ABSTRACT
Multiple layers of regulation are required to ensure appropriate patterns of gene expression for accurate cell differentiation. Interphase chromatin is non-randomly distributed within the nucleus, with highly compacted, transcriptionally silent heterochromatin enriched at the nuclear and nucleolar periphery. Whether this spatial organization serves a function in organismal physiology, rather than simply being a byproduct of chromatin metabolism, is a fundamental question. Recent work performed in C. elegans embryos characterized the molecular mechanisms that drive the perinuclear anchoring of heterochromatin. Moreover, for the first time it was shown that heterochromatin sequestration helps to restrict cell differentiation programs, while sustaining commitment to a specified fate. Here, we describe and comment on these findings, placing them in a broader context.
KEYWORDS: C. elegans, CEC-4, cell fate, H3K9 methylation, Heterochromatin, nuclear periphery
Heterochromatin and euchromatin
Almost as long light microscopy has existed, imaging has revealed 2 distinct “types” of chromatin in the nucleus. Indeed, already in 1928, Emil Heitz described a carmine acetic acid-stained heterochromatin that persisted in a condensed form throughout interphase and a euchromatic fraction that would decondense after mitosis to become invisible to this stain.1 Following these initial observations, many studies have elaborated on the key molecular features of these 2 chromatin types. Heterochromatin is generally transcriptionally inert and the DNA within it is packed at high density, whereas euchromatin contains transcriptionally active genes and appears less condensed.2
Nucleosomes, the fundamental units of chromatin, harbor histone tails that are subject to post-translational modifications, some of which correlate specifically with euchromatin or heterochromatin. For example, on the N-terminal tail of histone H3, methylation states of lysines 9 and 27 correlate with heterochromatin, while the methylation states of lysines 4 and 36 on the same histone tail, are enriched in euchromatin.3 Strikingly, not only are euchromatin and heterochromatin functionally distinct, but they also tend to occupy distinct zones within interphase nuclei, strongly suggesting that the 3D structure of the nucleus is intimately linked to its function. The question that has persisted for a very long time, however, is whether this spatial segregation of chromatin types contributes to the regulation of the genome during cell type specification.
Heterochromatin at the nuclear periphery
The spatial segregation of euchromatin and heterochromatin means that active domains generally shift away from the nucleolus and the nuclear periphery, where heterochromatin is enriched. Conversely, heterochromatic domains, which are rich in repetitive LINE elements, tend to associate with the nuclear lamina (and not nuclear pores), and are excluded from open internal zones.4,5,6 The separation of chromatin types is less prominent in pluripotent embryonic stem cells, yet as cells differentiate heterochromatin accumulates and is increasingly sequestered at the nuclear edge.7, 8 Large-scale reorganization, as well as specific single gene repositioning, is well-documented in a number of species.9-14 This can be monitored as a gain or loss of interaction with the nuclear lamina, given that the nuclear lamins are one of the few non-diffusing bona fide structural proteins of the nucleus.15,6 This topological organization raises 2 questions: what drives tissue-specific promoters toward the nuclear core as they are induced? And what triggers the anchoring of silent domains at the nuclear periphery? Recent data obtained in the model organism C. elegans provide compelling answers to the second question, and start to link nuclear architecture and cell fate stability.16 These findings are the topic of this Extraview.
Pathways of heterochromatin sequestration at the nuclear periphery
In a large and elegant study, Solovei et al.17 probed perinuclear proteins for their involvement in heterochromatin sequestration at the nuclear rim in mouse. The authors identified 2 integral components of the nuclear lamina, Lamin B Receptor (LBR) and lamin A/C, whose simultaneous loss leads to the accumulation of heterochromatin at the inner nuclear core. These factors act on 2 distinct pathways, which function largely in early (LBR) or late (lamin A/C) stages of differentiation.17 Although these 2 proteins are clearly implicated in chromatin distribution, it was unclear which marks on the chromatin were being recognized by the lamina as heterochromatic marks. Because selective chromatin recognition could not be proven in this system, it remained possible that the loss of LBR and lamin A/C perturbed chromatin distribution by displacing some other protein that specifically bridges to chromatin. Although LBR binds the heterochromatic factors HP1α and HP1γ in vitro,18 these 2 proteins were also found associated with genomic regions that were located far from the nuclear periphery.19 In addition, LBR was reported to recognize another histone mark, histone H4 bearing dimethylation on lysine 20 (H4K20me2)20 which unfortunately is broadly distributed in the genome with no particular enrichment in heterochromatin.21 Similar doubts persisted for lamin A/C, which can bind DNA and histone dimers in vitro,22 yet shows no specific affinity for repressive histone modifications. Indeed, given that ectopic expression of Lamin A/C did not restore heterochromatin sequestration in Lbr−/− Lmna−/− mice,17 Lamin A/C is unlikely anchor heterochromatin alone. Taken together, these data argued that unknown additional players were likely to be involved in the tethering of heterochromatin to the nuclear envelope.
Addressing this genetically, the Gasser laboratory took an untargeted approach to find factors that are necessary for the sequestration of heterochromatin at the nuclear lamina. They performed a genome-wide RNAi screen in C. elegans, scoring first for heterochromatin de-repression, and secondarily for its de-localization away from the nuclear periphery. Whereas many factors were implicated in repression, only the loss of a pair of closely related enzymes, the SAMS-3/4 S-adenosylmethionine (SAM) synthases, led to heterochromatin release from the nuclear lamina. Since SAM is the unique methyl-group donor, the authors hypothesized that histone methylation might be crucial for heterochromatin sequestration, and indeed, the ablation of 2 histone methyl transferases, MET-2 and SET-25, similarly allowed heterochromatin to shift inwards.23 Specifically, it was shown that the mono-, di- or tri-methylation of lysine 9 on histone H3, were key signals for the perinuclear anchoring of chromatin.
Anchoring H3K9 methylated chromatin at the nuclear envelope: CEC-4
How does H3K9 methylation promote perinuclear sequestration? The simplest hypothesis is that this signal can be specifically recognized by factor(s) that are at the same time capable of binding the nuclear envelope. In order to identify these, Gonzalez-Sandoval et al.16 performed a targeted RNAi screen in C. elegans embryos knocking down all validated and putative histone modification readers, one-by-one. Out of 65 Tudor-, chromo-, MBT (Malignant Brain Tumor)- and PHD (Plant HomeoDomain)-domain-containing proteins in worms, the loss of a single uncharacterized gene called cec-4, mimicked the effects of loss of H3K9 methylation: down-regulation of CEC-4 alone led to the displacement of a heterochromatic reporter from the nuclear periphery.
CEC-4 contains a chromodomain (CD) that binds H3K9me1, me2 and me3 almost exclusively, with an affinity similar to those reported for fly, human and mouse HP1.24 Its loss leads to a release of heterochromatin from the periphery in the nuclei of C. elegans embryos, perfectly fulfilling the initial experimental hypothesis. Furthermore, analysis of CEC-4 localization revealed that it is concentrated at the nuclear rim. A double point mutation that interferes with H3K9me-recognition, but which does not alter its subnuclear distribution, was sufficient to relocate a heterochromatic reporter. This showed that CEC-4 is the molecule that links heterochromatin to the nuclear envelope in worm embryos. However, the story did not end there.
Over the last years, one topic of major debate in the field of nuclear organization has been whether subnuclear localization regulates gene expression, or not. Although the nuclear periphery is generally a repressive environment that can favor the silencing of some promoters,25,26 it is also clear that the nuclear envelope is permissive to transcription and that gene de-repression per se is not sufficient to cause internalization.15,23 Similarly, and astoundingly, although loss of CEC-4 allowed heterochromatin to shift inwards, it did not increase transcription levels. In other words, CEC-4 is a highly specialized H3K9me-reader whose function is to anchor, but not to repress, heterochromatin. This enabled a clean genetic separation of transcriptional repression from peripheral sequestration, even though both phenomena are mediated by the H3K9me mark. Other H3K9me readers, namely worm HP1 homologues and LIN-61, a MBT domain protein with selective affinity for H3K9me2/me3, led to silencing of a heterochromatic reporter, but their loss did not affect its spatial segregation. Thus different “readers” bind the same posttranslational modification on histone H3 to achieve different phenotypes: peripheral sequestration or transcriptional silencing.
CEC-4 helps suppress other differentiation pathways during cell-fate induction
It is well-established that heterochromatin sequestration at the nuclear envelope increases with cell differentiation.27 Therefore, it was possible that any function served by the anchoring of heterochromatin, might only be manifest during the process of tissue differentiation. The first indications suggested this was unlikely: cec-4 mutant embryos differentiated normally under standard laboratory conditions. However, at the L1 larval stage other pathways of heterochromatin anchoring, which work in the absence of CEC-4, were induced. To circumvent this problem of compensation and to be able to test for a regulatory function of perturbed chromatin segregation, the authors induced the muscle differentiation program in all cells of mid-stage embryos by overexpressing the muscle master regulator MyoD (HLH-1 in worms). Whereas wild-type embryos are plastic enough to commit fully to this cell fate upon HLH-1 induction, cec-4 mutants failed to do so. In contrast, larvae-like individuals expressing non-muscle markers, such as an intestine gene, emerged after MyoD induction (Fig. 1). This suggests that the perinuclear sequestration of heterochromatin by CEC-4 may indeed help stabilize a specified cell fate.16
Figure 1.
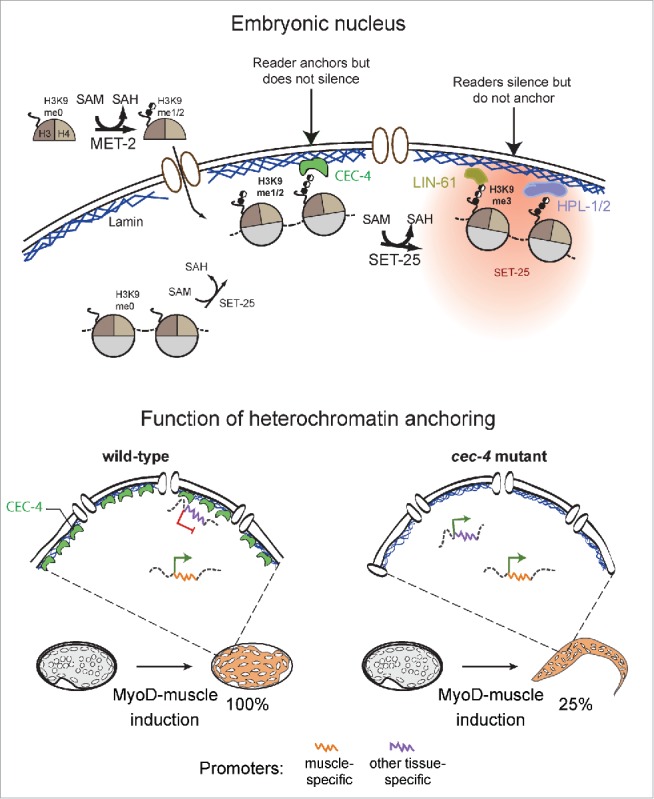
Model of heterochromatin anchoring function. CEC-4 autonomously localizes to the nuclear envelope to read all forms of H3K9 methylated histones. In worms, MET-2 and SET-25 are the enzymes responsible for the deposition of H3K9me1, me2 and me3 (upper half). CEC-4 is a H3K9me reader that anchors but does not silence, while the HP1 homologues, HPL-1 and HPL-2, and the MBT domain protein LIN-61, silence but do not anchor. Cells of wild-type and cec-4 mutant embryos were forced into the muscle differentiation program by inducing the muscle master regulator MyoD (HLH-1 in worms). While 100% of wild-type embryos commit to a muscle fate, 25% of embryos lacking CEC-4 resist full commitment and manage to hatch into larvae-like structures that express markers of other tissues (modified from16). These data represent the first evidence for functionality of a factor solely dedicated to chromatin anchoring in a multicellular organism, and argue that the perinuclear sequestration of heterochromatin promotes the stabilization of a specified cell fate.
Heterochromatin segregation in other species and redundant pathways
In mammals it is very likely that H3K9 methylation (particularly H3K9me2) is also a signal for peripheral localization, as 80% of mammalian Lamina Associated Domains (LADs) are enriched for H3K9me2/me3.6,28 Moreover, reducing the levels of H3K9me2 by knocking down or inhibiting the relevant histone methyl transferase (G9a) led to a decreased interaction with the nuclear lamina.29-31 Although there seems to be no direct homolog of CEC-4 in non-nematode species, it is highly likely that a similar mechanism of heterochromatin recognition and anchoring will be found in mammals, possibly mediated by more than one factor or more than one mark. It is also likely that other pathways of anchoring exist both in worms and vertebrates. Indeed, the complete loss of H3K9 methylation does not affect heterochromatin sequestration at the nuclear envelope in the differentiated tissues of worm larvae.23
Recently the repressive histone mark deposited by and read by Polycomb proteins, H3K27me3, has been shown to contribute to the anchoring of sequences at the borders of LADs in mouse cells.31 Whereas the loss of H3K27me3 did not affect the peripheral localization of a heterochromatic reporter in C. elegans embryos23, nor in larvae (DSC, unpublished data), there may still be specific sequences in worms, equivalent to the edges of LADs in mammals, that depend on Polycomb mediated modifications for perinuclear sequestration.
In addition, an alternative, sequence-specific mechanism of binding to the nuclear envelope was documented in a study on the IgH locus in mouse.32 This pathway involves DNA recognition by cKrox, which binds to GAGA motifs, and the interaction of cKrox with the histone deacetylase HDAC3 and the nuclear lamina protein Lap2β.32 HDAC3 appears to be a potentially interesting factor in perinuclear localization as it also binds the nuclear envelope protein Emerin.33 Even though it is very unlikely that a transcription factor-dependent mechanism can account for widespread anchoring of all heterochromatic domains, a role for histone deacetylation is an attractive option, given that heterochromatin is enriched in hypoacetylated histones and histone deacetylases are involved in heterochromatin formation.34
Moving toward the study of tissue-specific heterochromatin segregation
CEC-4 is the first CD-containing protein documented to localize autonomously to the nuclear envelope. The characterization of its function demonstrates that within a multicellular organism the spatial segregation of chromatin types is an active process. Intriguingly, as embryos develop into larvae the expression of CEC-4 becomes regulated in a tissue-specific fashion. In particular, CEC-4 levels are very high in muscle and very low in intestine. Since it seems unlikely that differentiation changes the specificity of CEC-4 for H3K9 methylated chromatin, one must ask why it is subject to differential regulation?
One possibility is that different tissues require different degrees of nuclear architectural regulation to achieve optimal functionality, with muscle needing higher levels of heterochromatin anchoring. With this in mind, it is interesting to note that muscle is one of the key tissues affected by laminopathies, which are genetic diseases caused by mutations in nuclear lamina components. Examples of this are Emery-Dreifuss muscular dystrophy,35 where mutations in LMNA lead specifically to muscle disease, despite the fact that the gene is widely expressed. We propose that this tissue-specific aspect of lamin (dys)function may extend to CEC-4 as well, although this remains to be shown.
A second possibility is that different tissues induce specific pathways of heterochromatin segregation as they differentiate, and that CEC-4 remains more functional in muscle than in other tissues such as intestine, despite the presence of other anchors that anchor heterochromatin in its absence. Indeed, the absence of CEC-4 or H3K9 methylation does not impair perinuclear anchoring of a heterochromatic reporter in intestine or hypoderm cells of L1 larvae.16,23 Interestingly, work performed in mammals identified tissue-specific nuclear envelope transmembrane proteins which are involved in chromosome peripheral positioning,36 further supporting the notion that additional anchors contribute to cell-type specific nuclear architecture. It is now crucial to identify the pathways of anchoring in differentiated tissues and to determine their common features. Do they act broadly or only in one tissue and to what extent are they redundant with anchors that recognize H3K9 methylation?
High throughput tissue-specific approaches to address questions of nuclear organization represent a challenge that will require the use of model organisms. The advantages of working with C. elegans include a lower degree of redundancy in chromatin modifiers and histone mark readers than in mammals. Yet the general principles of chromatin segregation appear to be conserved across species. Thus, the future looks bright for forthcoming chromatin studies in worm development.
Abbreviations
- CD
chromodomain
- EDMD
Emery-Dreifuss muscular dystrophy
- HP1
heterochromatin protein 1
- LAD
lamina associated domain
- LBR
lamin B receptor
- LINE
long interspersed nuclear element
- MBT
malignant brain tumor
- PHD
plant homeodomain
- SAM
S-adenosylmethionine
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Adriana Gonzalez-Sandoval and Jennifer C. Harr for comments on the manuscript.
Funding
D.S.C. was supported by Marie Curie Intra-European and EMBO Fellowships. S.M.G thanks the Novartis Research Foundation and Swiss National Science Foundation for support.
References
- [1].Heitz E. Das heterochromatin der moose. Jahrb Wiss Botanik 1928:762-818 [Google Scholar]
- [2].Frenster JH, Allfrey VG, Mirsky AE. Repressed and Active Chromatin Isolated from Interphase Lymphocytes. Proc Natl Academy of Sci USA 1963; 50:1026-32; http://dx.doi.org/ 10.1073/pnas.50.6.1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res 2011; 21:381-95; PMID:21321607; http://dx.doi.org/ 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al.. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008; 453:948-51; PMID:18463634; http://dx.doi.org/ 10.1038/nature06947 [DOI] [PubMed] [Google Scholar]
- [5].Ikegami K, Egelhofer TA, Strome S, Lieb JD. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol 2010; 11:R120; PMID:21176223; http://dx.doi.org/ 10.1186/gb-2010-11-12-r120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al.. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 2010; 38:603-13; PMID:20513434; http://dx.doi.org/ 10.1016/j.molcel.2010.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rozman C, Woessner S, Feliu E, Lafuente R, Berga L. Cell Ultrastructure for Hematologists (ed EdicionesDoyma S A; Barcelona, Spain: ) 1993 [Google Scholar]
- [8].Ugarte F, Sousae R, Cinquin B, Martin EW, Krietsch J, Sanchez G, Inman M, Tsang H, Warr M, Passegue E, et al.. Progressive Chromatin Condensation and H3K9 Methylation Regulate the Differentiation of Embryonic and Hematopoietic Stem Cells. Stem Cell Reports 2015; 5:728-40; PMID:26489895; http://dx.doi.org/ 10.1016/j.stemcr.2015.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 2002; 296:158-62; PMID:11935030; http://dx.doi.org/ 10.1126/science.1068768 [DOI] [PubMed] [Google Scholar]
- [10].Yang Q, Riblet R, Schildkraut CL. Sites that direct nuclear compartmentalization are near the 5′ end of the mouse IG heavy-chain locus. Mol Cell Biol 2005; 25:6021-30; PMID:15988016; http://dx.doi.org/ 10.1128/MCB.25.14.6021-6030.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Williams RR, Azuara V, Perry P, Sauer S, Dvorkina M, Jorgensen H, Roix J, McQueen P, Misteli T, Merkenschlager M, et al.. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J cell sci 2006; 119:132-40; PMID:16371653; http://dx.doi.org/ 10.1242/jcs.02727 [DOI] [PubMed] [Google Scholar]
- [12].Szczerbal I, Foster HA, Bridger JM. The spatial repositioning of adipogenesis genes is correlated with their expression status in a porcine mesenchymal stem cell adipogenesis model system. Chromosoma 2009; 118:647-63; PMID:19585140; http://dx.doi.org/ 10.1007/s00412-009-0225-5 [DOI] [PubMed] [Google Scholar]
- [13].Yao J, Fetter RD, Hu P, Betzig E, Tjian R. Subnuclear segregation of genes and core promoter factors in myogenesis. Genes & Dev 2011; 25:569-80; PMID:21357673; http://dx.doi.org/ 10.1101/gad.2021411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, Chandra V, Bossen C, Glass CK, Murre C. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol 2012; 13:1196-204; PMID:23064439; http://dx.doi.org/ 10.1038/ni.2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Meister P, Towbin BD, Pike BL, Ponti A, Gasser SM. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes & Dev 2010; 24:766-82; PMID:20395364; http://dx.doi.org/ 10.1101/gad.559610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gonzalez-Sandoval A, Towbin BD, Kalck V, Cabianca DS, Gaidatzis D, Hauer MH, Geng L, Wang L, Yang T, Wang X, et al.. Perinuclear Anchoring of H3K9-Methylated Chromatin Stabilizes Induced Cell Fate in C. elegans Embryos. Cell 2015; 163:1333-47; PMID:26607792; http://dx.doi.org/ 10.1016/j.cell.2015.10.066 [DOI] [PubMed] [Google Scholar]
- [17].Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al.. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 2013; 152:584-98; PMID:23374351; http://dx.doi.org/ 10.1016/j.cell.2013.01.009 [DOI] [PubMed] [Google Scholar]
- [18].Ye Q, Worman HJ. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J biol chem 1996; 271:14653-6; PMID:8663349; http://dx.doi.org/ 10.1074/jbc.271.25.14653 [DOI] [PubMed] [Google Scholar]
- [19].Minc E, Allory Y, Worman HJ, Courvalin JC, Buendia B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma 1999; 108:220-34; PMID:10460410; http://dx.doi.org/ 10.1007/s004120050372 [DOI] [PubMed] [Google Scholar]
- [20].Hirano Y, Hizume K, Kimura H, Takeyasu K, Haraguchi T, Hiraoka Y. Lamin B receptor recognizes specific modifications of histone H4 in heterochromatin formation. J biol chem 2012; 287:42654-63; PMID:23100253; http://dx.doi.org/ 10.1074/jbc.M112.397950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes & Dev 2004; 18:1251-62; PMID:15145825; http://dx.doi.org/ 10.1101/gad.300704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wilson KL, Foisner R. Lamin-binding Proteins. Cold Spring Harb Perspect Biol 2010; 2:a000554; PMID:20452940; http://dx.doi.org/ 10.1101/cshperspect.a000554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Towbin BD, Gonzalez-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 2012; 150:934-47; PMID:22939621; http://dx.doi.org/ 10.1016/j.cell.2012.06.051 [DOI] [PubMed] [Google Scholar]
- [24].Steffen PA, Fonseca JP, Ringrose L. Epigenetics meets mathematics: towards a quantitative understanding of chromatin biology. BioEssays 2012; 34:901-13; PMID:22911103; http://dx.doi.org/ 10.1002/bies.201200076 [DOI] [PubMed] [Google Scholar]
- [25].Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 2008; 452:243-7; PMID:18272965; http://dx.doi.org/ 10.1038/nature06727 [DOI] [PubMed] [Google Scholar]
- [26].Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet 2008; 4:e1000039; PMID:18369458; http://dx.doi.org/ 10.1371/journal.pgen.1000039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Talamas JA, Capelson M. Nuclear envelope and genome interactions in cell fate. Front Genet 2015; 6:95; PMID:25852741; http://dx.doi.org/ 10.3389/fgene.2015.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet 2009; 41:246-50; PMID:19151716; http://dx.doi.org/ 10.1038/ng.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. Single-cell dynamics of genome-nuclear lamina interactions. Cell 2013; 153:178-92; PMID:23523135; http://dx.doi.org/ 10.1016/j.cell.2013.02.028 [DOI] [PubMed] [Google Scholar]
- [30].Bian Q, Khanna N, Alvikas J, Belmont AS. beta-Globin cis-elements determine differential nuclear targeting through epigenetic modifications. J cell biol 2013; 203:767-83; PMID:24297746; http://dx.doi.org/ 10.1083/jcb.201305027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Harr JC, Luperchio TR, Wong X, Cohen E, Wheelan SJ, Reddy KL. Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J cell biol 2015; 208:33-52; PMID:25559185; http://dx.doi.org/ 10.1083/jcb.201405110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zullo JM, Demarco IA, Pique-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, et al.. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell 2012; 149:1474-87; PMID:22726435; http://dx.doi.org/ 10.1016/j.cell.2012.04.035 [DOI] [PubMed] [Google Scholar]
- [33].Demmerle J, Koch AJ, Holaska JM. The nuclear envelope protein emerin binds directly to histone deacetylase 3 and activates HDAC3 activity. J biol chem 2012; 287:22080-8; PMID:22570481; http://dx.doi.org/ 10.1074/jbc.M111.325308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Murakami Y. Histone deacetylases govern heterochromatin in every phase. EMBO J 2013; 32:2301-3; PMID:23832177; http://dx.doi.org/ 10.1038/emboj.2013.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Helbling-Leclerc A, Bonne G, Schwartz K. Emery-Dreifuss muscular dystrophy. European Jl human genetics 2002; 10:157-61; http://dx.doi.org/ 10.1038/sj.ejhg.5200744 [DOI] [PubMed] [Google Scholar]
- [36].Zuleger N, Boyle S, Kelly DA, de las Heras JI, Lazou V, Korfali N, Batrakou DG, Randles KN, Morris GE, Harrison DJ, et al.. Specific nuclear envelope transmembrane proteins can promote the location of chromosomes to and from the nuclear periphery. Genome Biol 2013; 14:R14; PMID:23414781; http://dx.doi.org/ 10.1186/gb-2013-14-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]


