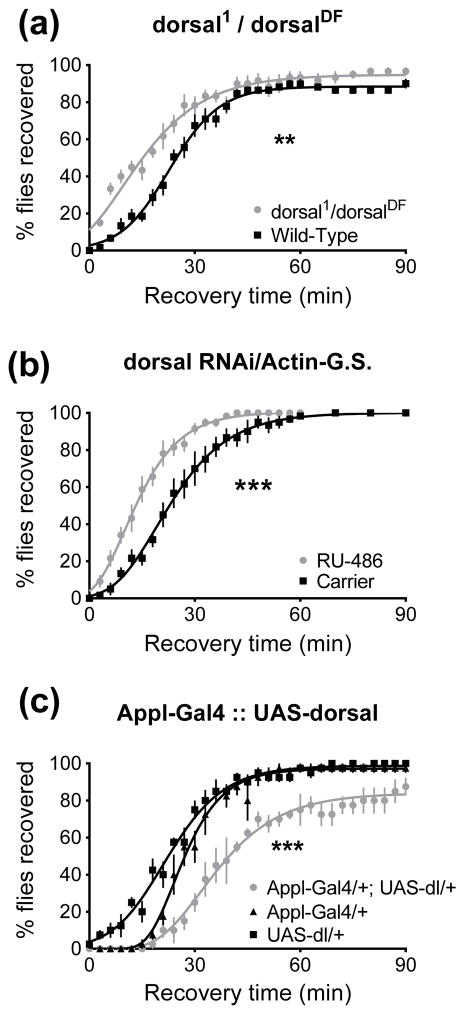
dorsal is another NF-κB transcription factor, and while Dif and Dorsal are both stimulated by Toll pathway activation and are redundant in some contexts, they have some distinct roles. dorsal, but not Dif is required for embryonic development. On the other hand Dif plays the greater role in the adult innate immune system (see Discussion). Nevertheless we also examined the role of dorsal on ethanol resistance. When we tested dorsal null animals – transheterozygotes carrying a null allele of dorsal (dorsal1) and a deficiency removing dorsal (dorsalDF) – the mutants were more resistant than the wild-type control (p=0.0079, Fig. 8A). Knockdown of dorsal with an RNAi transgene also led to increased resistance (p<0.0001, Fig. 8B). Conversely, when a dorsal transgene (Yagi & Ip, 2005) was driven in the nervous system using the Appl-Gal4 driver we observed that the animals had reduced resistance to ethanol (p=0.003 vs. Appl-Gal4 alone, p<0.0001 vs. UAS-dorsal alone, Fig. 8C). However, dorsal overexpression might have reduced ethanol resistance because the animals are less fit and have physical defects. We observed that these flies had crumpled, unexpanded wings. Furthermore, overexpressing dorsal using a tubulin-Gal4 was lethal. We also observed lethality when the UAS-dorsal was combined with the Actin-GeneSwitch driver. Lethality occurred even when the animals were raised without inducer at 18°C to minimize expression from the transgenes (Duffy, 2002).
Figure 8.
Loss of dorsal increases resistance, while overexpression of dorsal decreases resistance in a recovery from ethanol sedation assay. 0 minutes denotes the beginning of the recovery period. A) dorsal-null animals carrying a dorsal null allele and a deficiency uncovering dorsal are more resistant than wild type animals (**, p=0.0079). B) Knockdown of dorsal expression in the adult increases resistance (***, p<0.0001). C) Overexpression of a UAS-dorsal transgene in neurons caused decreased resistance (***, p=0.003 vs. Appl-Gal4 alone, p<0.0001 vs. UAS-dorsal alone).
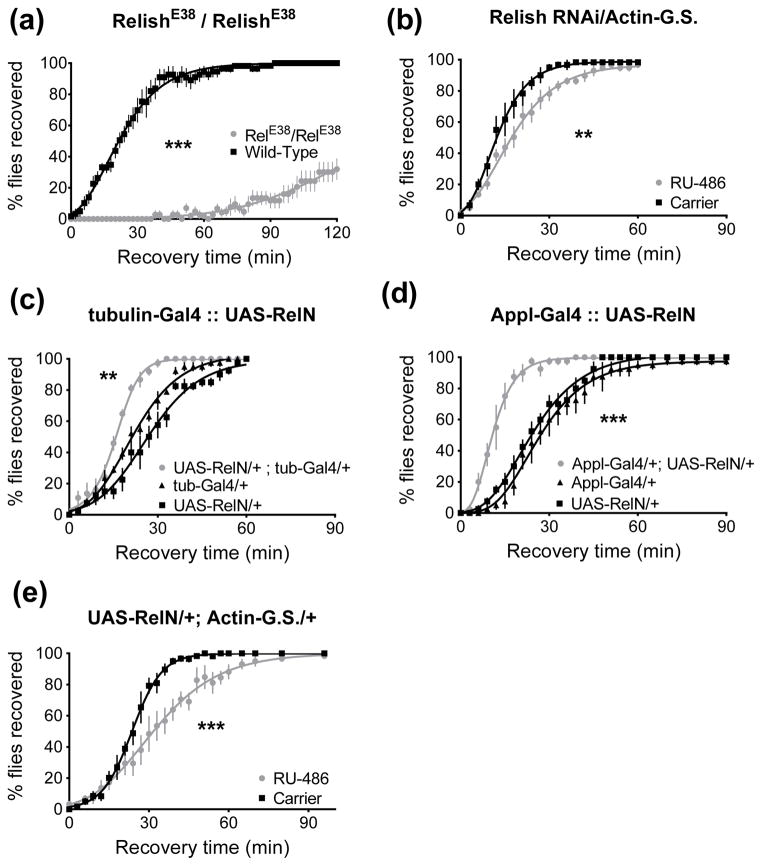
Relish is a third NF-κB family transcription factor in Drosophila. It is primarily activated by the IMD innate immune pathway, which responds to infection by Gram-negative bacteria. We included Relish in this study because there is evidence of cross-talk between the Toll and IMD pathways, and because Relish has been shown to heterodimerize with both Dif and Dorsal (Tanji et al., 2010; Dushay et al., 1996; Hedengren et al., 1999). Animals homozygous for the loss-of-function RelishE38 allele (Hedengren et al., 1999) showed reduced resistance to ethanol (p<0.0001, Fig. 9A). Knockdown of Relish in adults using RNAi and the Actin-GeneSwitch driver also reduced resistance (p=0.0023, Fig. 9B). In contrast to Dif and Dorsal, where functional regulation is achieved via sequestration by Cactus protein, Relish has an autoinhibitory domain that is cleaved after IMD pathway activation (Stöven et al., 2000). Overexpression of the active form of Relish (UAS-RelN, Yagi & Ip, 2005) with tubulin-Gal4 or Appl-Gal4 increases resistance (tubulin-Gal4: p<0.001, Fig. 9C; Appl-Gal4: p<0.0001, Fig. 9D). However, three day overexpression with the GeneSwitch system significantly reduces resistance (p<0.0001, Fig. 9E).
Figure 9.
Relish affects resistance to ethanol in a recovery from ethanol sedation assay. 0 minutes denotes the beginning of the recovery period. A) Homozygous loss-of-function RelE38 animals recover more slowly than wild type animals (***, p<0.0001). All animals eventually recover, this graph is clipped. B) Three day RNAi knockdown of Relish using Actin-GeneSwitch reduces resistance (**, p=0.0023). C) Expression of the N-terminal active domain of Relish (RelN) using the tubulin-Gal4 driver increases resistance (**, p<0.0001 vs. UAS-RelN alone, p=0.001 vs. tubulin-Gal4 alone). D) Overexpression of RelN in the nervous system increases resistance (***, p<0.0001 vs. either parental). E) Three day overexpression of RelN using Actin-GeneSwitch reduces resistance (***, p<0.0001).
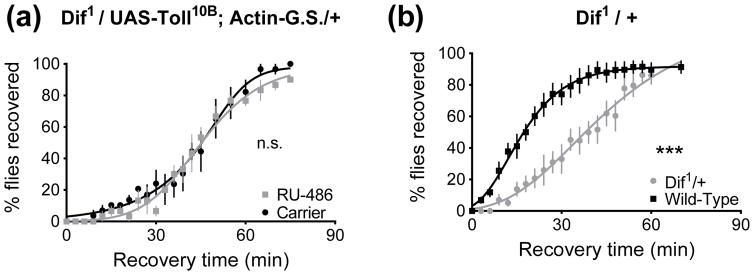
To establish that overexpression of Toll is acting through the canonical signaling pathway to increase resistance, we overexpressed a constitutively active Toll in a Dif mutant background. To do so we generated Dif1/UAS-Toll10B; Actin-GeneSwitch/+ animals and fed them RU-486 inducer for three days. Feeding inducer had no effect on resistance, indicating that for the increased resistance response, Toll acts through Dif (p=0.38, Fig. 10A). This experiment utilized animals heterozygous for the Dif1 loss-of-function allele, and here we show that animals heterozygous for Dif1 over a wild-type chromosome are more sensitive to ethanol (p<0.0001, Fig. 10B), mimicking the phenotype of the homozygote reported in Fig. 7A.
Figure 10.
Increased Toll activity produces ethanol resistance in a Dif-dependent manner. 0 minutes denotes the beginning of the recovery period. A) A Dif1 mutation blocks the effect of overexpressing constitutively active UAS-Toll10B using Actin-GeneSwitch (p=0.38). Overexpressing Toll10B with functional Dif increases resistance (see Fig. 3E). B) Dif is haploinsufficient with respect to ethanol resistance: Dif1 heterozygotes are less resistant to ethanol sedation (p<0.0001). The animals in Fig. 10A are heterozygous for Dif1, whereas the Dif1 mutants reported in Fig. 7A were homozygous.
The innate immune system is stimulated by ethanol sedation
To determine whether the Drosophila Toll innate immune signaling pathway is activated after ethanol sedation we performed RNA-seq 30 minutes after ethanol sedation. Activation of the Toll pathway results in increased transcription of antimicrobial genes. We observed a change in abundance of 137 mRNAs purified from adult heads (Table S1). This includes eight antimicrobial peptide genes that represent 38.1% of the microbial peptide genes encoded in Drosophila (p<0.0001; Table S1; Hetru et al., 2003). Furthermore, the top two DAVID gene ontology clusters were stress-response genes and innate immune genes (Table S1; Huang et al., 2009b; Huang et al., 2009a). In addition to the enriched subset of innate immune genes, we saw induction of genes linked to heat shock response, stress response, programmed cell death, and calcium sensing. However, there is a dearth of genes whose ontology is neural specific or behavioral specific.
The Toll signaling pathway is not required for animals to acquire 24h ethanol tolerance
The observation that genetic manipulation of the Toll-signaling pathway affects resistance and the observation that ethanol sedation activates the Toll-signaling pathway led us to hypothesize that Toll signaling might be a trigger for the production of ethanol tolerance. Ethanol tolerance is a reduced response to an effect of ethanol caused by prior ethanol exposure (ethanol-induced ethanol resistance). We tested for the capacity to acquire ethanol tolerance by comparing the rate of recovery from ethanol sedation in animals that are recovering from their first ethanol sedation to the rate of recovery of animals recovering from their second ethanol sedation (24h between first and second sedation; Cowmeadow 2005). The activity of the Toll pathway was manipulated in the same way in both first and second sedation animals. We activated the Toll pathway using the constitutively active Toll receptor (Actin-GeneSwitch driven expression of UAS-Toll10B) or blocked signaling from the pathway with loss-of-function mutations in the Dif transcription factor. Whereas both manipulations altered innate ethanol resistance, neither affected the capacity to acquire 24h tolerance (Supplementary Fig. S2).
Discussion
In this study we examined resistance to ethanol sedation because baseline resistance to the effects of ethanol can be used as a real world predictor of drinking problems in humans. In a 25 year study, the baseline resistance of college-aged participants was a strong predictor of alcohol use disorders later in life (Schuckit & Smith, 2011). An individual’s level of response to alcohol has been shown to have a strong genetic component (Heath et al., 1999; Schuckit et al., 2004). Individuals who have a lower response to alcohol have to drink more to experience the pleasurable effects of alcohol and can also drink longer. As a result, they expose themselves to higher levels of alcohol, which in turn promotes addiction and increases their risk for alcohol toxicity. Here we show that the Toll innate-immune signaling pathway can profoundly influence resistance to ethanol sedation in adult Drosophila.
Consistent with the hypothesis that ethanol sedation rapidly promotes signaling down the Toll innate immune signaling pathway, 30 minutes after ethanol sedation, we observed increased expression of a number of antimicrobial peptide genes—these genes are outputs of innate immune signaling pathways. Eight of the 21 canonical antimicrobial peptide genes were upregulated. Previously, Kong et al. (2010) reported that the genes encoding core members of the innate immune signaling pathways, Toll, Myd88, cactus, Imd, and Relish, are induced less than two fold about 90 minutes after the start of ethanol exposure and return to baseline within three hours. We did not observe induction of these pathway genes in our analysis and Kong et al. only reported dro5 antimicrobial peptide gene induction. This difference may be because we did not accept changes that were less than two fold, or because of differences in the treatment protocol. Stimulation of the signaling pathway and induction of expression of the signaling pathway genes are fundamentally distinct events. However, upregulation of the pathway genes themselves is also interesting because it could sensitize this pathway to future ethanol or inflammatory stimuli and might contribute to acute tolerance (see below).
Every member of the Toll signaling pathway that we tested had an effect on resistance to ethanol in at least one paradigm. Suppressing Toll pathway signaling by mutation or by knockdown of pathway members decreases resistance to ethanol sedation, while increasing Toll pathway activity increases resistance to ethanol sedation (Table 1) with one notable exception — dorsal. Whereas, the effects of manipulating Dif expression fit the model that increased and decreased Toll pathway activity increases and decreases ethanol resistance, respectively, the manipulation of dorsal produced the opposite result. Overexpression of dorsal, however, also caused developmental defects affecting adult fitness while overexpression of Toll, Dif, or Relish did not obviously affect fitness. Furthermore, there is strong evidence that Dif and dorsal play distinct roles in Drosophila embryos and adults (Rutschmann et al., 2000; Lemaitre et al., 1996; Lemaitre et al., 1995; Gross et al., 1996; Meng et al., 1999). An absence of Dorsal protein in the embryo lethally disrupts dorsal-ventral patterning while the absence of Dif protein does not perturb embryogenesis. Whereas, in the innate immune system Dif is a strong regulator of antifungal genes, dorsal is not required for normal immune function. For instance, dorsal mutations do not affect the induction of Drosomycin after infection, while Dif mutants show a substantial reduction in the ability to induce Drosomycin after infection (Rutschmann et al., 2000; Lemaitre et al., 1996). In Drosophila, the fat body is a major hub of immune signaling activity, and fat body explants have been used to assay Toll and IMD pathway activity after exposure to various pathogens and pathogen components. In dissected fat bodies, the nuclear translocation of Dif protein can be stimulated by bacterial coat components, but the movement of Dorsal into the nucleus requires components of the hemolymph (Bettencourt et al., 2004). In at least one context Dorsal was shown to have the opposite effect of Dif. In the case of Cecropin transcriptional control, Dorsal was shown to suppress gene activation by Dif, reducing expression of a Cecropin reporter construct when co-expressed with Dif (Petersen et al., 1995). Finally, mutations that constitutively activate Toll signaling, such as Toll10B or cactus loss-of-function alleles, cause the formation of melanotic tumors in larvae. dorsal is not involved in this process, as null mutations in dorsal do not block the appearance of tumors (Lemaitre et al., 1995). Together these studies show that dorsal and Dif have distinct and sometimes opposing roles, and our results extend these observations to include effects on ethanol resistance.
Table 1.
Summary of Behavior Experiments
| Overexpression
|
|||||
|---|---|---|---|---|---|
| Gene | Mutant Allele | Inducible RNAi | Tub-Gal4 | Appl-Gal4 | Actin-G.S. |
| spätzle | Sensitive | ||||
| Toll | Sensitive | Sensitive | Resistant | Resistant | Resistant |
| Myd88 | Sensitive | ||||
| tube | Sensitive | Sensitive | |||
| pelle | Sensitive | Sensitive | |||
| cactus | Resistant | Resistant | |||
| Dif | Sensitive | Sensitive | Resistant | Resistant | n.s. |
| dorsal | Resistant | Resistant | Lethal | Sensitive | Lethal |
| Relish | Sensitive | Sensitive | Resistant | Resistant | Sensitive |
Our experiments using Actin-GeneSwitch to drive overexpression of either Dif or Relish yielded results that did not match the results obtained when the UAS transgene was driven by tubulin-Gal4 or Appl-Gal4. Dif overexpression led to resistance when driven by the two Gal4 drivers, but had no effect when driven by the Actin-GeneSwitch driver, and Relish overexpression driven by Actin-GeneSwitch decreased resistance, as opposed to increased resistance when driven by the Gal4 constructs. This conflict may result from poor GeneSwitch induction of the Dif and Rel transgenes or may indicate that the overexpression phenotype for these transcription factors has a developmental component.
The observations that Toll signaling modulates resistance, and that the innate immune system is rapidly activated by ethanol exposure led us to speculate that the Toll pathway might be involved in the generation of ethanol rapid tolerance (rapid tolerance has been defined as the tolerance manifest after ethanol clearance). However, in our experiments perturbation of Toll pathway activity did not affect the ability to acquire 24h rapid tolerance, indicating that the Toll pathway is not necessary for producing 24h rapid alcohol tolerance despite the fact that modulating its activity affects ethanol resistance. This is not unusual in that mechanisms that produce resistance and rapid tolerance have been shown to be distinct before. Genes that contribute to resistance are not necessarily required for the acquisition of rapid tolerance, and genes necessary for the acquisition of rapid tolerance do not always affect baseline resistance. For instance, measurement of the magnitude of resistance and rapid tolerance in 205 inbred, sequenced Drosophila lines did not show a correlation between the magnitudes of resistance and rapid tolerance (see Fig.1 in Morozova et al., 2015). Furthermore, Drosophila experiments describing circadian fluctuation in ethanol-induced behaviors also exposed a disconnect between resistance and rapid tolerance. In a loss of righting reflex assay, baseline resistance oscillated in a circadian manner, peaking in the early evening, but rapid tolerance did not oscillate—the magnitude of rapid tolerance was the same regardless of time of day (van der Linde & Lyons, 2011). Although the innate immune signaling pathway does not appear to have a role in producing 24h rapid tolerance, our data predicts that it may contribute to the production of a transient form of tolerance called acute tolerance (defined as tolerance that appears during a drug experience). Expression data suggests that ethanol causes a sudden activation of innate immune signaling. Because increased Toll activity increases resistance, we would expect animals to become ethanol resistant during immune activation. This increase in resistance (acute tolerance) is perhaps later subsumed by a distinct mechanism responsible for 24h rapid tolerance.
Our findings mesh well with how Drosophila interact with ethanol in their natural environment. The findings in Milan et al. (2012) demonstrate that Drosophila seek ethanol-containing food after becoming infected by parasitic wasps, and that ethanol consumption helps kill the invading wasp. In order to promote maximum fitness, an animal that is driven to self medicate with ethanol might be expected to increase its resistance to the sedative effects of the drug, lest it become intoxicated and become easy prey. It has been shown that infection by endoparasitic wasps activates the Toll Pathway, so perhaps increasing resistance to ethanol arose as an adaptive response in anticipation of ethanol consumption. While it has not been shown that the Drosophila Toll pathway modulates drinking in the adult fly, in mammals innate immune signaling through pathways related to Drosophila Toll such as IL-1 and TLR4 have been shown to increase drinking (Robinson et al., 2014). Here we have shown that in Drosophila the innate immune system regulates how the animal responds to ethanol sedation, and that innate immune signaling is rapidly induced by ethanol exposure.
Supplementary Material
Figure S1: Additional Controls. Manipulation of Toll expression does not affect ethanol absorption or metabolism (panels A & B). A) Overexpression of a constitutively active Toll10B isoform does not affect ethanol absorption or metabolism (2 way ANOVA treatment effect p=0.5499, n.s. difference at each point). B) Knockdown of Toll expression with a UAS-driven RNAi construct targeting Toll driven by Actin-GeneSwitch does not affect ethanol absorption or metabolism (2 way ANOVA treatment effect p=0.0520, n.s. difference at each point). Treatment with RU-486 inducer does not affect alcohol resistance but does induce a GFP reporter (panels C & D). C) Actin-GeneSwitch animals were crossed to the TRiP RNAi background stock, and tested for effects on alcohol resistance in the presence of RU-486 or carrier. No difference in resistance was seen (p=0.4289). D) Actin-GeneSwitch was crossed to our wild type Canton S strain once and tested for effects on alcohol resistance. RU-486 feeding caused no change (p=0.8477). Feeding adults RU-486 induces target gene expression. E) UAS-GFP; Actin-GeneSwitch animals were tested for induction of GFP after drug feeding. Fluorescence is notably brighter in dissected brains of inducer fed animals (RU-486, top) than in controls (Carrier, bottom).
Figure S2: Manipulation of Toll pathway activity does not block the ability to acquire 24h tolerance. In these tolerance tests, on the day prior to data collection half of the animals received a sedating dose of ethanol and the control group received no ethanol. 24h later both groups were sedated and the rate of recovery recorded. A) Overexpression of the constitutively active Toll10B allele using the ubiquitous Actin-GeneSwitch driver does not block the acquisition of tolerance, as can be seen in the faster recovery for the group receiving its second sedation (***, p<0.0001). All animals were fed RU-486 inducer for three days prior to the experiment. B) Dif1 homozygous loss-of-function animals are able to acquire tolerance, as can be seen in the faster recovery for the group receiving its second sedation. (**, p=0.0012).
Table S1: Findings from our RNA-seq study. List of genes significantly regulated 30 min after ethanol sedation and findings from gene ontology analysis performed using DAVID.
Table S2: Drosophila stocks used in this study. Details on the full genotype of stocks used, as well as the source stocks of alleles and transgenes.
Acknowledgments
We thank the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing transgenic RNAi fly stocks used in this study. We thank Dr. Y. T. Ip for providing the UAS-driven transgenes for Toll, Dif, dorsal and Relish. The authors claim no conflict of interest. This work was supported by NIH grant number 1F31AA021326-01 to BRT, NIH grant number 2R01AA018037-06A1 to NSA, and NIH grant numbers K08AA017481 and R01AA017920 to AZP.
References
- Anderson KV, Nüsslein-Volhard C. Information for the dorsal–ventral pattern of the Drosophila embryo is stored as maternal mRNA. Nature. 1984;311:223–227. doi: 10.1038/311223a0. [DOI] [PubMed] [Google Scholar]
- Ausubel F. Current Protocols in Molecular Biology. Wiley; New York, NY: 1994. [Google Scholar]
- Bajo M, et al. Innate immune factors modulate ethanol interaction with GABAergic transmission in mouse central amygdala. Brain, behavior, and immunity. 2014;40:191–202. doi: 10.1016/j.bbi.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt R, et al. Hemolymph-dependent and -independent responses in Drosophila immune tissue. Journal of cellular biochemistry. 2004;92:849–63. doi: 10.1002/jcb.20123. [DOI] [PubMed] [Google Scholar]
- Bouchery EE, et al. Economic costs of excessive alcohol consumption in the U.S., 2006. American journal of preventive medicine. 2011;41:516–24. doi: 10.1016/j.amepre.2011.06.045. [DOI] [PubMed] [Google Scholar]
- Brazma A, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nature genetics. 2001;29:365–71. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Buchon N, Silverman N, Cherry S. Immunity in Drosophila melanogaster — from microbial recognition to whole-organism physiology. Nature Reviews Immunology. 2014;14:796–810. doi: 10.1038/nri3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowmeadow RB, Krishnan HR, Atkinson NS. The slowpoke gene is necessary for rapid ethanol tolerance in Drosophila. Alcoholism, clinical and experimental research. 2005;29:1777–86. doi: 10.1097/01.alc.0000183232.56788.62. [DOI] [PubMed] [Google Scholar]
- Crews FT, et al. High mobility group box 1/Toll-like receptor danger signaling increases brain neuroimmune activation in alcohol dependence. Biological psychiatry. 2013;73:602–12. doi: 10.1016/j.biopsych.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C, Shurtleff D, Harris RA. Neuroimmune mechanisms of alcohol and drug addiction. International review of neurobiology. 2014;118:1–12. doi: 10.1016/B978-0-12-801284-0.00001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy JB. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis (New York, NY : 2000) 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- Dushay MS, Asling B, Hultmark D. Origins of immunity: Relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:10343–7. doi: 10.1073/pnas.93.19.10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, et al. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA. Genetic influences on the development of alcoholism. Current psychiatry reports. 2013;15:412. doi: 10.1007/s11920-013-0412-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Soares L, Bonini NM. Design and implementation of in vivo imaging of neural injury responses in the adult Drosophila wing. Nature Protocols. 2013;8:810–819. doi: 10.1038/nprot.2013.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris SP, et al. Transcriptome organization for chronic alcohol abuse in human brain. Molecular psychiatry. 2014 doi: 10.1038/mp.2014.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatscher-Bader T, et al. Alcohol-responsive genes in the frontal cortex and nucleus accumbens of human alcoholics. Journal of neurochemistry. 2005;93:359–70. doi: 10.1111/j.1471-4159.2004.03021.x. [DOI] [PubMed] [Google Scholar]
- Geisler R, et al. cactus, a gene involved in dorsoventral pattern formation of Drosophila, is related to the IκB gene family of vertebrates. Cell. 1992;71:613–621. doi: 10.1016/0092-8674(92)90595-4. [DOI] [PubMed] [Google Scholar]
- Grant BF, et al. Epidemiology of DSM-5 Alcohol Use Disorder: Results From the National Epidemiologic Survey on Alcohol and Related Conditions III. JAMA psychiatry. 2015 doi: 10.1001/jamapsychiatry.2015.0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross I, et al. Drosophila Immunity: A Comparative Analysis of the Rel Proteins Dorsal and Dif in the Induction of the Genes Encoding Diptericin and Cecropin. Nucleic Acids Research. 1996;24:1238–1245. doi: 10.1093/nar/24.7.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, et al. Identification of lipoteichoic acid as a ligand for draper in the phagocytosis of Staphylococcus aureus by Drosophila hemocytes. Journal of immunology (Baltimore, Md : 1950) 2009;183:7451–60. doi: 10.4049/jimmunol.0901032. [DOI] [PubMed] [Google Scholar]
- Heath AC, et al. Genetic differences in alcohol sensitivity and the inheritance of alcoholism risk. Psychological medicine. 1999;29:1069–81. doi: 10.1017/s0033291799008909. [DOI] [PubMed] [Google Scholar]
- Hecht PM, Anderson KV. Genetic characterization of tube and pelle, genes required for signaling between Toll and dorsal in the specification of the dorsal-ventral pattern of the Drosophila embryo. Genetics. 1993;135:405–17. doi: 10.1093/genetics/135.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedengren M, et al. Relish, a Central Factor in the Control of Humoral but Not Cellular Immunity in Drosophila. Molecular Cell. 1999;4:827–837. doi: 10.1016/s1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- Hetru C, Troxler L, Hoffmann JA. Drosophila melanogaster antimicrobial defense. The Journal of infectious diseases. 2003;187(Suppl):S327–34. doi: 10.1086/374758. [DOI] [PubMed] [Google Scholar]
- Hu X, et al. Multimerization and interaction of Toll and Spätzle in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9369–74. doi: 10.1073/pnas.0307062101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang HR, et al. Endocytic pathway is required for Drosophila Toll innate immune signaling. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8322–7. doi: 10.1073/pnas.1004031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imler JL. Overview of Drosophila immunity: a historical perspective. Developmental and comparative immunology. 2014;42:3–15. doi: 10.1016/j.dci.2013.08.018. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, et al. Decreased expression of NEFH and PCP4/PEP19 in the prefrontal cortex of alcoholics. Neuroscience research. 2004;49:379–85. doi: 10.1016/j.neures.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Kacsoh BZ, et al. Fruit flies medicate offspring after seeing parasites. Science (New York, NY ) 2013;339:947–50. doi: 10.1126/science.1229625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong EC, et al. Ethanol-regulated genes that contribute to ethanol sensitivity and rapid tolerance in Drosophila. Alcoholism, clinical and experimental research. 2010;34:302–16. doi: 10.1111/j.1530-0277.2009.01093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan HR, et al. A role for dynamin in triggering ethanol tolerance. Alcohol Clin Exp Res. 2012;36:24–34. doi: 10.1111/j.1530-0277.2011.01587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau GW, et al. The Drosophila melanogaster toll pathway participates in resistance to infection by the gram-negative human pathogen Pseudomonas aeruginosa. Infection and immunity. 2003;71:4059–66. doi: 10.1128/IAI.71.7.4059-4066.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, et al. Functional analysis and regulation of nuclear import of dorsal during the immune response in Drosophila. The EMBO journal. 1995;14:536–45. doi: 10.1002/j.1460-2075.1995.tb07029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, et al. The Dorsoventral Regulatory Gene Cassette Controls the Potent Antifungal Response in Drosophila Adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Letsou A, et al. Genetic and molecular characterization of tube, a Drosophila gene maternally required for embryonic dorsoventral polarity. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:810–4. doi: 10.1073/pnas.88.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewohl JM, et al. Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcoholism, clinical and experimental research. 2000;24:1873–82. [PubMed] [Google Scholar]
- van der Linde K, Lyons LC. Circadian modulation of acute alcohol sensitivity but not acute tolerance in Drosophila. Chronobiol Int. 2011;28:397–406. doi: 10.3109/07420528.2011.577921. [DOI] [PubMed] [Google Scholar]
- Liu J, et al. Altered gene expression profiles in the frontal cortex of cirrhotic alcoholics. Alcoholism, clinical and experimental research. 2007;31:1460–6. doi: 10.1111/j.1530-0277.2007.00444.x. [DOI] [PubMed] [Google Scholar]
- Liu J, et al. Gene expression profiling of individual cases reveals consistent transcriptional changes in alcoholic human brain. Journal of neurochemistry. 2004;90:1050–8. doi: 10.1111/j.1471-4159.2004.02570.x. [DOI] [PubMed] [Google Scholar]
- Liu J, et al. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2006;31:1574–82. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- Luce-Fedrow A, et al. Use of Drosophila S2 cells as a model for studying Ehrlichia chaffeensis infections. Applied and environmental microbiology. 2008;74:1886–91. doi: 10.1128/AEM.02467-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield BE, et al. Exploration of host-pathogen interactions using Listeria monocytogenes and Drosophila melanogaster. Cellular Microbiology. 2003;5:901–911. doi: 10.1046/j.1462-5822.2003.00329.x. [DOI] [PubMed] [Google Scholar]
- Marek LR, Kagan JC. Phosphoinositide binding by the Toll adaptor dMyD88 controls antibacterial responses in Drosophila. Immunity. 2012;36:612–22. doi: 10.1016/j.immuni.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield RD, et al. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. Journal of neurochemistry. 2002;81:802–13. doi: 10.1046/j.1471-4159.2002.00860.x. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Mao Z, Davis RL. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Science’s STKE: signal transduction knowledge environment. 2004;2004:pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- Meng X, Khanuja BS, Ip YT. Toll receptor-mediated Drosophila immune response requires Dif, an NF-kappaB factor. Genes & development. 1999;13:792–7. doi: 10.1101/gad.13.7.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan NF, Kacsoh BZ, Schlenke TA. Alcohol consumption as self-medication against blood-borne parasites in the fruit fly. Current biology: CB. 2012;22:488–93. doi: 10.1016/j.cub.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncrieffe MC, Grossmann JG, Gay NJ. Assembly of oligomeric death domain complexes during Toll receptor signaling. The Journal of biological chemistry. 2008;283:33447–54. doi: 10.1074/jbc.M805427200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova TV, et al. Polymorphisms in early neurodevelopmental genes affect natural variation in alcohol sensitivity in adult drosophila. BMC genomics. 2015;16:865. doi: 10.1186/s12864-015-2064-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova TV, Anholt RRH, Mackay TFC. Phenotypic and transcriptional response to selection for alcohol sensitivity in Drosophila melanogaster. Genome biology. 2007;8:R231. doi: 10.1186/gb-2007-8-10-r231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova TV, Mackay TFC, Anholt RRH. Transcriptional networks for alcohol sensitivity in Drosophila melanogaster. Genetics. 2011;187:1193–205. doi: 10.1534/genetics.110.125229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Osterwalder T, et al. A conditional tissue-specific transgene expression system using inducible GAL4. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12596–601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddibhatla I, et al. Role for sumoylation in systemic inflammation and immune homeostasis in Drosophila larvae. PLoS pathogens. 2010;6:e1001234. doi: 10.1371/journal.ppat.1001234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen UM, et al. The dorsal-related immunity factor, Dif, is a sequence-specific trans-activator of Drosophila Cecropin gene expression. The EMBO journal. 1995;14:3146–58. doi: 10.1002/j.1460-2075.1995.tb07317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, et al. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32:1884–97. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G, et al. Neuroimmune pathways in alcohol consumption: evidence from behavioral and genetic studies in rodents and humans. International review of neurobiology. 2014;118:13–39. doi: 10.1016/B978-0-12-801284-0.00002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman G, et al. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12602–7. doi: 10.1073/pnas.221303998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S, et al. cactus, a maternal gene required for proper formation of the dorsoventral morphogen gradient in Drosophila embryos. Development (Cambridge, England) 1991;112:371–88. doi: 10.1242/dev.112.2.371. [DOI] [PubMed] [Google Scholar]
- Rutschmann S, et al. The Rel protein DIF mediates the antifungal but not the antibacterial host defense in Drosophila. Immunity. 2000;12:569–80. doi: 10.1016/s1074-7613(00)80208-3. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics (Oxford, England) 2004;20:3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Schlenke TA, et al. Contrasting infection strategies in generalist and specialist wasp parasitoids of Drosophila melanogaster. PLoS pathogens. 2007;3:1486–501. doi: 10.1371/journal.ppat.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz H, Franz M, Heberlein U. The hangover gene defines a stress pathway required for ethanol tolerance development. Nature. 2005;436:845–847. doi: 10.1038/nature03864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA. Low level of response to alcohol as a predictor of future alcoholism. The American journal of psychiatry. 1994;151:184–9. doi: 10.1176/ajp.151.2.184. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL. Onset and course of alcoholism over 25 years in middle class men. Drug and alcohol dependence. 2011;113:21–8. doi: 10.1016/j.drugalcdep.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Kalmijn J. The search for genes contributing to the low level of response to alcohol: patterns of findings across studies. Alcoholism, clinical and experimental research. 2004;28:1449–58. doi: 10.1097/01.alc.0000141637.01925.f6. [DOI] [PubMed] [Google Scholar]
- Small C, et al. An introduction to parasitic wasps of Drosophila and the antiparasite immune response. Journal of visualized experiments: JoVE. 2012:e3347. doi: 10.3791/3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov BP, et al. Transcription profiling reveals mitochondrial, ubiquitin and signaling systems abnormalities in postmortem brains from subjects with a history of alcohol abuse or dependence. Journal of neuroscience research. 2003;72:756–67. doi: 10.1002/jnr.10631. [DOI] [PubMed] [Google Scholar]
- Song X, et al. The evolution and origin of animal Toll-like receptor signaling pathway revealed by network-level molecular evolutionary analyses. PloS one. 2012;7:e51657. doi: 10.1371/journal.pone.0051657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino RP, Melk JP, Govind S. Genetic analysis of contributions of dorsal group and JAK-Stat92E pathway genes to larval hemocyte concentration and the egg encapsulation response in Drosophila. Genetics. 2004;166:1343–56. doi: 10.1534/genetics.166.3.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöven S, et al. Activation of the Drosophila NF-kappaB factor Relish by rapid endoproteolytic cleavage. EMBO reports. 2000;1:347–52. doi: 10.1093/embo-reports/kvd072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji T, et al. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Molecular and cellular biology. 2007;27:4578–88. doi: 10.1128/MCB.01814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji T, Yun EY, Ip YT. Heterodimers of NF-kappaB transcription factors DIF and Relish regulate antimicrobial peptide genes in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14715–20. doi: 10.1073/pnas.1009473107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauszig-Delamasure S, et al. Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nature Immunology. 2001;3:91–97. doi: 10.1038/ni747. [DOI] [PubMed] [Google Scholar]
- Torroja L, et al. Neuronal overexpression of APPL, the Drosophila homologue of the amyloid precursor protein (APP), disrupts axonal transport. Current Biology. 1999;9:489–493. doi: 10.1016/s0960-9822(99)80215-2. [DOI] [PubMed] [Google Scholar]
- Towb P, Bergmann A, Wasserman SA. The protein kinase Pelle mediates feedback regulation in the Drosophila Toll signaling pathway. Development. 2001;128:4729–4736. doi: 10.1242/dev.128.23.4729. [DOI] [PubMed] [Google Scholar]
- Turvey SE, Broide DH. Innate immunity. The Journal of allergy and clinical immunology. 2010;125:S24–32. doi: 10.1016/j.jaci.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetreno RP, Crews FT. Current hypotheses on the mechanisms of alcoholism. Handbook of clinical neurology. 2014;125:477–97. doi: 10.1016/B978-0-444-62619-6.00027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber ANR, et al. Binding of the Drosophila cytokine Spätzle to Toll is direct and establishes signaling. Nature immunology. 2003;4:794–800. doi: 10.1038/ni955. [DOI] [PubMed] [Google Scholar]
- Yagi Y, Ip YT. Helicase89B is a Mot1p/BTAF1 homologue that mediates an antimicrobial response in Drosophila. EMBO reports. 2005;6:1088–94. doi: 10.1038/sj.embor.7400542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, et al. Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6626–31. doi: 10.1073/pnas.1018514108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou JY, Crews FT. Release of Neuronal HMGB1 by Ethanol through Decreased HDAC Activity Activates Brain Neuroimmune Signaling. In: Block ML, editor. PLoS ONE. Vol. 9. 2014. p. e87915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Additional Controls. Manipulation of Toll expression does not affect ethanol absorption or metabolism (panels A & B). A) Overexpression of a constitutively active Toll10B isoform does not affect ethanol absorption or metabolism (2 way ANOVA treatment effect p=0.5499, n.s. difference at each point). B) Knockdown of Toll expression with a UAS-driven RNAi construct targeting Toll driven by Actin-GeneSwitch does not affect ethanol absorption or metabolism (2 way ANOVA treatment effect p=0.0520, n.s. difference at each point). Treatment with RU-486 inducer does not affect alcohol resistance but does induce a GFP reporter (panels C & D). C) Actin-GeneSwitch animals were crossed to the TRiP RNAi background stock, and tested for effects on alcohol resistance in the presence of RU-486 or carrier. No difference in resistance was seen (p=0.4289). D) Actin-GeneSwitch was crossed to our wild type Canton S strain once and tested for effects on alcohol resistance. RU-486 feeding caused no change (p=0.8477). Feeding adults RU-486 induces target gene expression. E) UAS-GFP; Actin-GeneSwitch animals were tested for induction of GFP after drug feeding. Fluorescence is notably brighter in dissected brains of inducer fed animals (RU-486, top) than in controls (Carrier, bottom).
Figure S2: Manipulation of Toll pathway activity does not block the ability to acquire 24h tolerance. In these tolerance tests, on the day prior to data collection half of the animals received a sedating dose of ethanol and the control group received no ethanol. 24h later both groups were sedated and the rate of recovery recorded. A) Overexpression of the constitutively active Toll10B allele using the ubiquitous Actin-GeneSwitch driver does not block the acquisition of tolerance, as can be seen in the faster recovery for the group receiving its second sedation (***, p<0.0001). All animals were fed RU-486 inducer for three days prior to the experiment. B) Dif1 homozygous loss-of-function animals are able to acquire tolerance, as can be seen in the faster recovery for the group receiving its second sedation. (**, p=0.0012).
Table S1: Findings from our RNA-seq study. List of genes significantly regulated 30 min after ethanol sedation and findings from gene ontology analysis performed using DAVID.
Table S2: Drosophila stocks used in this study. Details on the full genotype of stocks used, as well as the source stocks of alleles and transgenes.