ABSTRACT
The nucleolus is a nuclear subcompartment for tightly regulated rRNA production and ribosome subunit biogenesis. It also acts as a cellular stress sensor and can release enriched factors in response to cellular stimuli. Accordingly, the content and structure of the nucleolus change dynamically, which is particularly evident during cell cycle progression: the nucleolus completely disassembles during mitosis and reassembles in interphase. Although the mechanisms that drive nucleolar (re)organization have been the subject of a number of studies, they are only partly understood. Recently, we identified Alu element-containing RNA polymerase II transcripts (aluRNAs) as important for nucleolar structure and rRNA synthesis. Integrating these findings with studies on the liquid droplet-like nature of the nucleolus leads us to propose a model on how RNA polymerase II transcripts could regulate the assembly of the nucleolus in response to external stimuli and during cell cycle progression.
KEYWORDS: Alu repeat-containing RNA, cell cycle, intracellular phase separation, nuclear bodies, RNA-protein interactions, RNA-protein droplets-like structures
The nucleolus is the cellular factory for producing rRNAs (rRNAs) and assembling ribosome subunits. In response to cellular stress and environmental cues its activity and structure can change, and enriched factors involved in cellular functions such as cell cycle regulation, DNA repair or mRNA processing can be released.1-6 The dynamic nature of nucleolar structure and function is particularly evident during cell cycle progression as reviewed previously6,7: At the onset of mitosis nucleoli completely disappear and rRNA production is turned off. Following cell division, the nucleolus reassembles and rRNA production is restarted.
To rationalize the plastic conformation of the nucleolus, the model of a liquid droplet-like structure established via a liquid-liquid phase separation process has been proposed.8-10 This description accounts for the self-assembly of the nucleolus as well as that of PML and Cajal bodies, splicing speckles and other nuclear bodies. The partitioning of the nucleus into organelles with specific activities cannot be explained by viewing the nucleus simply as a membrane-confined container for the nucleoplasm – an aqueous solution with high concentrations of proteins and nucleic acids macromolecules. At the same time, the above nuclear subcompartments are variable in size and shape and their constituting protein and RNA components are in rapid exchange with factors from the nucleoplasm.11 Thus, like in a liquid, molecules in these nuclear subcompartments constantly rearrange. The apparent similarity of this aspect of nuclear organization with two liquids that do not mix and form separate phases, as for example oil drops in water, has led to the view that features of nuclear subcompartments within the nucleoplasm can be described by a liquid-liquid phase separation process as reviewed previously.12 Another equivalent term used in this context is that of a liquid-demixing phase separation.13 Within a cell, this process frequently involves partially unstructured proteins,14 and can be regulated by their interaction with RNA.15,16 When considered in the context of this physico-chemical framework, our recent findings on the role of Alu element-containing RNA polymerase (Pol) II transcripts in nucleolus structure and function have a number of implications.17 They suggest that the interaction of RNA Pol II transcripts with unstructured nucleolar protein shifts the equilibrium between the two liquid phases, the nucleolus and the nucleoplasm and acts as a factor that affects the (dis)assembly of the nucleolus. Based on these findings, we discuss here how these RNA-protein interactions could be modulated in the cell to provide an additional regulatory layer for controlling structure and function of the nucleolus.
RNA-driven nuclear subcompartment assembly and maintenance
In the past few years, several studies have illustrated the ability of specific coding and non-coding RNA transcripts to initiate the formation of nuclear bodies. In a number of these, cell lines that had lacO operator repeats stably integrated into their genome were used to tether specific RNAs to these genomic loci. The RNA transcripts containing several MS2 stem-loops were bound to the lacO arrays via the high affinity MS2 coat protein fused to lac repressor. For example, it was reported that tethering the replication-dependent histone gene H2B RNA to lacO operator repeats array induced the formation of histone locus bodies-associated Cajal bodies and the accumulation of coilin, a Cajal body-associated protein.18 The same study found that β-globin transcripts tethered to lacO arrays formed de novo speckles as identified by the enrichment of the serine-arginin rich splicing factor SC35, a nuclear speckle-associated protein. In addition, the lacO-bound non-coding RNA NEAT1 triggered the formation of de novo paraspeckles, which were associated to paraspeckle-specific proteins such as PSP1, NONO and PSF.18,19 Likewise, lacO-tethered non-coding satellite III repeat RNA recruited several proteins associated to nuclear stress bodies, for example heat shock-specific transcription factor HSF1 or the splicing factor SF2/ASF.18 These results suggest that specific RNA transcripts can serve as a structural scaffold for the assembly of various nuclear subcompartments.
In our recent work, a related RNA-mediated assembly process was proposed for the organization of the nucleolus.17,20 Both the inhibition of Pol I and II induced structural changes in the nucleolus. However, a dispersion of nucleolar domains throughout the nucleoplasm was specific for Pol II inhibition. By analyzing the RNA content of the nucleolus, we found that it was enriched in Alu element-containing RNA transcripts (aluRNAs) originating from intronic regions of Pol II transcribed genes. We also found that depletion of aluRNAs led to the same dispersion phenotype of the nucleolus as Pol II inhibition. Furthermore, overexpression of aluRNAs resulted in larger nucleoli suggesting that the nucleolus size and the available amount of Pol II-produced aluRNAs were linked. Inhibition of Pol II or depletion of aluRNAs not only resulted in nucleoli dispersion, but also induced a strong reduction of Pol I transcriptional activity. This supports the view that nucleolus structure and function are closely connected as discussed previously.21 In addition, our study points to an RNA polymerase ‘cross-talk’ where the product of Pol II transcription influences the transcriptional activity of Pol I and the maintenance of the nucleolar subcompartment.
RNA-driven liquid-liquid phase separation during nucleolus formation
To describe the principles underlying the dynamic organization of the nucleolus the model of a liquid-liquid phase separation has been applied, where liquid droplet-like structures become separated from the nucleoplasm.8-10 Furthermore, a mechanism has been proposed according to which this type of liquid-liquid phase separation involves unstructured domains in RNA-binding proteins.16 The latter are referred to as low complexity domains or intrinsically disordered domains.16,22 Although liquid droplet-like structures form also in the absence of RNA, it was recently shown that the presence of RNA could induce such a phase separation at much lower protein concentration. The presence of a short RNA sequence from the promoter region of DNMT3b efficiently induced FUS (fused in sarcoma) protein self-assembly, a process that was also observed at much higher concentrations of FUS in absence of RNA.23 In addition, a study by Zhang et al. showed that specific mRNAs drive Whi3 (a known RNA-binding protein and regulator of the cell cycle) assembly into droplets with distinct biophysical properties that were dependent on the mRNA transcript.24 Thus, RNAs binding to an unstructured protein domain can promote liquid droplet-like assembly by shifting the equilibrium toward protein association.
As shown in our recent work, aluRNAs interacts with at least three nucleolar proteins, nucleolin (NCL), nucleophosmin (NPM) and fibrillarin, which are essential for nucleolus structure and function.25-27 All three of them contain unstructured domains. The C-terminal region of NCL28 and fibrillarin29 both contain a low complexity domain, a glycin-arginine rich (GAR) domain, which is reported to interact with RNA transcripts.30 NPM contains basic and acidic sequences, which were reported to be intrinsically disordered and to regulate its interaction with RNA.31 For NCL and NPM, their unstructured domains were linked to the ability of the proteins to self-associate,29,32 which is an essential part of the process of liquid droplet formation.13,16 These known properties of NCL, NPM and fibrillarin led us to propose that aluRNAs support nucleolar assembly by interacting with those proteins and promoting a liquid-liquid phase separation. In support of this view, recent in vitro work has shown the ability of fibrillarin to phase-separate at lower concentrations if yeast RNA was present.10 It was also demonstrated that the RNA-binding domains and the GAR domain of NCL are important for its localization to the nucleolus.33,34 This could mean that those domains are important for the protein to phase-separate in an RNA-dependent manner.
Remarkably, some specific proteins found in the RNA-enriched nuclear bodies mentioned above also contain disordered domains as for example coilin in Cajal bodies,35 which was also reported to self-associate.36 In paraspeckles, the N- and C-terminal regions of PSP1, NONO, and PSF are predicted to be disordered,37 and in speckles, SC35 can self-associate36 and contains a serine-arginine rich domain,38 which like the GAR domain is a low complexity domain. Interestingly, a recent study by Frege et al.39 reported the presence of intrinsically disordered proteins in many membrane-less nuclear subcompartments. This suggests that a mechanism based on RNA-protein interaction-mediated liquid-liquid phase separation could also promote the assembly of other RNA-containing nuclear bodies.
RNA pol II transcripts during cell cycle-dependent nucleolus (dis)assembly
In our recent work, we focused on the effect of depleting or overexpressing aluRNAs on nucleoli during the interphase of the cell cycle. This raises the question whether aluRNAs or other Pol II transcripts could also be relevant for the disassembly of nucleoli at the onset of mitosis and their reassembly at the end of cell division. By tracing the location of the NCL and NPM nucleolar marker proteins during the cell-cycle the (dis)assembly of nucleoli has been dissected in a number of studies, e.g. refs. 26,40 In Figure 1 we have reproduced these experiments via simultaneous visualization of the NCL and NPM distributions in HeLa cells to illustrate that this process involves droplet-like nucleolar substructures. At telophase, the NCL and NPM images indicated that nucleoli were still disrupted. During cytokinesis some accumulation into areas was observed for NCL and to a lesser extend also for NPM, while fully assembled nucleoli with co-localization of NCL and NPM were only present during interphase.
Figure 1.
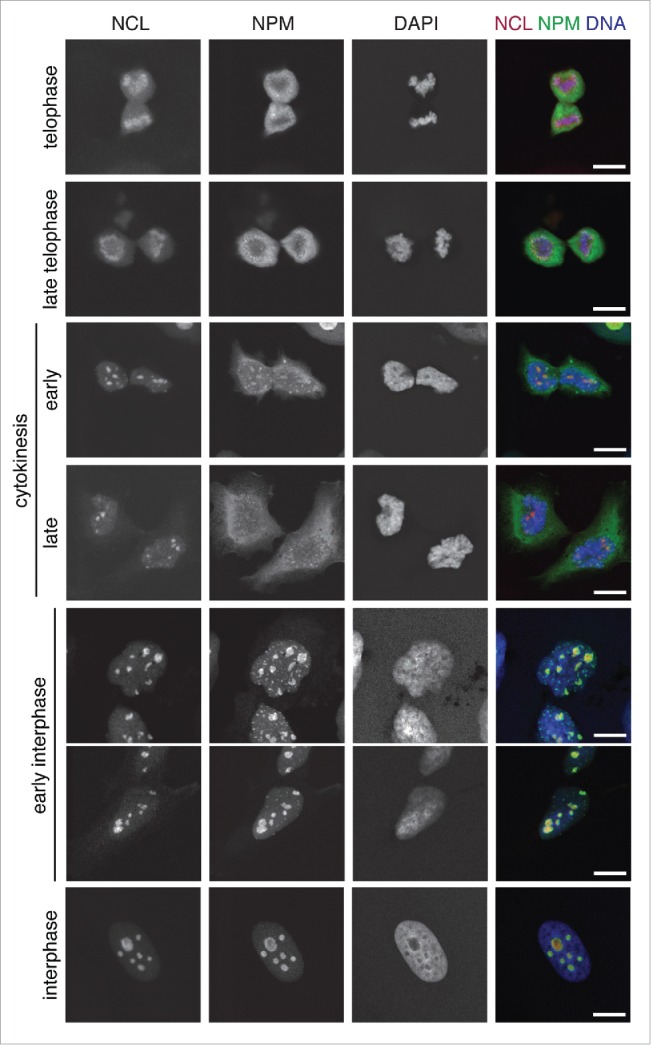
Cell cycle-dependent structural changes of the nucleolus. Confocal laser scanning microscopy (CLSM) images showing the nucleolar marker proteins nucleolin (NCL, red, stably expressed RFP-NCL) and nucleophosmin (NPM, green, immunofluorescence) with DNA (DAPI, blue) counterstaining in U2OS cells at different stages of the cell cycle. As evident from the NCL and NPM distribution, nucleoli are still completely disrupted during telophase and fully assembled during interphase. Scale bars, 10 µm.
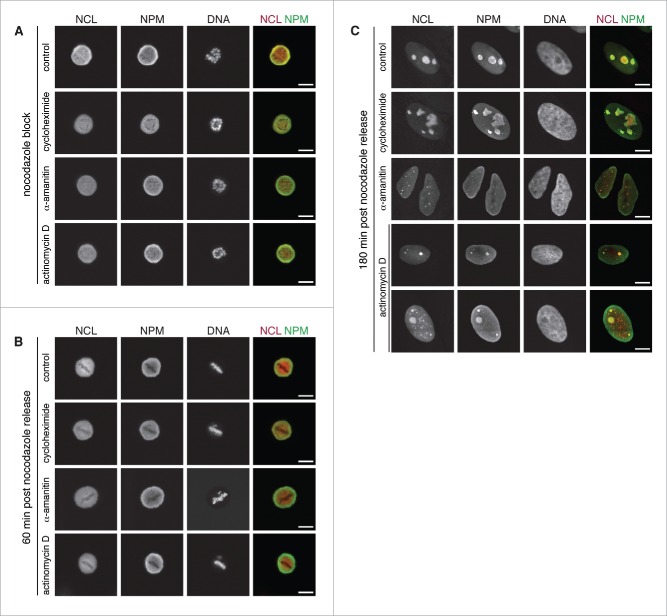
If Pol II transcripts are essential for nucleolus formation, the inhibition of Pol II during mitosis and early interphase should prevent the proper assembly of nucleoli after mitosis completion. To test this hypothesis, we synchronized cell by thymidine block41 followed by a nocodazole block. During nocodazole block, cells were treated with specific Pol I (low concentration of actinomycin D) and Pol II (low concentration of α-amanitin) inhibitors for 5 hours as well as with the protein translation inhibitor cycloheximide. During this treatment, cells remained blocked in prometaphase, the chromosomes were condensed but not aligned due to the lack of microtubules. Without inhibitor as well as in presence of actinomycin D, α-amanitin, and cycloheximide, the nucleoli remained disassembled and the NCL and NPM marker proteins redistributed to the chromatin free regions of the cell (Fig. 2A). Next, nocodazole was washed out and cells were further incubated with the respective Pol I or Pol II inhibitors, or translation inhibitor. The cells progressed normally to form the metaphase plate (Fig. 2B). The inhibition of Pol I did not prevent the formation of nucleoli, as reported previously,42 which still contained both NCL and NPM (Fig. 2C). For some of the cells, additional NCL dots formed in the nucleoplasm, which were not co-localizing with NPM. These nucleoplasmic foci might originate from aberrant accumulation of NCL that could not be integrated into nucleoli due to Pol I inhibition. In contrast to this relatively minor structural phenotype, cells treated with a Pol II inhibitor completely failed to form nucleoli after release of the nocodazole block and nucleolar particles remained dispersed (Fig. 2C). In addition, a previous study reported that roscovitine, another inhibitor of Pol II transcription,43 impaired post-mitotic nucleolus assembly.44 The phenotype of persistent dispersed nucleolar subdomains observed upon Pol II inhibition did not appear to be due to a lack of protein synthesis (Fig. 2C). The inhibition of protein translation for several hours did not give rise to disintegrated nucleolar subdomains. Taken together, these observations suggest that Pol II transcription and the resulting Pol II transcripts such as aluRNAs could be important for the post-mitotic assembly of nucleoli. However, to corroborate this conclusion one would have to dissect the role of specific Pol II transcripts in further experiments to exclude indirect effects of the global Pol II transcription.
Figure 2.
Impaired post mitosis nucleolus assembly through Pol II inhibition. (A) CLSM images showing NCL (red, stably expressed RFP-NCL), NPM (green, immunofluorescence) and DNA (DAPI) in U2OS cells treated with nocodazole (100 ng/ml) and either actinomycin D (50 ng/ml), α-amanitin (50 µg/ml) or cycloheximide (50 µg/ml) for 5 h. (B) Cells 60 min after they were released from the nocodazole block. (C) Cells 180 min after they were released from the nocodazole block. Scale bars, 10 µm.
Regulation of RNA-driven liquid-liquid phase separation
For a phase separation process, the concentration of the various components is a critical parameter as illustrated by studies showing that droplet-like assembly only occurred above a critical concentration.10,23,24,45 This parameter could be regulated in the cell by nucleocytoplasmic transport processes and cellular turnover of the macromolecules involved and potentially competing binding partners. For RNA-driven liquid demixing phase separation systems, the interaction of RNA transcripts with specific protein partners might be required to reach critical phase separation conditions in vivo, e.g., to allow for droplet formation and assembly at physiological protein concentrations lower than those needed in in vitro studies. In such a system, any perturbation of RNA-protein interaction would result in the dispersion of the droplets, their dissolution and redistribution of the liquid phases.
At the onset of mitosis, the nuclear envelope breaks down, nucleoplasm and cytoplasm mix rapidly and nuclear factors become diluted. Furthermore, Pol II transcription is strongly reduced during mitosis.46 Thus, the concentration of nuclear protein and RNA components might drop below the critical concentration required for a liquid-liquid phase separation process. In addition, nuclear proteins become more accessible and can be bound at their nuclear localization signal47 (NLS) by proteins of the importin family.48 It is well established that interaction of importins with the NLS part of a protein can interfere with the function of the protein49 and possibly also with its RNA-binding ability. As the nuclear envelope reassembles, NLSs are released from importins in the nucleus and proteins are no longer inhibited from performing their nuclear functions. In support of this view, the nucleolus assembly correlates with post-mitotic nuclear envelope formation.50
Another factor that could influence droplet formation and assembly are posttranslational modifications of the proteins involved. Phosphorylation, for example, has been shown to regulate the interaction of intrinsically disordered domains with RNA13,51 and to influence the ability of intrinsically disordered protein to phase separate.16,52 Moreover, other posttranscriptional modifications like acetylation, methylation or sumoylation could alter the RNA binding potential of a protein53 or modify its localization.54 Such a regulatory mechanism might apply also for the nucleolus. It was proposed that phosphorylation of an abundant nucleolar protein may drive the cell cycle nucleolus (dis-)assembly dynamics.52 The major cell cycle switch at the onset of mitosis is the activation of the cyclin-dependent kinase CDC2.55 Interestingly, both NCL and NPM are strongly regulated via phosphorylation and are phosphorylated by CDC2 at the onset of mitosis.56-58 Furthermore, it was reported that phosphorylation controls the localization of NCL with translocation of mitotic phosphorylated NCL to the cytoplasm59 and that phosphorylation of NPM prevents the protein to interact with RNA.57 This suggests that mitotic phosphorylation of these proteins by CDC2 could inhibit nucleolar assembly by either acting negatively on the ability of NCL and NPM to associate to RNA or by blocking further association of these proteins when bound to RNA.
Model for RNA-dependent nucleolar assembly
A model for an aluRNA-dependent mechanism that drives changes of nucleolar structure including the disassembly at mitosis and reassembly at interphase is depicted in Figure 3. At the onset of mitosis, the nucleolus disassembles, which correlates with the arrest of Pol I transcriptional activity. However, the absence of Pol I activity during mitosis and the restart of rRNA production at the end of cell division are not sufficient to explain these structural changes. Indeed, nucleoli assembly can occur in the presence of Pol I specific inhibitors as shown in Figure 2A and as previously reported.42 This process is strongly inhibited by treatment with Pol II inhibitors17 and during mitosis (Fig. 2C) where the assembly stops at the stage of nucleolar droplets. For the nucleation of such droplets, rRNA may be crucial as suggested in recent studies.10,60 However, these relatively small prenucleolar bodies containing NCL and NPM appear to require the association with Pol II aluRNA transcripts as the “glue” to assemble them into larger domains. In the absence of the latter RNAs the coalescence step required to achieve the liquid-liquid phase separation does not occur (Fig. 3A) and prevents proper post-mitotic reassembly of the nucleoli (Fig. 2C). Accordingly, we propose that the interaction of nucleolar proteins as for example NCL and NPM with RNA Pol II transcripts drives nucleolar assembly in a cell cycle-dependent manner (Fig. 3B). This process could involve phosphorylation-dependent RNA-protein interactions.
Figure 3.
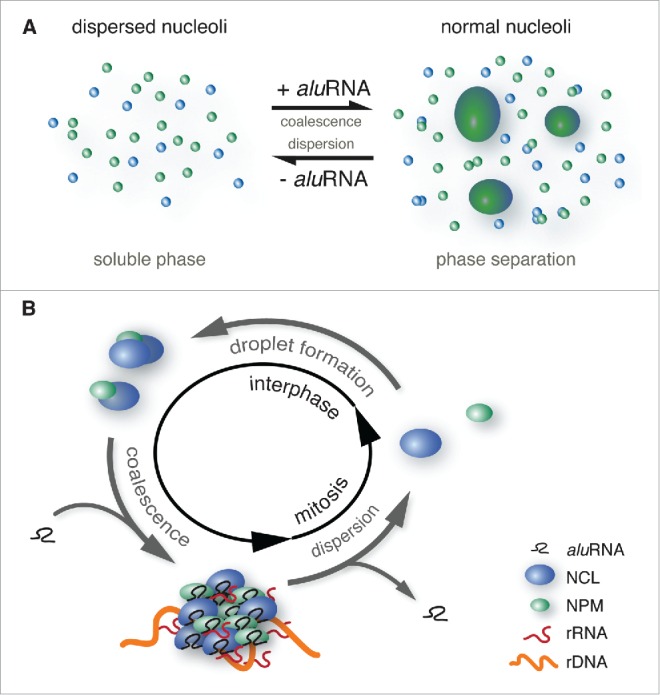
Model for RNA and cell cycle-dependent (dis)assembly of the nucleolus. (A) Scheme of RNA-driven liquid-liquid phase separation of the nucleolus. The interaction of nucleolar proteins like NCL or NPM (blue and green respectively) with aluRNAs might trigger a conformational change of their unstructured regions that drives the assembly of nucleolar subdomains via a liquid-liquid phase separation process into larger domains representing functional nucleoli. Depletion of aluRNAs induces a change of this equilibrium back into the dispersed state with small nucleolar droplets. (B) As illustrated in the enclosed scheme, assembly and disassembly of the nucleolus are dependent on the interactions between RNA transcripts (aluRNAs) and proteins (NCL, NPM). The RNA-protein assemblies form nucleolar domains that associate with rDNA and efficiently support Pol I transcriptional activity. Protein-RNA interactions could be dependent on the cell cycle state, which affects posttranslational modifications of the proteins, changes in their concentration and/or exposition to competing interacting species, like for example binding of importins to their NLS. Thereby, the formation of RNA-protein assemblies could be coupled to specific cell cycle phases.
Concluding remarks
Increasing evidence points to RNA as an important factor for proper formation of nuclear subcompartments and intact chromatin structure.61-63 The assembly of an RNA-dependent nuclear scaffold induces the local enrichment of effector molecules and their associated activities for efficient and controlled reactions in the nucleus.64 The concept of (dis)assembly of the nucleolus driven by RNA interaction with intrinsically disordered domains of nucleolar proteins might also apply to other nuclear bodies.39,65,66 Modulating these RNA-protein interactions during the cell cycle appears to be a regulation principle that is relevant not only for the nucleolus but also in other systems: (i) Phosphorylation affects coilin protein activity and its association with RNA in Cajal bodies differently in mitosis as compared to interphase.67,68 (ii) The assembly of histone locus bodies occurs in an RNA-dependent manner and changes from G1 to S phase as reviewed recently.69 (iii) Phosphorylation of the PRC2 (Polycomb Repressive Complex 2) component EZH2 is cell cycle-regulated and up-regulates its ability to bind RNA.70 This could be relevant for the formation and composition of distinct subnuclear foci containing PRC complexes and referred to as polycomb bodies.71
In addition, the RNA-dependent composition of nuclear subcompartments can be modulated independently of the cell cycle. Indeed, external stimuli like for example growth signals as demonstrated previously by Yang et al.72 can have similar effects. In the latter study it was found that the activity of polycomb bodies and interchromatin granules/SC35-containing splicing domains is controlled by the RNA-binding specificities of the polycomb complex Pc2 protein in dependence of its methylation state. The switch of Pc2 interactions between the TUG1 and MALALT1/NEAT2 non-coding RNAs affected the three-dimensional location of transcription units and the coordinated regulation of gene expression programs.
Deregulation of nuclear subcompartment organization is also linked to pathological phenotypes including cancer and neurodegenerative disorders.6,73,74 For example, highly proliferating tumor cells harbor larger and more active nucleoli for increased rRNAs and ribosome production.75 On the other hand, cells from patients suffering from neurodegenerative diseases often present less active nucleoli with structural aberrations.76 Accordingly, there is a renewed interest for using the nucleolus as a potential target for therapeutic treatments of cancer and other diseases.77,78 It becomes clear that further elucidating the regulation of RNA-protein interactions is crucial to understand the mechanisms underlying the assembly dynamics and the activities of the nucleolus as well as those of other nuclear bodies. Thus, further work in this direction will allow making progress in the identification of key regulators, risk factors and potential therapeutic targets.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Ingrid Grummt and Attila Németh for helpful discussions and for critical reading of the manuscript.
References
- [1].Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell 2010; 40:216-27; PMID:20965417; http://dx.doi.org/ 10.1016/j.molcel.2010.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J 2003; 22:6068-77; PMID:14609953; http://dx.doi.org/ 10.1093/emboj/cdg579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Olson MO. Sensing cellular stress: another new function for the nucleolus? Sci STKE 2004; 224:pe10.. [DOI] [PubMed] [Google Scholar]
- [4].Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D. Nucleolus: the fascinating nuclear body. Histochem Cell Biol 2008; 129:13-31; PMID:18046571; http://dx.doi.org/ 10.1007/s00418-007-0359-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol 2007; 8:574-85; PMID:17519961; http://dx.doi.org/ 10.1038/nrm2184 [DOI] [PubMed] [Google Scholar]
- [6].Tsai RY, Pederson T. Connecting the nucleolus to the cell cycle and human disease. FASEB J 2014; 28:3290-6; PMID:24790035; http://dx.doi.org/ 10.1096/fj.14-254680 [DOI] [PubMed] [Google Scholar]
- [7].Hernandez-Verdun D. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2011; 2:189-94; PMID:21818412; http://dx.doi.org/ 10.4161/nucl.2.3.16246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhu L, Brangwynne CP. Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr Opin Cell Biol 2015; 34:23-30; PMID:25942753; http://dx.doi.org/ 10.1016/j.ceb.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hyman AA, Simons K. Cell biology. Beyond oil and water–phase transitions in cells. Science 2012; 337:1047-9; PMID:22936764; http://dx.doi.org/ 10.1126/science.1223728 [DOI] [PubMed] [Google Scholar]
- [10].Berry J, Weber SC, Vaidya N, Haataja M, Brangwynne CP. RNA transcription modulates phase transition-driven nuclear body assembly. Proc Natl Acad Sci U S A 2015; 112:E5237-45; PMID:26351690; http://dx.doi.org/ 10.1073/pnas.1509317112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wachsmuth M, Caudron-Herger M, Rippe K. Genome organization: Balancing stability and plasticity. Biochim Biophys Acta 2008; 1783:2061-79; PMID:18722483; http://dx.doi.org/ 10.1016/j.bbamcr.2008.07.022 [DOI] [PubMed] [Google Scholar]
- [12].Hyman AA, Weber CA, Julicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol 2014; 30:39-58; PMID:25288112; http://dx.doi.org/ 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed] [Google Scholar]
- [13].Li P, Banjade S, Cheng H-C, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al.. Phase transitions in the assembly of multivalent signalling proteins. Nature 2012; 483:336-40; PMID:22398450; http://dx.doi.org/ 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Uversky VN, Kuznetsova IM, Turoverov KK, Zaslavsky B. Intrinsically disordered proteins as crucial constituents of cellular aqueous two phase systems and coacervates. FEBS Lett 2015; 589:15-22; PMID:25436423; http://dx.doi.org/ 10.1016/j.febslet.2014.11.028 [DOI] [PubMed] [Google Scholar]
- [15].Lin Y, Protter DS, Rosen MK, Parker R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell 2015; 60:208-19; PMID:26412307; http://dx.doi.org/ 10.1016/j.molcel.2015.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell 2012; 149:1188-91; PMID:22682242; http://dx.doi.org/ 10.1016/j.cell.2012.05.022 [DOI] [PubMed] [Google Scholar]
- [17].Caudron-Herger M, Pankert T, Seiler J, Németh A, Voit R, Grummt I, Rippe K. Alu element-containing RNAs maintain nucleolar structure and function. EMBO J 2015; 34:2758-74; PMID:26464461; http://dx.doi.org/ 10.15252/embj.201591458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol 2011; 13:167-73; PMID:21240286; http://dx.doi.org/ 10.1038/ncb2157 [DOI] [PubMed] [Google Scholar]
- [19].Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol 2011; 13:95-101; PMID:21170033; http://dx.doi.org/ 10.1038/ncb2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Carmo-Fonseca M. Assembly of the nucleolus: in need of revision. EMBO J 2015; 34:2731-2; PMID:26471727; http://dx.doi.org/ 10.15252/embj.201593185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].McKeown PC, Shaw PJ. Chromatin: linking structure and function in the nucleolus. Chromosoma 2009; 118:11-23; PMID:18925405; http://dx.doi.org/ 10.1007/s00412-008-0184-2 [DOI] [PubMed] [Google Scholar]
- [22].Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al.. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012; 149:753-67; PMID:22579281; http://dx.doi.org/ 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schwartz JC, Wang X, Podell ER, Cech TR. RNA seeds higher-order assembly of FUS protein. Cell Rep 2013; 5:918-25; PMID:24268778; http://dx.doi.org/ 10.1016/j.celrep.2013.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS. RNA controls PolyQ protein phase transitions. Mol Cell 2015; 60:220-30; PMID:26474065; http://dx.doi.org/ 10.1016/j.molcel.2015.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ugrinova I, Monier K, Ivaldi C, Thiry M, Storck S, Mongelard F, Bouvet P. Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. BMC Mol Biol 2007; 8:66-81; PMID:17692122; http://dx.doi.org/ 10.1186/1471-2199-8-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Amin MA, Matsunaga S, Uchiyama S, Fukui K. Depletion of nucleophosmin leads to distortion of nucleolar and nuclear structures in HeLa cells. Biochem J 2008; 415:345-51; PMID:18729828; http://dx.doi.org/ 10.1042/BJ20081411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Amin MA, Matsunaga S, Ma N, Takata H, Yokoyama M, Uchiyama S, Fukui K. Fibrillarin, a nucleolar protein, is required for normal nuclear morphology and cellular growth in HeLa cells. Biochem Biophys Res Commun 2007; 360:320-6; PMID:17603021; http://dx.doi.org/ 10.1016/j.bbrc.2007.06.092 [DOI] [PubMed] [Google Scholar]
- [28].Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J Cell Sci 1999; 112:761-72; PMID:10036227 [DOI] [PubMed] [Google Scholar]
- [29].Rakitina DV, Taliansky M, Brown JWS, Kalinina NO. Two RNA-binding sites in plant fibrillarin provide interactions with various RNA substrates. Nucleic Acids Res 2011; 39:8869-80; PMID:21785141; http://dx.doi.org/ 10.1093/nar/gkr594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ghisolfi L, Kharrat A, Joseph G, Amalric F, Erard M. Concerted activities of the RNA recognition and the glycine-rich C-terminal domains of nucleolin are required for efficient complex formation with pre-ribosomal RNA. Eur J Biochem 1992; 209:541-8; PMID:1425660; http://dx.doi.org/ 10.1111/j.1432-1033.1992.tb17318.x [DOI] [PubMed] [Google Scholar]
- [31].Hisaoka M, Nagata K, Okuwaki M. Intrinsically disordered regions of nucleophosmin/B23 regulate its RNA binding activity through their inter- and intra-molecular association. Nucleic Acids Res 2014; 42:1180-95; PMID:24106084; http://dx.doi.org/ 10.1093/nar/gkt897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hanakahi LA, Bu Z, Maizels N. The C-terminal domain of nucleolin accelerates nucleic acid annealing. Biochemistry 2000; 39:15493-9; PMID:11112535; http://dx.doi.org/ 10.1021/bi001683y [DOI] [PubMed] [Google Scholar]
- [33].Pellar GJ, DiMario PJ. Deletion and site-specific mutagenesis of nucleolin's carboxy GAR domain. Chromosoma 2003; 111:461-9; PMID:12707784; http://dx.doi.org/ 10.1007/s00412-003-0231-y [DOI] [PubMed] [Google Scholar]
- [34].Créancier L, Prats H, Zanibellato C, Amalric F, Bugler B. Determination of the functional domains involved in nucleolar targeting of nucleolin. Mol Biol Cell 1993; 4:1239-50; http://dx.doi.org/ 10.1091/mbc.4.12.1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Machyna M, Neugebauer KM, Stanek D. Coilin: The first 25 years. RNA Biol 2015; 12:590-6; PMID:25970135; http://dx.doi.org/ 10.1080/15476286.2015.1034923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hebert MD, Matera AG. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell 2000; 11:4159-71; PMID:11102515; http://dx.doi.org/ 10.1091/mbc.11.12.4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lee M, Passon DM, Hennig S, Fox AH, Bond CS. Construct optimization for studying protein complexes: obtaining diffraction-quality crystals of the pseudosymmetric PSPC1-NONO heterodimer. Acta Crystallogr D Biol Crystallogr 2011; 67:981-7; PMID:22101825; http://dx.doi.org/ 10.1107/S0907444911039606 [DOI] [PubMed] [Google Scholar]
- [38].Fu XD, Maniatis T. Isolation of a complementary DNA that encodes the mammalian splicing factor SC35. Science 1992; 256:535-8; PMID:1373910; http://dx.doi.org/ 10.1126/science.1373910 [DOI] [PubMed] [Google Scholar]
- [39].Frege T, Uversky VN. Intrinsically disordered proteins in the nucleus of human cells. Biochem Biophys Rep 2015; 1:33-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ma N, Matsunaga S, Takata H, Ono-Maniwa R, Uchiyama S, Fukui K. Nucleolin functions in nucleolus formation and chromosome congression. J Cell Sci 2007; 120:2091-105; PMID:17535846; http://dx.doi.org/ 10.1242/jcs.008771 [DOI] [PubMed] [Google Scholar]
- [41].Sirri V, Roussel P, Hernandez-Verdun D. The mitotically phosphorylated form of the transcription termination factor TTF-1 is associated with the repressed rDNA transcription machinery. J Cell Sci 1999; 112 (Pt 19):3259-68; PMID:10504331 [DOI] [PubMed] [Google Scholar]
- [42].Dousset T, Wang C, Verheggen C, Chen D, Hernandez-Verdun D, Huang S. Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol Biol Cell 2000; 11:2705-17; PMID:10930464; http://dx.doi.org/ 10.1091/mbc.11.8.2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ljungman M, Paulsen MT. The cyclin-dependent kinase inhibitor roscovitine inhibits RNA synthesis and triggers nuclear accumulation of p53 that is unmodified at Ser15 and Lys382. Mol Pharmacol 2001; 60:785-9; PMID:11562441 [PubMed] [Google Scholar]
- [44].Sirri V, Hernandez-Verdun D, Roussel P. Cyclin-dependent kinases govern formation and maintenance of the nucleolus. J Cell Biol 2002; 156:969-81; PMID:11901165http://dx.doi.org/ 10.1083/jcb.200201024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mitrea DM, Kriwacki RW. Phase separation in biology; functional organization of a higher order. Cell Commun Signal 2016; 14:1; PMID:26727894; http://dx.doi.org/ 10.1186/s12964-015-0125-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Parsons GG, Spencer CA. Mitotic repression of RNA polymerase II transcription is accompanied by release of transcription elongation complexes. Mol Cell Biol 1997; 17:5791-802; PMID:9315637; http://dx.doi.org/ 10.1128/MCB.17.10.5791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Emmott E, Hiscox JA. Nucleolar targeting: the hub of the matter. EMBO Rep 2009; 10:231-8; PMID:19229283; http://dx.doi.org/ 10.1038/embor.2009.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gorlich D, Prehn S, Laskey RA, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell 1994; 79:767-78; PMID:8001116; http://dx.doi.org/ 10.1016/0092-8674(94)90067-1 [DOI] [PubMed] [Google Scholar]
- [49].Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW. Ran induces spindle assembly by reversing the inhibitory effect of importin α on TPX2 activity. Cell 2001; 104:83-93; PMID:11163242; http://dx.doi.org/ 10.1016/S0092-8674(01)00193-3 [DOI] [PubMed] [Google Scholar]
- [50].Leung AK, Gerlich D, Miller G, Lyon C, Lam YW, Lleres D, Daigle N, Zomerdijk J, Ellenberg J, Lamond AI. Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J Cell Biol 2004; 166:787-800; PMID:15353547; http://dx.doi.org/ 10.1083/jcb.200405013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Aumiller WM Jr, Keating CD. Phosphorylation-mediated RNA/peptide complex coacervation as a model for intracellular liquid organelles. Nat Chem 2016; 8:129-37; PMID:26791895 [DOI] [PubMed] [Google Scholar]
- [52].Dimario PJ. Cell and molecular biology of nucleolar assembly and disassembly. Int Rev Cytol 2004; 239:99-178; PMID:15464853; http://dx.doi.org/ 10.1016/S0074-7696(04)39003-0 [DOI] [PubMed] [Google Scholar]
- [53].Calabretta S, Richard S. Emerging Roles of Disordered Sequences in RNA-Binding Proteins. Trends Biochem Sci 2015; 40:662-72; PMID:26481498; http://dx.doi.org/ 10.1016/j.tibs.2015.08.012 [DOI] [PubMed] [Google Scholar]
- [54].Liu X, Liu Z, Jang SW, Ma Z, Shinmura K, Kang S, Dong S, Chen J, Fukasawa K, Ye K. Sumoylation of nucleophosmin/B23 regulates its subcellular localization, mediating cell proliferation and survival. Proc Natl Acad Sci U S A 2007; 104:9679-84; PMID:17535915; http://dx.doi.org/ 10.1073/pnas.0701806104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lee MG, Nurse P. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature 1987; 327:31-5; PMID:3553962; http://dx.doi.org/ 10.1038/327031a0 [DOI] [PubMed] [Google Scholar]
- [56].Peter M, Nakagawa J, Dorée M, Labbé JC, Nigg EA. Identification of major nucleolar proteins as candidate mitotic substrates of cdc2 kinase. Cell 1990; 60:791-801; PMID:2178776; http://dx.doi.org/ 10.1016/0092-8674(90)90093-T [DOI] [PubMed] [Google Scholar]
- [57].Okuwaki M, Tsujimoto M, Nagata K. The RNA binding activity of a ribosome biogenesis factor, nucleophosmin/B23, is modulated by phosphorylation with a cell cycle-dependent kinase and by association with its subtype. Mol Biol Cell 2002; 13:2016-30; PMID:12058066; http://dx.doi.org/ 10.1091/mbc.02-03-0036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Belenguer P, Caizergues-Ferrer M, Labbé JC, Dorée M, Amalric F. Mitosis-specific phosphorylation of nucleolin by p34cdc2 protein kinase. Mol Cell Biol 1990; 10:3607-18; PMID:2192260; http://dx.doi.org/ 10.1128/MCB.10.7.3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Schwab MS, Gossweiler U, Dreyer C. Subcellular distribution of distinct nucleolin subfractions recognized by two monoclonal antibodies. Exp Cell Res 1998; 239:226-34; PMID:9521840; http://dx.doi.org/ 10.1006/excr.1997.3878 [DOI] [PubMed] [Google Scholar]
- [60].Falahati H, Pelham-Webb B, Blythe S, Wieschaus E. Nucleation by rRNA dictates the precision of nucleolus assembly. Curr Biol 2016; 26:277-85; PMID:26776729; http://dx.doi.org/ 10.1016/j.cub.2015.11.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bergmann JH, Spector DL. Long non-coding RNAs: modulators of nuclear structure and function. Curr Opin Cell Biol 2014; 26:10-8; PMID:24529241; http://dx.doi.org/ 10.1016/j.ceb.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Caudron-Herger M, Rippe K. Nuclear architecture by RNA. Curr Opin Genet Dev 2012; 22:179-87; PMID:22281031; http://dx.doi.org/ 10.1016/j.gde.2011.12.005 [DOI] [PubMed] [Google Scholar]
- [63].Mercer TR, Mattick JS. Understanding the regulatory and transcriptional complexity of the genome through structure. Genome Res 2013; 23:1081-8; PMID:23817049; http://dx.doi.org/ 10.1101/gr.156612.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Hancock R. A role for macromolecular crowding effects in the assembly and function of compartments in the nucleus. J Struct Biol 2004; 146:281-90; PMID:15099570; http://dx.doi.org/ 10.1016/j.jsb.2003.12.008 [DOI] [PubMed] [Google Scholar]
- [65].Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol 2003; 4:605-12; ; http://dx.doi.org/ 10.1038/nrm1172 [DOI] [PubMed] [Google Scholar]
- [66].Dundr M, Misteli T. Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol 2010; 2:a000711; PMID:21068152; http://dx.doi.org/ 10.1101/cshperspect.a000711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Carmo-Fonseca M, Ferreira J, Lamond AI. Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis–evidence that the coiled body is a kinetic nuclear structure. J Cell Biol 1993; 120:841-52; PMID:7679389; http://dx.doi.org/ 10.1083/jcb.120.4.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Broome HJ, Carrero ZI, Douglas HE, Hebert MD. Phosphorylation regulates coilin activity and RNA association. Biol Open 2013; 2:407-15; PMID:23616925; http://dx.doi.org/ 10.1242/bio.20133863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Romeo V, Schumperli D. Cycling in the nucleus: regulation of RNA 3′ processing and nuclear organization of replication-dependent histone genes. Curr Opin Cell Biol 2016; 40:23-31; PMID:26895140; http://dx.doi.org/ 10.1016/j.ceb.2016.01.015 [DOI] [PubMed] [Google Scholar]
- [70].Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev 2010; 24:2615-20; PMID:21123648; http://dx.doi.org/ 10.1101/gad.1983810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Pirrotta V, Li HB. A view of nuclear Polycomb bodies. Curr Opin Genet Dev 2012; 22:101-9; PMID:22178420; http://dx.doi.org/ 10.1016/j.gde.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 2011; 147:773-88; PMID:22078878; http://dx.doi.org/ 10.1016/j.cell.2011.08.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sleeman JE, Trinkle-Mulcahy L. Nuclear bodies: new insights into assembly/dynamics and disease relevance. Curr Opin Cell Biol 2014; 28:76-83; PMID:24704702; http://dx.doi.org/ 10.1016/j.ceb.2014.03.004 [DOI] [PubMed] [Google Scholar]
- [74].Zimber A, Nguyen QD, Gespach C. Nuclear bodies and compartments: functional roles and cellular signalling in health and disease. Cell Signal 2004; 16:1085-104; PMID:15240004; http://dx.doi.org/ 10.1016/j.cellsig.2004.03.020 [DOI] [PubMed] [Google Scholar]
- [75].Derenzini M, Trere D, Pession A, Govoni M, Sirri V, Chieco P. Nucleolar size indicates the rapidity of cell proliferation in cancer tissues. J Pathol 2000; 191:181-6; PMID:10861579; http://dx.doi.org/ 10.1002/(SICI)1096-9896(200006)191:2%3c181::AID-PATH607%3e3.0.CO;2-V [DOI] [PubMed] [Google Scholar]
- [76].Parlato R, Kreiner G. Nucleolar activity in neurodegenerative diseases: a missing piece of the puzzle? J Mol Med (Berl) 2013; 91:541-7; PMID:23179684; http://dx.doi.org/ 10.1007/s00109-012-0981-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Drygin D, O'Brien SE, Hannan RD, McArthur GA, Von Hoff DD. Targeting the nucleolus for cancer-specific activation of p53. Drug Discov Today 2014; 19:259-65; PMID:23993916; http://dx.doi.org/ 10.1016/j.drudis.2013.08.012 [DOI] [PubMed] [Google Scholar]
- [78].Burger K, Mühl B, Harasim T, Rohrmoser M, Malamoussi A, Orban M, Kellner M, Gruber-Eber A, Kremmer E, Hölzel M, et al.. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J Biol Chem 2010; 285:12416-25; PMID:20159984; http://dx.doi.org/ 10.1074/jbc.M109.074211 [DOI] [PMC free article] [PubMed] [Google Scholar]