ABSTRACT
A concern in the field of genomics is the proper interpretation of large, high-throughput sequencing datasets. The use of DNA FISH followed by high-content microscopy is a valuable tool for validation and contextualization of frequently occurring gene pairing events at the single-cell level identified by deep sequencing. However, these techniques possess certain limitations. Firstly, they do not permit the study of colocalization of many gene loci simultaneously. Secondly, the direct assessment of the relative position of many clustered gene loci within their respective chromosome territories is impossible. Thus, methods are required to advance the study of higher-order nuclear and cellular organization. Here, we describe a multiplexed DNA FISH technique combined with indirect immunofluorescence to study the relative position of 6 distinct genomic or cellular structures. This can be achieved in a single hybridization step using spectral imaging during image acquisition and linear unmixing. Here, we detail the use of this method to quantify gene pairing between highly expressed spliceosomal genes and compare these data to randomly positioned in silico simulated gene clusters. This is a potentially universally applicable approach for the validation of 3C-based technologies, deep imaging of spatial organization within the nucleus and global cellular organization.
KEYWORDS: chromosome territories, Cajal bodies, DNA FISH, genome organization, gene positioning, nuclear bodies, spliceosomal U snRNA genes, spectral imaging
Introduction
The cell nucleus is a highly ordered but non-homogeneous stochastic organelle. Within its boundaries reside extensive genetic information, transcribed RNA and dynamic protein complexes that function in unison to coordinate essential nuclear processes, such as DNA replication, DNA repair, gene transcription, RNA processing, and mRNA transport. These complexes are the basis for a number of interwoven but structurally defined sub-nuclear compartments. These include chromosomes, which are contained within distinct chromosome territories (CTs),1 as well as protein- and RNA-based structures that lack defining membranes, known as nuclear bodies (NBs).2 Examples of well-studied NBs include the nucleolus, Cajal bodies (CBs),3 histone locus bodies,4 PML nuclear bodies,5 nuclear speckles,6 paraspeckles,7 transcription factories,8 the perinucleolar compartment,9 nuclear stress bodies,10 and numerous other “orphan” NBs.11 These structures occupy a substantial volume of the cell nucleus and require further functional characterization. Interest in the cooperation between NB function and genome organization began in the 1990s. Early efforts at characterizing various NB-genome interfaces, including identification of spliceosome U snRNA genes as regions that frequently associate with the CB,4,12-14 hinted at the possibility of wider genomic organization by NBs. However, these early studies, although vital for the field, were limited by the single number of genes that could be visualized simultaneously and the lack of suitable genome-wide mapping tools.
Fortunately, tremendous effort has been directed into contextualizing the data and technologies that originated from the Human Genome Project. This includes the characterization of epigenetic marks but also the higher-order organization of the genome in the 3D space within the cell nucleus. Indeed, since the development of chromosome conformation capture (3C),15 a wave of sequencing-based reports that indicate the 3D organization of the human genome have been published.16 This has culminated in the identification of highly ordered linear units of chromatin by Hi-C,17 called Topologically-Associated Domains (TADs).18 Improvements in mass data storage and analytical technologies will improve our understanding of this biologically essential topological phenomenon. Intriguingly, several sequencing-based studies have recently indicated that NBs assemble as a direct consequence of specific gene expression activity and form characteristic long-lived contact interactions with a defined cohort of genomic loci and CTs.19-21
Nonetheless, the limitations of these population-based studies using millions of mostly asynchronous cells are becoming apparent. Despite indications that genomic domains are known to form and maintain numerous spatial associations with both near (kBp) and distant (MBp) genomic locations,22 it is unknown how many physical gene pairing events occur simultaneously. Furthermore, due to data acquisition sensitivity and bioinformatic normalization, Hi-C gene pairing events are limited to ∼3.5 MBp linear distances23 and many low frequency but biologically relevant contact events may be excluded from genome-wide contact maps.
The genomic visualization technique DNA Fluorescent In Situ Hybridization (FISH) is often used to validate these sequencing-based data sets and has undergone substantial developments to expand the number of distinguishable visualization targets in a single FISH experiment. For example, 3 methods have been implemented using multicolor probes to karyotype the chromosomes present in a single cell. The first, multiplex FISH (M-FISH) uses fluorophore-specific narrow band pass optical filters for imaging of each fluorescent dye.24,25 Data are analyzed by M-FISH software which classifies each chromosome based on the combination of colocalized fluorescent signals. The second method is spectral karyotyping (SKY) which uses interferometry. In this approach, chromosome-specific libraries are labeled with 5 different fluorophores or different combinations of these flourophores. During imaging, the emitted light is passed through an interferometer which creates a fluorophore-specific difference in optical path with unique spectral characteristics. Spectral combinatorial karyotyping of 24 human chromosomes is classified by the software which analyzes spectral emission data.26 The third, recently published, method uses spectral imaging and linear unmixing with 4 distinct fluorophores and combinatorial color code to distinguish all human and mouse chromosomes.27 All these karyotyping methods depend on highly reproducible labeling which might be a problem in highly variable large probe sets. However, karyotyping DNA FISH also can be applied to visualize CTs in the interphase nucleus. Additionally, combinations of fluorophores have allowed the co-visualization of up to 11 mRNA transcripts.28 However, this approach still only used 4 distinct fluorophores and required pseudocoloring of colocalized combinatorial fluorescent foci. Imaging of more fluorophores, as described here, combined with pseudocoloring could allow even greater numbers of genes to be co-visualized.
Typically, however, the DNA FISH approach for detecting gene loci in the interphase nucleus enables the simultaneous imaging and assessment of up to 4 gene loci (3 if you include the visualization of the entire nucleus by DAPI staining in your protocol). Great strides have been made in the realm of high-content DNA FISH microscopy approaches (hiFISH), which utilize automated image acquisition and analysis tools to interrogate DNA probe libraries.21,29,30 Potentially, this approach can accurately quantify gene pairing events that occur across larger linear genomic distances and between chromosomes at high-resolution but current high-content microscopy experiments are still limited to only 4 distinct fluorophores for visualization (one of which must be dedicated to a nuclear stain for automated image analysis). Therefore, current methods are inadequate for the co-visualization of multiple genes in conjunction with co-detection of chromosome territories, sub-nuclear structures and/or distinct epigenetic marks.
To overcome the problems associated with characterizing the genomic context in which NBs, and other nuclear structures and compartments, reside, we have developed a novel multi-color DNA FISH/immunofluorescence (DNA FISH/IF) visualization approach. We have implemented this method, which we have named spectraFISH, for the study of 5 distinct genes or CTs with the CB, a major NB involved in spliceosomal snRNP biogenesis31-33 and found frequently in cancer cells.21 This nuclear structure actively coordinates34 a network of gene pairing interactions with major and minor spliceosome U snRNA genes,12-14,35 and other highly active intron-encoded U snoRNA/scaRNA genes,21,36 but also histone gene clusters (e.g. HIST1) via the physically-associated histone locus body.37,38
Additionally, there is a diverse range of available sequencing and high-content imaging data which are also suitable for higher-order microscopic validation using spectraFISH (summarized in Table 1). In the data presented below, we have visualized the genomic context where CBs are localized and the frequency of association between these CB-dependent genes are clustered. We achieved this by selecting genes of interest from a circular chromosome conformation capture (4C)-seq dataset21 (using the RNU1 gene as a bait region) and co-visualized with up to 5 of these genes (or 4 CTs) with the CB marker protein, coilin. Alternatively, spectraFISH could also be modified to assess colocalization of DNA FISH foci with NBs or other nuclear structures.
Table 1.
Potential validation strategies for genomic data sets using spectraFISH.
| Genomic Dataset | Probe Selection Strategy | Potential Target Validation | Refs. |
|---|---|---|---|
| Hi-C | High interaction frequency with shared gene loci | 3D topology of TADs or whole chromosomes and relation to nuclear structures | 18,39 |
| 4C-seq | High interaction frequency | Frequency of long-range gene interaction between >2 genes/structures | |
| Loss of interaction upon NB disassembly | 15,21 | ||
| hiFISH | Known direct interaction with NBs or genes | Frequency of long-range gene interaction between >2 genes/structures | 21,29,40 |
| NB-centric deep sequencing assays (e.g. Immuno-TRAP) | Specific co-localization with structure | Higher-order gene interaction with target nuclear structure | 19,20 |
| ChIP-seq / DamID | Shared NB/compartment marker protein or transcription factor binding sites | Gene position within CTs or relative to NBs/transcription factories | 33,41 |
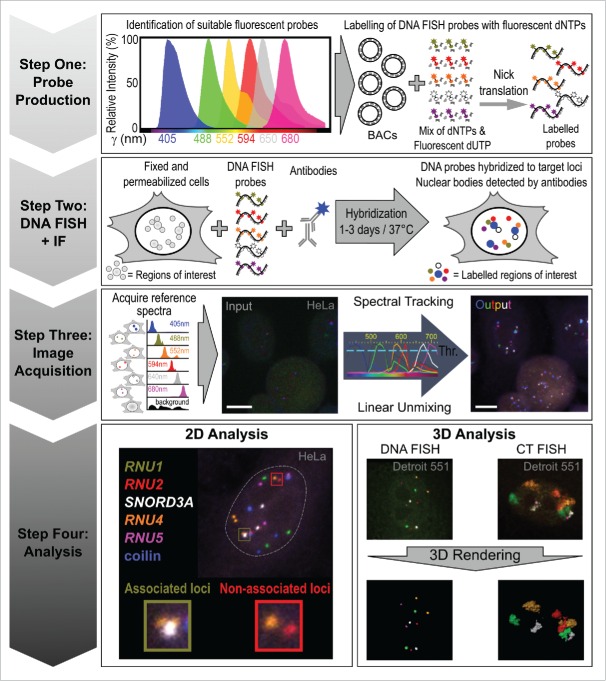
SpectraFISH is built upon ongoing progress in direct fluorescent labeling of DNA probes and microscopic multicolor innovations that allow the generation and imaging of biological samples labeled with increasing numbers of distinct fluorophores. Here, we fully describe this technique which allows researchers to assess, validate and challenge phenomena from large genomic data sets which are typically difficult to co-visualize on a single-cell and single-allele level. We detail how to select suitable fluorescent dyes for DNA probe production, perform in situ hybridization and acquire multichannel images using spectral imaging and linear unmixing (summarized in Fig. 1). We also provide an example of the technique by examining the clustering of spliceosomal U snRNA genes, including a comparison to random theoretical gene associations, and include some suggestions for additional analyses.
Figure 1.
Overview of the SpectraFISH method.
Protocol
Materials
Absolute Ethanol (Molecular biology grade)
Bacterial artificial chromosomes (BACs), oligopaint probes, chromosome paints or other DNA probes for visualization of genomic loci [See: Troubleshooting]
Coilin antibody (Rabbit) (Cat no.sc-32860, Santa Cruz Biotechnology) [Note: For detection of CBs]
Cultured Cells, such as human adherent HeLa cervical carcinoma or Detroit 551 skin fibroblast cells
Dextran Sulfate 50% solution (Cat no. S4030, EMD Millipore)
Dry ice
DyLight 405 nm Donkey anti-rabbit IgG (Cat no. 711-475-152, Jackson Immunoresearch)
Fluorescently labeled nucleotides
Green: 488 nm dUTP (Cat no. 02N32-050, Abbott Laboratories)
Orange: 552 nm Amersham CyDye Cy3 dUTP (Cat no. PA53022, GE Healthcare)
Deep-red: 594 nm CF594-dUTP (Cat no. 40006, Biotium)
Far-red: 640 nm Amersham CyDye Cy5 dUTP (Cat no. PA55022, GE Healthcare)
Near-infrared: 680 nm CF680R-dUTP (Cat no. 40003, Biotium)
Formamide (deionized) (Cat no. F9037, Sigma)
Human COT DNA (Cat no. 11581074001, Roche)
Hydrochloric acid
Nick Translation kit from commercial sources (e.g., Roche, Sigma, GE Healthcare, ThermoFisher etc.) or non-commercially-sourced protocols (e.g. Meaburn (2010)42)
Nuclease-free water (Cat no. AM9932, Ambion)
Paraformaldehyde (PFA, 4% final concentration) (Cat no. 15710, Electron Microscopy Sciences)
Phosphate Buffered Saline (PBS)
ProLong antifade mounting reagent without DAPI (Cat no. P36930, ThermoFisher)
Rubber Cement
Sodium Acetate (3M pH 5.2) (Cat no. 351-035-721), Quality Biological)
SSC (Saline Sodium Citrate) (Cat no. BF-170-1000, Baltimore Bioworks)
Triton X-100 (Cat no. T8787, Sigma)
Tween-20 (Cat no. P2287, Sigma)
Yeast tRNA (10 mg/ml) (Cat no. AM7119, Sigma)
Equipment
Boekel Scientific Slide Moat Incubator (ThermoFisher)
Centrifuge (Capable of cooling to +4°C)
Coplin jars
Cover slips (0.13 to 0.17 mm thick)
Microscopic slides (1 mm thick)
Water bath
Zeiss laser-scanning confocal microscopes LSM710 and LSM780 (Carl Zeiss Microimaging, Jena, Germany) equipped with 405 nm Diode laser, Argon/2 (458, 488, 514 nm), 561 nm Diode laser, 594 nm Diode laser, 633 nm HeNe laser (or equivalent)
Zeiss Zen Black 2012 edition Imaging Software (or equivalent)
Instructions
1. Production of fluorescently labeled DNA probes
1.1. Fluorescent probe selection
Fluorescent probes should be selected that possess distinguishable emission spectra and minimal spectral overlap. However, these emission profiles can often share the same spectral region. Researchers should preview these spectra and assess their suitability using a number of web-based tools, including the ThermoFisher SpectraViewer (https://www.thermofisher.com/us/en/home/life-science/cell-analysis/labeling-chemistry/fluorescence-spectraviewer.html), Chroma Spectra Viewer (https://www.chroma.com/spectra-viewer) and the Biolegend Fluorescence Spectra Analyzer (http://www.biolegend.com/spectraanalyzer). The distinct, desirable and relatively narrow theoretical emission spectra for the 6 fluorescent markers used in developing this technique are shown in Figure 1 and listed in Materials. Users should examine the amplitude and crosstalk between fluorophores of interest using these online tools by viewing the theoretical emission spectra produced after selecting the laser wavelengths that most closely replicate those integrated into their confocal microscope system. Other alternative fluorophore combinations with similar emission spectra characteristics that can be used are: Alexa Fluor 405, 488, 555, 594, 635 and 680 (ThermoFisher), or highly photostable dyes from ATTO-TEC (Jena Bioscience GmbH): ATTO-425, ATTO-488, ATTO-550, ATTO-594, ATTO-635 and ATTO-680. Other combinations of dyes from several sources can be used: Pacific Blue, FITC, Cy3, Texas Red, Alexa Fluor 635 and Alexa Fluor 680. Users must ensure that the fluorophores selected can a) be excited using the laser lines available on their confocal microscope, and, b) the confocal microscope to be used for imaging must have an adequate number and range of detectors to acquire an image with 6 channels. Additionally, users must ensure that the imaging software provided by the microscope manufacturer is capable of performing spectral imaging (the freely-available, open-source software ImageJ (NIH, Bethesda, MD) can also perform linear unmixing).
1.2 In accordance with previous protocols,42 perform nick translation of BACs that target distinct genomic loci using the fluorescently tagged dUTPs identified in Sections 1.1 and 1.2. [Note: From this point onwards the exposure of the probes to light, whether free or hybridized to cellular DNA, should be limited]
1.3 After the nick translation reaction has stopped, use probes immediately (while maintaining the labeled probes on ice and in the dark) or store in the dark at −20°C.
2. DNA and/or CT FISH
Note: Perform all wash steps in Coplin jars or other appropriate vessels. All wash steps in this protocol should be performed at room temperature unless stated otherwise.
-
2.1
Wash cells grown on 12 mm glass circle coverslips with PBS and fix with 4% PFA in PBS for 10 min.
-
2.2
After washing with PBS, permeabilize cells using 0.5% Triton X-100 in PBS for 20 min on ice and wash with PBS. [Note: performing this step on ice improves the visualization of CBs by limiting disassembly of the structure.]
-
2.3
Incubate cells in 0.1N HCl for 15 min and wash 2 times in 2xSSC for 10 min.
-
2.4
Equilibrate cells in 50% formamide / 2xSSC for at least 30 min.
-
2.5
For one 12 mm round coverslip, combine 150 ng of DNA or CT FISH probe for each fluorescent dye and precipitate with 3 µg Cot-1 DNA and 1 µg yeast tRNA in 2 volumes of ice-cold absolute ethanol. Include 1/10 volume of 3 M sodium acetate and incubate on dry ice or in deep freezer (−80°C) for at least 15 min.
-
2.6
Centrifuge precipitation mixture for 30 min at ≥12,000 rpg at 4°C and then use a lyophilizer to remove ethanol from the pellet of probe mix.
-
2.7
Resuspend the semidry DNA pellet in 7 µl of hybridization solution (10% dextran sulfate / 50% formamide pH 7.0 / 2xSSC / 1% Tween-20). Incubate at 37°C to aid DNA pellet resuspension.
-
2.8
Denature the cocktail at 85°C for 5 min and place briefly on ice.
-
2.9
Add probe cocktail to cells, seal with rubber cement and incubate at 85°C for 5 min using a Slide Moat incubator.
-
2.10
Hybridize at 37°C in a humidified chamber containing absorbent paper soaked in 2xSSC overnight (DNA FISH) or for 72 h (CT FISH)
-
2.11
After hybridization, prepare Coplin jars containing 50% formamide/2xSSC at 45°C (Buffer A), 1xSSC at 60°C (Buffer B) and PBS (Buffer C).
-
2.12
Wash cells 3 times in Buffer A for 5 min, followed by 3 times in Buffer B for 5 min and finally 3 times in Buffer C for 5 min.
3. Immunofluorescent detection of nuclear bodies
-
3.1
Incubate cells with rabbit polyclonal antibody against coilin (or other NB-specific marker protein) for 1 h.
-
3.2
Wash cells 3 times with PBS.
-
3.3
Incubate cells with donkey anti-rabbit secondary antibody conjugated to CF 405 S dye (or another species-appropriate secondary antibody conjugated to a fluorophore that absorbs at 405 nm) for 1 h.
-
3.4
Wash cells 3 times with PBS.
-
3.5
Mount cells with ProLong antifade mounting reagent without DAPI (ThermoFisher) [Note: The exclusion of DAPI from the mounting medium at this step is crucial. Otherwise, visualization of NBs is completely abolished.]
4. Generation of spectraFISH images and analysis
The imaging technique known as spectral imaging, coupled with subsequent image processing using linear unmixing, is used to separate mixed fluorescent emission signals in samples and clearly resolve the spatial contribution of each fluorophore (often referred to as Emission Fingerprinting) to each pixel of the image.43
4.1. Acquisition of reference spectra
Each pixel in the raw image must be assigned to the appropriate channel depending on its fluorescent composition. To achieve this, a reference spectrum is generated for each fluorophore. Cells, maintained under the same experimental conditions, are labeled with each fluorophore individually and imaged using identical microscopy settings. These are then used for efficient separation of mixed/overlapping emission profiles in the final spectraFISH image (which contains all 6 fluorophores) in Section 4.2. Each individually-labeled specimen is visualized by spectral imaging with illumination of all excitation wavelengths simultaneously using the same objective and consistent detector settings. Next, using appropriate microscopy software (e.g., Zeiss Zen Black) a region of interest (ROI) for a specific DNA FISH signal is drawn. The average intensity over all pixels in the ROI generates a reference spectrum that is recorded in a spectral database. In addition, the ROI of the background outside of cells is recorded to a database for background correction. Recorded individual reference spectra signatures may be repeatedly reused in various samples under same experimental and visualization settings.
4.2. Spectral imaging and unmixing into separate channels
Using the laser scanning confocal microscopes Zeiss LSM710 and 780 coupled with a spectral imaging 34-channel QUASAR detection unit, the entire spectrum of fluorophores used in a sample is illuminated and recorded (63× 1.40 NA objective using z-sectioning (z = 250−300 nm)). However, in theory, this approach can be performed using any laser scanning confocal microscope capable of simultaneous spectral imaging. Z-stacked 6-color DNA/CT FISH raw spectral imaging datasets have the background noise subtracted and are linearly unmixed using Zen software with our recorded spectra database (acquired in Section 4.1) to separate individual fluorescent emission signals. Appropriate channels are then assigned for each fluorophore and the channels are combined to produce a multicolor image. Initially, many foci are not detectable in a raw acquired image (Fig. 1, Step 3), although nuclei with some gene loci can clearly be visualized to allow accurate adjustment of the field of view for imaging and z-stacking. Spectral tracking and linear unmixing generates the desired 6-color DNA or CT spectraFISH multi-channel images in which each channel contains the fluorescent signal from a single fluorophore. These images can be analyzed as maximum intensity projections or as 3D renderings using software, such as Zeiss Zen or Imaris (Bitplane), respectively (Section 6).
5. Troubleshooting
The BACs and chromosome painting probes used in this protocol were purchased from BACPAC (https://bacpac.chori.org/) and ID Labs Inc. (http://www.idlabs.com) respectively. This technique is also amenable to the use of plasmids, Oligopaint-derived probe libraries44 and high-throughput oligo synthesis strategies.45 We strongly recommend that researchers confirm the specificity of their probes using PCR, metaphase spreads and localization in diploid cells and prior to usage in this protocol. Only probes that show strong and specific signals should be selected for use in SpectraFISH.
To maximize target gene visualization, 6 DNA FISH probes, rather than 5, can be used if an appropriate dUTP is selected (e.g. γ = 405 nm). Due to commercially-available fluorophore-labeled CTs, we were only capable to co-visualize 4 CTs (plus 1 gene loci of interest and CB detection). Visualization of more CTs is now possible as 5 complementary CT probes (with absorption peaks at 415 nm, 495 nm, 556 nm, 616 nm and 647 nm) have become commercially available.
Although the spatial resolution of the technique is limited by the laser scanning confocal microscope used, we have successfully imaged and distinguished between gene loci using BAC probes that are separated by 2 Mbp (SNORD112 and DAB1, both on human chromosome 1) using a 63× 1.4NA objective.21
We do not recommend the inclusion of a blocking step prior to immunodetection of NBs but this may be necessary depending on the level of off-target binding displayed by the primary antibody selected for IF.
CB staining was achieved by antibody staining following DNA FISH. Certain nuclear targets may benefit from performing IF first, followed by a second round of fixation prior to DNA FISH.
To maximize the number of structures/loci detected, we omitted the use of DAPI. For manual-scoring applications, we have found the nuclear periphery visualized through DNA or CT FISH background staining (DNA FISH) to be sufficient to distinguish between neighboring cells (Figs. 2 and 3). For high-content automated imaging applications, users may consider the inclusion of DAPI staining to simplify analysis by automated nuclear detection segmentation programs prior to DNA FISH foci detection.
It is imperative that all spectra are correctly defined for optimal linear unmixing to permit the clear separation of fluorescence signals. This includes consideration of sample autofluorescence, particularly a problem with clinical tissue samples, as well as proper spectral annotation to avoid probe mis-assignment. However, some crosstalk between channels is occasionally observed because some fluorophores are significantly brighter than others. Typically, this can be avoided by adjusting the brightness of the affected channels with different probe concentrations. If the problem persists, bleed-through regions are easily identifiable in the maximum intensity projection overlay image.
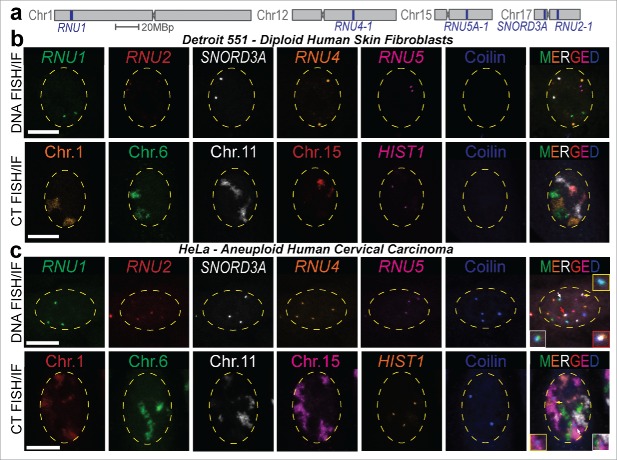
Figure 2.
Visualization of 6 distinct nuclear structures using SpectraFISH. (A) Chromosomal positions of BAC probes detecting spliceosome U sn/snoRNA gene loci used in this study, (B, C) SpectraFISH microscopy single pseudochannel images in normal diploid (Detroit 551, B) and aneuploid (HeLa, C) nuclei (arrows indicate CB-gene/CT colocalization).
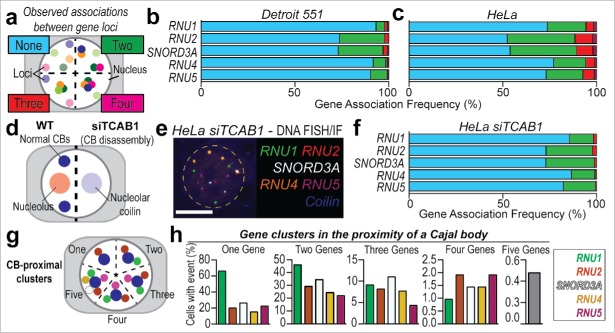
Figure 3.
Higher-order gene pairing analysis using SpectraFISH in diploid and aneuploid cells. (A) Quantitation strategy for colocalized genes, (B) Gene co-localization frequency (%, normalized to total number of alleles scored) for non-clustered alleles (“None,” blue), pairs of U snRNA and snoRNA genes (“Two,” green) and higher-order complexes (“Three,” red; “Four,” magenta) in Detroit 551 cells, and, (C) HeLa cells, (D) Schematic depicting effect of TCAB1 depletion by siRNA (siTCAB1, 60 h) upon CB integrity and coilin distribution in HeLa cells, (E) SpectraFISH microscopy single pseudochannel images in HeLa nuclei following TCAB1 siRNA treatment, (F) Gene co-localization frequency (%) for pairs of U snRNA and snoRNA genes in HeLa cells following TCAB siRNA treatment, (G) Quantitation strategy for CB-proximal gene clustering in HeLa cells, (H) Frequency (%) of 2, 3, 4 or 5 genes clustering proximal to a CB, normalized to total cell number. N = 2 biological replicates, Average number of alleles scored per cell: HeLa – RNU1: 3.02, RNU2: 2.94, SNORD3A: 3.09, RNU4: 3.89, RNU5: 3.08; Detroit 551 fibroblasts – Only cells containing 2 alleles of each gene were scored. White scale bar = 5 µm, nucleus false annotated by yellow dashed oval.
6.1. Implementation of spectraFISH
6.1.1. Example 2D analysis
Here, for the first time, we report the inter-chromosomal clustering frequency of 4 of the major spliceosome U snRNA genes (RNU1, RNU2, RNU4, RNU5) and the most abundant nucleolar snRNA, SNORD3A (U3 snoRNA). These genes are all located on different chromosomes, aside from RNU2 and SNORD3A, which are both located on chromosome 17. Gene pairing events were simultaneously quantified from the same HeLa and primary Detroit 551 fibroblast cells, rather than separate microscopy slides (Fig. 2a–c). This represents an unbiased investigation of single and higher-order gene pairing complexes (Fig. 3a). Gene pairing events occur infrequently in primary diploid cells (“Two genes,” Fig. 3b) and are more frequent in HeLa cells, which is likely to be a result of aneuploidy and the high proliferative status of cancer cells (Fig. 3c). Simultaneous colocalization of 3 or more genes was a rarer event but was still more frequently observed in aneuploid HeLa cells than in diploid fibroblast cells (8.17±1.22% vs. 1.94±0.30%, pooled average). These data indicate that, although infrequent, higher-order interchromosomal gene clustering events do occur in cell culture models. Induction of CB disassembly by TCAB1 knockdown (an obligate CB component) by siRNA results in CB depletion and redistribution of coilin, the CB structural component and marker protein, into nucleoli46 (Fig. 3d–e). In this scenario, CB disassembly coincides with a reduction in both inter- and intrachromosomal gene clusters between 3 or more spliceosome U snRNA genes (Fig. 3f). The observed frequency of association between 4 genes was very low, even in HeLa cells (<1.5%, pooled average) and no clusters that contained all 5 genes were recorded. However, quantification in HeLa cells indicated that these higher-order gene clustering events which occur in the near-proximity of a CB are present in 5–10% of cells (Fig. 3g–h).
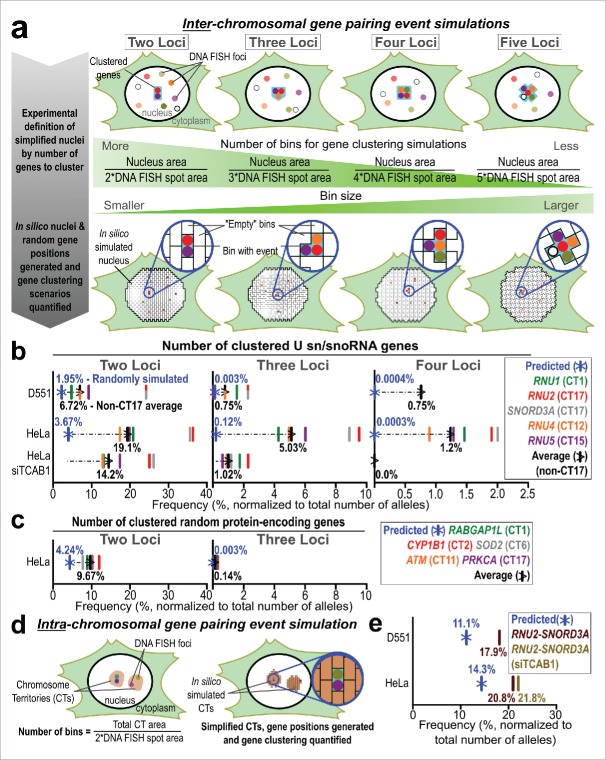
To contextualize our data, we sought to estimate the rate of gene clustering if the positioning of these gene loci was completely random. Published methods to estimate random gene pairing do not allow users to cluster many gene loci, limit the analysis to within a single CT or require biophysical modeling parameters.47,48 Therefore, to predict the observed frequency of random gene associations when using SpectraFISH, we developed a simple computational approach. Initially, we calculated the average nuclear area for each condition based on our microscopy images, as well as the average DNA FISH spot area. Using these data, we then simplified the cell nucleus into a number of gene-pairing “bins” (Fig. 4a and Supplementary Table 1). The total number of bins was dependent on whether the frequency of 2, 3, 4 or 5 gene loci clustered in close proximity to one another was to be estimated (bin number for each calculation is summarized in Supplementary Table 1). Thus, the number of bins when estimating the frequency of 2 genes clustering is higher than when comparing 5 genes. Using these simplified scenarios, in which any gene can be assumed to occupy any 2D nuclear position, each bin was assigned a different number and random bin coordinates were generated in silico for each gene (2 alleles per gene in diploid Detroit 551 cells and 3 to 4 alleles per gene in aneuploid HeLa cells). Clustering was considered to occur when the same bin number was shared by several gene loci. The random gene association frequency was normalized to the total number of simulated alleles (2 × 107 in diploid Detroit 551 cells).
Figure 4.
Computational approach to predicting higher-order random gene clustering events. (A) Predicted random inter-chromosomal gene pairing strategy. Nuclei were simplified into a defined number of regions (bins), dependent on the size of the nucleus and number of genes co-visualized. Nuclear and DNA FISH spot areas were quantified using LSM Image Browser. Detroit 551: 10 columns, assigned to 5 genes with 2 alleles each, and HeLa: 16 columns, assigned to 5 genes with 3 alleles (1 gene = 4 alleles to replicate the in vivo situation) each; 1×107 rows; Bin number range: randomly-generated integers between 1 and n, where n = Nucleus area divided by 2, 3, 4, or 5 * DNA FISH spot area, depending on the number of clustering genes to estimate. Tables were populated with randomly-assigned bin numbers (as a proxy for nuclear coordinates) using RStudio. Clustering was considered to have occurred when exactly 2, 3, 4 or 5 alleles from different genes shared the same bin number, (B) Frequency of 2, 3 and 4 U sn/snoRNA genes predicted to stochastically cluster (blue) in Detroit 551, HeLa and HeLa siTCAB1 cells compared to observed and non-CT17 average clustering frequencies (black), (C) Frequency of 2 or 3 unrelated genomic loci predicted to stochastically cluster (blue) in HeLa cells compared to observed and average clustering frequencies (black). Simulated association frequency was adjusted to compensate for additional alleles of PRKCA and RABGAP1L (5 and 4, respectively) (D) Predicted, random intra-chromosomal gene associations simulation strategy. The area of CT17 was quantified using LSM Image Browser to estimate the association between RNU2 and SNORD3A. Detroit 551 and HeLa: 2 columns, assigned to 2 genes with 1 allele each; 1×107 rows; Bin number range: randomly-generated integers between 1 and n, where n = Individual CT17 area divided by 2*DNA FISH spot area, (E) Predicted in silico association frequency between RNU2 and SNORD3A (blue) in Detroit 551, HeLa (brown) and HeLa siTCAB1 cells (light brown).
Intuitively, random gene clustering was higher in HeLa cells than Detroit 551 cells, due to aneuploidy, and the frequency of stochastic gene pairings decreased as additional genes were added to the clustering area for both cell types (Fig. 4b). We observed a more similar association frequency (inter-chromosomal clustering) between the simulated 2 gene clustering scenarios and Detroit 551 cells compared to HeLa cells (1.95% of alleles vs 6.72% and 3.67% vs. 19.1%, respectively). The most striking difference between the predicted and observed association frequencies was displayed by 3 and 4 clustered loci in HeLa cells (5.03% vs. 0.12% and 1.2% vs. 0.0003%, respectively). The predicted association frequency for 5 clustered genes located on different chromosomes was <0.00001% in both HeLa and Detroit 551 cells. Accordingly, no clusters that possessed 5 genes were observed when manually scoring microscopy images. Disassembly of CBs by depletion of TCAB1 by siRNA in HeLa cells decreased the frequency of all gene pairing events. However, 2 gene pairing remained comparatively high, whereas clustering between 3 or 4 genes was decreased and more similar to the expected association frequency. We also compared the association of 5 independent protein-encoding genes located on different chromosomes (Fig. 4c) and found a closer association between the predicted stochastic 2 gene association rate and the observed association rate, event when compensating for aneuploidy (9.67% vs. 4.24%). Very few instances of 3 genes or more clustering were observed in this scenario. For comparison, predicted simulated random gene clustering normalized on a per-cell basis is provided in Supplementary Figure 1.
We observed an increased pairing frequency for RNU2 and SNORD3A in both HeLa and Detroit 551 cells (Fig. 3b–c, f). These genes are located on opposite arms of chromosome 17, which we believe to be responsible for this phenomenon. Therefore, we queried whether this higher association frequency was solely driven by the increased random chance of these 2 genes clustering, as a product of their limited nuclear positioning compared to the other genes assessed, or if other factors were involved. We repeated the same computational process as detailed above, but limited the simulated clustering region to the physical area accessed by CT17 genes in each cell type and only assessed clustering between 2 genes (Fig. 4d–e). Here, we observed a stronger agreement between the expected random association within CT17 and the observed data in both Detroit 551 cells (11.1% vs. 17.9%), and in HeLa cells (14.3% vs. 20.8%).
6.1.2. Example 2D analysis - conclusions
Here, we describe a number of higher-order gene clusters that are formed between spliceosomal U sn/snoRNA genes. Without analysis of higher-order nuclear organization using spectraFISH microscopy, these multi-gene clusters may otherwise be missed or misestimated by conventional analyses. Further investigation is required to characterize the biological significance of these particular topological genomic conformations. This may include more efficient and expedited biogenesis of spliceosome U snRNPs by consolidating transcription, co-transcriptional 3′-end processing and initial assembly for multiple snRNPs in a central location around the CB in HeLa cells, where transcriptional demands are higher than in non-transformed, slower proliferating cells.
The discrepancy between the expected 2 gene association frequencies and the observed in silico predictions is possibly related to the transcriptional activity of these genes. All 5 are highly transcribed genes and are likely to be retained in active chromatin domains at the periphery of their respective chromosome territories. This may decrease the nuclear area that they can occupy and decrease the number of bins used in our in silico clustering predictions. Hence, the co-localization of protein-encoding genes that are not functionally related and display lower transcriptional activity (and are free of CB-dependent 4C-seq contact regions21) associate less frequently in comparison. These observations support the view CBs maintain higher-order gene clustering events by retaining these target genes in non-random nuclear positions and therefore increase their association frequency in HeLa cells. CBs have been suggested to augment the transcriptional activity of these genes, perhaps by providing an optimized platform for their expression and co-transcriptional 3-end RNA processing, which coincides with increased association between these genes. When this gene clustering platform is absent, such as in diploid cells or following CB disassembly, there is a greater similarity between the predicted and observed association frequencies, which may indicate that these genes can occupy more nuclear positions or possess a more constrained topological arrangement.
These data may also imply that when 2 genes are on the same chromatin fiber, e.g., within smaller regions such as chromosome 17, stochasticity is a stronger determinant for gene pairing than other factors, such as chromatin state or transcriptional activity. Further work will be required to assess whether this trend is true for all chromosomes and to identify similarities or differences between gene-rich and gene-poor chromosomes. Additionally, the contribution of transcriptional activity, chromatin status, and other factors, including nuclear body proximity, to gene associations should be assessed.
6.2. Other potential image analysis strategies
Analysis of 2D maximally-projected spectraFISH images is a potentially useful tool in furthering our understanding of genomic organization. However, it does possess certain limitations. Primarily, collapsing a 3D structure (the cell nucleus) into a flat 2D image is likely to result in loss of cellular context and increase the chance of observing false-positive gene pairing events. Fortunately, there is a growing appreciation for analysis of cellular organization in 3D space49,50 and the strengths of this technique are likely to beneficial for 3D analysis.51 Linear unmixing yields a very high signal-to-noise ratio, which should improve proximity analysis by reducing non-specific foci. Additionally, the inclusion of 6 intermingling genomic regions or cellular structures will result in a visually-striking and informative image. Analyses performed using 2D and 3D scoring methods have previously been shown to correlate strongly.52 Example 3D rendering images produced using Imaris (Bitplane) are shown in Figure 1, Step 4. SpectraFISH images could also be segregated into different cell cycle stages using a DAPI-based integration strategy for subsequent analysis.53 With appropriate modifications, including the use of a nuclear stain, these data could also be analyzed using software to automate morphometric and colocalization analysis, such as CellProfiler, which is capable of automatically identifying nuclei and sub-nuclear structures for subsequent colocalization analysis.54 This may be especially useful in systems where substantial changes in cell cycle progression55 (e.g., cancer) or nuclear morphology (e.g. laminopathies56) is expected and may interfere with normal gene positioning and NB biology.
Final remarks and outlook
Here, we detail a spectral imaging technique to visualize higher-order genomic organization (spectraFISH). This microscopic tool allows researchers to simultaneously co-visualize 5 distinct genes, chromosomes, NBs or other nuclear structures. However, this methodology can also be applied to better understand the relationship between specific genomic loci or regions and epigenetic marks, or the dynamic manner in which multiple NBs associate with target genes under various physiological and experimental conditions. Further progress and development of near-infrared and far-infrared lasers, super-resolution microscopy approaches, detectors and photostable fluorophores will allow efficient, crosstalk-free co-visualization of more than 6 fluorophores. Indeed, a similar strategy has been demonstrated recently for protein-based structures in different cellular compartments.57 The data presented here indicates that obvious differences exist between inter- and intra-chromosomal gene associations that require further investigation. This is a flexible, high-resolution visualization tool for the dissection of higher-order genome dynamics which cannot typically be quantified using high-throughput genomic sequencing or automated deep imaging methodologies.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
Confocal microscopy was performed using the National Cancer Institute Confocal Microscopy Core Facility (Bethesda, MD). We thank Zeiss Specialist Elise Shumsky-Glier and Dr. Keisuke Hasegawa for their advice during the initial stages of developing this technique. Detroit 551 cells were kindly donated by Dr. Dominik Duelli (Abbott Molecular, IL). We are especially thankful to Dr. Gordon Hager (National Cancer Institute, Bethesda, MD) for sharing his thoughts and criticisms throughout the development of this technique and his continuing support of our studies.
Funding
This work was funded by NIH grant R01GM090156 from NIGMS.
References
- [1].Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol 2010; 2:a003889; PMID:20300217; http://dx.doi.org/ 10.1101/cshperspect.a003889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Dundr M. Nuclear bodies: multifunctional companions of the genome. Curr Opin Cell Biol 2012; 24:415-22; PMID:22541757; http://dx.doi.org/ 10.1016/j.ceb.2012.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gall JG. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol 2000; 16:273-300; PMID:11031238; http://dx.doi.org/ 10.1146/annurev.cellbio.16.1.273 [DOI] [PubMed] [Google Scholar]
- [4].Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci U S A 1995; 92:5915-9; PMID:7597053; http://dx.doi.org/ 10.1073/pnas.92.13.5915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR α in acute promyelocytic leukemia cells. Cell 1994; 76:345-56; PMID:8293468; http://dx.doi.org/ 10.1016/0092-8674(94)90341-7 [DOI] [PubMed] [Google Scholar]
- [6].Huang S, Spector DL. Nascent pre-mRNA transcripts are associated with nuclear regions enriched in splicing factors. Genes Dev 1991; 5:2288-302; PMID:1748285; http://dx.doi.org/ 10.1101/gad.5.12a.2288 [DOI] [PubMed] [Google Scholar]
- [7].Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI. Paraspeckles: a novel nuclear domain. Curr Biol 2002; 12:13-25; PMID:11790299; http://dx.doi.org/ 10.1016/S0960-9822(01)00632-7 [DOI] [PubMed] [Google Scholar]
- [8].Davidson S, Macpherson N, Mitchell JA. Nuclear organization of RNA polymerase II transcription. Biochem Cell Biol 2013; 91:22-30; PMID:23442138; http://dx.doi.org/ 10.1139/bcb-2012-0059 [DOI] [PubMed] [Google Scholar]
- [9].Huang S, Deerinck TJ, Ellisman MH, Spector DL. The dynamic organization of the perinucleolar compartment in the cell nucleus. J Cell Biol 1997; 137:965-74; PMID:9166399; http://dx.doi.org/ 10.1083/jcb.137.5.965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Biamonti G, Vourc'h C. Nuclear stress bodies. Cold Spring Harb Perspect Biol 2010; 2:a000695; PMID:20516127; http://dx.doi.org/ 10.1101/cshperspect.a000695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Carmo-Fonseca M, Berciano MT, Lafarga M. Orphan nuclear bodies. Cold Spring Harb Perspect Biol 2010; 2:a000703; PMID:20610547; http://dx.doi.org/ 10.1101/cshperspect.a000703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Smith KP, Carter KC, Johnson CV, Lawrence JB. U2 and U1 snRNA gene loci associate with coiled bodies. J Cell Biochem 1995; 59:473-85; PMID:8749717; http://dx.doi.org/ 10.1002/jcb.240590408 [DOI] [PubMed] [Google Scholar]
- [13].Jacobs EY, Frey MR, Wu W, Ingledue TC, Gebuhr TC, Gao L, Marzluff WF, Matera AG. Coiled bodies preferentially associate with U4, U11, and U12 small nuclear RNA genes in interphase HeLa cells but not with U6 and U7 genes. Mol Biol Cell 1999; 10:1653-63; PMID:10233169; http://dx.doi.org/ 10.1091/mbc.10.5.1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Frey MR, Matera AG. RNA-mediated interaction of Cajal bodies and U2 snRNA genes. J Cell Biol 2001; 154:499-509; PMID:11489914; http://dx.doi.org/ 10.1083/jcb.200105084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science 2002; 295:1306-11; PMID:11847345; http://dx.doi.org/ 10.1126/science.1067799 [DOI] [PubMed] [Google Scholar]
- [16].de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev 2012; 26:11-24; PMID:22215806; http://dx.doi.org/ 10.1101/gad.179804.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Belton JM, McCord RP, Gibcus JH, Naumova N, Zhan Y, Dekker J. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods 2012; 58:268-76; PMID:22652625; http://dx.doi.org/ 10.1016/j.ymeth.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al.. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009; 326:289-93; PMID:19815776; http://dx.doi.org/ 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ching RW, Ahmed K, Boutros PC, Penn LZ, Bazett-Jones DP. Identifying gene locus associations with promyelocytic leukemia nuclear bodies using immuno-TRAP. J Cell Biol 2013; 201:325-35; PMID:23589495; http://dx.doi.org/ 10.1083/jcb.201211097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Matheson TD, Kaufman PD. Grabbing the genome by the NADs. Chromosoma 2015; PMID:26174338.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang Q, Sawyer IA, Sung M-H, Sturgill D, Shevtsov SP, Pegoraro G, Hakim O, Baek S, Hager GL, Dundr M. Cajal bodies are linked to genome conformation. Nat Commun 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rowley MJ, Corces VG. The three-dimensional genome: principles and roles of long-distance interactions. Curr Opin Cell Biol 2016; 40:8-14; PMID:26852111; http://dx.doi.org/ 10.1016/j.ceb.2016.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tjong H, Li W, Kalhor R, Dai C, Hao S, Gong K, Zhou Y, Li H, Zhou XJ, Le Gros MA, Larabell CA, Chen L, Alber F. Population-based 3D genome structure analysis reveals driving forces in spatial genome organization. Proc Natl Acad Sci U S A 2016; 113(12):E1663-72; PMID:26951677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Speicher MR, Gwyn Ballard S, Ward DC. Karyotyping human chromosomes by combinatorial multi-fluor FISH. Nat Genet 1996; 12:368-75; PMID:8630489; http://dx.doi.org/ 10.1038/ng0496-368 [DOI] [PubMed] [Google Scholar]
- [25].Jalal SM, Law ME. Utility of multicolor fluorescent in situ hybridization in clinical cytogenetics. Genet Med 1999; 1:181-6; PMID:11256670; http://dx.doi.org/ 10.1097/00125817-199907000-00003 [DOI] [PubMed] [Google Scholar]
- [26].Schrock E, Blume C, Meffert MC, du Manoir S, Bersch W, Kiessling M, Lozanowa T, Thiel G, Witkowski R, Ried T, et al.. Recurrent gain of chromosome arm 7q in low-grade astrocytic tumors studied by comparative genomic hybridization. Genes Chromosomes Cancer 1996; 15:199-205; http://dx.doi.org/ 10.1002/(SICI)1098-2264(199604)15:4%3c199::AID-GCC1%3e3.0.CO;2-X [DOI] [PubMed] [Google Scholar]
- [27].Potapova TA, Unruh JR, Box AC, Bradford WD, Seidel CW, Slaughter BD, Sivagnanam S, Wu Y, Li R. Karyotyping human and mouse cells using probes from single-sorted chromosomes and open source software. BioTechniques 2015; 59:335-6, 338,, 340-332 passim [DOI] [PubMed] [Google Scholar]
- [28].Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science 2002; 297:836-40; PMID:12161654; http://dx.doi.org/ 10.1126/science.1072241 [DOI] [PubMed] [Google Scholar]
- [29].Shachar S, Voss TC, Pegoraro G, Sciascia N, Misteli T. Identification of gene positioning factors using high-throughput imaging mapping. Cell 2015; 162:911-23; PMID:26276637; http://dx.doi.org/ 10.1016/j.cell.2015.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Burman B, Misteli T, Pegoraro G. Quantitative detection of rare interphase chromosome breaks and translocations by high-throughput imaging. Genome Biol 2015; 16:146; PMID:26313373; http://dx.doi.org/ 10.1186/s13059-015-0718-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Novotny I, Blazikova M, Stanek D, Herman P, Malinsky J. In vivo kinetics of U4/U6.U5 tri-snRNP formation in Cajal bodies. Mol Biol Cell 2011; 22:513-23; PMID:21177826; http://dx.doi.org/ 10.1091/mbc.E10-07-0560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Novotny I, Malinova A, Stejskalova E, Mateju D, Klimesova K, Roithova A, Sveda M, Knejzlik Z, Stanek D. SART3-dependent accumulation of incomplete spliceosomal snRNPs in cajal bodies. Cell Rep 2015; 10:429-40; http://dx.doi.org/ 10.1016/j.celrep.2014.12.030 [DOI] [PubMed] [Google Scholar]
- [33].Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonne R, Luhrmann R. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell 2006; 17:3221-31; PMID:16687569; http://dx.doi.org/ 10.1091/mbc.E06-03-0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol 2007; 179:1095-103; PMID:18070915; http://dx.doi.org/ 10.1083/jcb.200710058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gao L, Frey MR, Matera AG. Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17p11.2 in a complex inverted repeat structure. Nucleic Acids Res 1997; 25:4740-7; PMID:9365252; http://dx.doi.org/ 10.1093/nar/25.23.4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Machyna M, Kehr S, Straube K, Kappei D, Buchholz F, Butter F, Ule J, Hertel J, Stadler PF, Neugebauer KM. The coilin interactome identifies hundreds of small noncoding RNAs that traffic through Cajal bodies. Mol Cell 2014; 56:389-99; PMID:25514182; http://dx.doi.org/ 10.1016/j.molcel.2014.10.004 [DOI] [PubMed] [Google Scholar]
- [37].Nizami Z, Deryusheva S, Gall JG. The Cajal body and histone locus body. Cold Spring Harb Perspect Biol 2010; 2:a000653; PMID:20504965; http://dx.doi.org/ 10.1101/cshperspect.a000653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Shopland LS, Byron M, Stein JL, Lian JB, Stein GS, Lawrence JB. Replication-dependent histone gene expression is related to cajal body (CB) association but does not require sustained CB contact. Mol Biol Cell 2001; 12:565-76; PMID:11251071; http://dx.doi.org/ 10.1091/mbc.12.3.565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Vieux-Rochas M, Fabre PJ, Leleu M, Duboule D, Noordermeer D. Clustering of mammalian Hox genes with other H3K27me3 targets within an active nuclear domain. Proc Natl Acad Sci U S A 2015; 112:4672-7; PMID:25825760; http://dx.doi.org/ 10.1073/pnas.1504783112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nandy K, Gudla PR, Meaburn KJ, Misteli T, Lockett SJ. Automatic nuclei segmentation and spatial FISH analysis for cancer detection. Conference proceedings : … Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering Med Biol Soc Annual Conference 2009; 2009:6718-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Meuleman W, Peric-Hupkes D, Kind J, Beaudry JB, Pagie L, Kellis M, Reinders M, Wessels L, van Steensel B. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res 2013; 23:270-80; PMID:23124521; http://dx.doi.org/ 10.1101/gr.141028.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Meaburn KJ. Fluorescence in situ hybridization on 3D cultures of tumor cells. Methods Mol Biol 2010; 659:323-36; PMID:20809324; http://dx.doi.org/ 10.1007/978-1-60761-789-1_25 [DOI] [PubMed] [Google Scholar]
- [43].Zimmermann T, Marrison J, Hogg K, O'Toole P. Clearing up the signal: spectral imaging and linear unmixing in fluorescence microscopy. Methods Mol Biol 2014; 1075:129-48; PMID:24052349; http://dx.doi.org/ 10.1007/978-1-60761-847-8_5 [DOI] [PubMed] [Google Scholar]
- [44].Beliveau BJ, Joyce EF, Apostolopoulos N, Yilmaz F, Fonseka CY, McCole RB, Chang Y, Li JB, Senaratne TN, Williams BR, Rouillard JM, Wu CT. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc Natl Acad Sci U S A 2012; 109:21301-6; PMID:23236188; http://dx.doi.org/ 10.1073/pnas.1213818110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Jensen M, Roberts L, Johnson A, Fukushima M, Davis R. Next generation 1536-well oligonucleotide synthesizer with on-the-fly dispense. J Biotechnol 2014; 171:76-81; PMID:24355807; http://dx.doi.org/ 10.1016/j.jbiotec.2013.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mahmoudi S, Henriksson S, Weibrecht I, Smith S, Soderberg O, Stromblad S, Wiman KG, Farnebo M. WRAP53 is essential for cajal body formation and for targeting the survival of motor neuron complex to cajal bodies. PLoS Biol 2010; 8:e1000521; PMID:21072240; http://dx.doi.org/ 10.1371/journal.pbio.1000521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Jost KL, Haase S, Smeets D, Schrode N, Schmiedel JM, Bertulat B, Herzel H, Cremer M, Cardoso MC. 3D-Image analysis platform monitoring relocation of pluripotency genes during reprogramming. Nucleic Acids Res 2011; 39:e113; PMID:21700670; http://dx.doi.org/ 10.1093/nar/gkr486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Strongin DE, Groudine M, Politz JC. Nucleolar tethering mediates pairing between the IgH and Myc loci. Nucleus 2014; 5:474-81; PMID:25482199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Dekker J, Mirny L. The 3D genome as moderator of chromosomal communication. Cell 2016; 164:1110-21; PMID:26967279; http://dx.doi.org/ 10.1016/j.cell.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, Trzaskoma P, Magalska A, Wlodarczyk J, Ruszczycki B, et al.. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell 2015; 163:1611-27; PMID:26686651; http://dx.doi.org/ 10.1016/j.cell.2015.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rueden CT, Eliceiri KW. Visualization approaches for multidimensional biological image data. Biotechniques 2007; 43:31, 33-36 [DOI] [PubMed] [Google Scholar]
- [52].Shachar S, Pegoraro G, Misteli T. HIPMap: A high-throughput imaging method for mapping spatial gene positions. Cold Spring Harb Symp Quant Biol 2015; PMID:26472748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Roukos V, Pegoraro G, Voss TC, Misteli T. Cell cycle staging of individual cells by fluorescence microscopy. Nat Protoc 2015; 10:334-48; PMID:25633629; http://dx.doi.org/ 10.1038/nprot.2015.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Bray MA, Vokes MS, Carpenter AE. Using cellProfiler for automatic identification and measurement of biological objects in images. Curr Protoc Mol Biol 2015; 109:14.17.11-14.17.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Gerard C, Goldbeter A. Dynamics of the mammalian cell cycle in physiological and pathological conditions. Wiley Interdiscip Rev Syst Biol Med 2016; 8:140-56; PMID:26613368 [DOI] [PubMed] [Google Scholar]
- [56].Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med 2005; 11:440-5; PMID:15750600; http://dx.doi.org/ 10.1038/nm1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Niehorster T, Loschberger A, Gregor I, Kramer B, Rahn HJ, Patting M, Koberling F, Enderlein J, Sauer M. Multi-target spectrally resolved fluorescence lifetime imaging microscopy. Nat Methods 2016; 13:257-62; PMID:26808668; http://dx.doi.org/ 10.1038/nmeth.3740 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.