Abstract
CD4+ T cell help is critical for optimal CD8+ T cell memory differentiation and maintenance in many experimental systems. In addition, many reports have identified reduced primary CD8+ T cell responses in the absence of CD4+ T cell help, which often coincides with reduced Ag or pathogen clearance. In this study, we demonstrate that absence of CD4+ T cells at the time of adenovirus vector immunization of mice led to immediate impairments in early CD8+ T cell functionality and differentiation. Unhelped CD8+ T cells exhibited a reduced effector phenotype, decreased ex vivo cytotoxicity, and decreased capacity to produce cytokines. This dysfunctional state was imprinted within 3 d of immunization. Unhelped CD8+ T cells expressed elevated levels of inhibitory receptors and exhibited transcriptomic exhaustion and anergy profiles by gene set enrichment analysis. Dysfunctional, impaired effector differentiation also occurred following immunization of CD4+ T cell–deficient mice with a poxvirus vector. This study demonstrates that following priming with viral vectors, CD4+ T cell help is required to promote both the expansion and acquisition of effector functions by CD8+ T cells, which is accomplished by preventing immediate dysfunction.
Introduction
CD4+ T cells are key regulators of the frequency and functionality of memory CD8+ T cells (1). There also appears a major role for CD4+ T cells in regulating primary CD8+ T cell responses, especially in the context of less inflammatory stimuli (2), as many reports identify decreased clearance of Ag in the absence of CD4+ T cells (3–10) and/or reduced frequency of IFN-γ–producing cells (5, 6, 9–14). Many of these studies report that the absence of CD4+ T cell help impairs the expansion of the primary CD8+ T cell response as measured by function-independent measures (MHC class I tetramers or frequency of TCR transgenic cells) (4–7, 11–15). Thus, the reduced Ag clearance observed may reflect the reduced frequency of primed CD8+ T cells, or these cells may also have inherent functional defects when primed in the absence of CD4+ T cells. Studies that have directly assessed the functionality of CD8+ T cells primed without CD4+ T cell help report, at most, minor functional alterations (12, 16–19). Thus, the extent to which CD4+ T cells promote functional primary CD8+ T cell responses independent of regulating the magnitude of the response remains to be clarified.
Following replication-incompetent adenovirus (Ad) vector immunization, unhelped CD8+ T cells fail to express effector phenotype markers (20) and display impaired primary CD8+ T cell expansion (13, 20–22). Viral vector vaccines, including Ad vectors, are being intensively studied as candidate vaccine platforms against an array of pathogens (23–30) and, thus, represent clinically relevant tools for probing immune regulatory pathways. Given the phenotypic alterations of unhelped Ad vector–elicited CD8+ T cell responses, we sought to determine to what degree this reflects functional and transcriptional alterations and to identify pathways regulating these Ad vector–elicited responses. Thus, further investigation in this area provides the opportunity to more clearly elucidate the role of CD4+ T cells in regulating CD8+ T cell effector differentiation.
Following vaccination or acutely controlled infection, CD8+ T cells differentiate into two distinct highly functional effector and memory populations (31). In contrast, when the stimulatory environment is not optimal, CD8+ T cells can become dysfunctional. In the context of chronic Ag exposure and inflammatory signals, CD8+ T cells become exhausted and progressively lose functionality (32). Or, if the priming environment lacks key signals, then CD8+ T cells immediately become anergic (33). These two dysfunctional states represent temporally distinct phenomena and are driven by distinct transcriptional programs (34), but both represent states of T cell hypofunctionality. Although atypical differentiation of Ad vector-elicited CD8+ T cells primed without CD4+ T cell help is observed (20), it is unclear when this initially occurs and how profoundly the functionality of these unhelped cells are altered compared with other well-described states of hypofunctionality. Thus, a more detailed investigation of the timing of when and how CD4+ T cells regulate Ad vector–elicited CD8+ T cell differentiation is required.
In this study, we sought to clarify the role of CD4+ T cells in immediate regulation of CD8+ T cell functionality by a thorough investigation of the functionality, transcriptional state, and phenotype of unhelped CD8+ T cells. CD4+ T cell help is needed at priming and absence of CD4+ T cells induces defects in CD8+ T cell differentiation that are observed within days of immunization. We demonstrate that in the absence of CD4+ T cell help CD8+ T cells induced by vaccination with replication-incompetent adenovirus and poxvirus vectors differentiate to a dysfunctional state, which involves hypoeffector functionality and mirrors many of the characteristics of CD8+ T cell exhaustion. CD8+ T cells express both exhaustion and anergy transcriptional signatures, which appears to be driven by excessive AP-1–independent NFAT signaling. Functionally, impaired IL-2 signaling in the absence of CD4+ T cells, which leads to elevated expression of programmed death-1 (PD-1), appears to contribute to the dysfunction of these CD8+ T cells. In sum, we identify an immediate need for CD4+ T cells in programming effector differentiation and preventing exhaustion-like dysfunction of CD8+ T cells following viral vector immunization.
Materials and Methods
Mice and vectors
Six- to ten-week-old C57BL/6, B6.SJL-ptprca (CD45.1+), B6.129S2-Cd4tm1Mak/J (CD4 knockout [KO]), B6.129S2-H2dlAb1-Ea/J (MHC II KO), C57BL/6-Tg(TcraTcrb)1100Mjb/J (OT-I), and B6.Cg-Tg(TcraTcrb)425Cbn/J (OT-II) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Thymectomized animals underwent adult thymectomy at The Jackson Laboratory. E1/E3 deleted Ad serotype 5 (Ad5)-SIINFEKL-Luc, Ad5-OVA, and Ad5HVR48(1–7)–SIVmac239 Gag (Gag) have been described previously (35–38). Mice were immunized i.m. with 109 viral particles of the indicated vector in the quadriceps using 100 μl divided equally between the two legs. Attenuated Copenhagen vaccinia strain–Env (NYVAC–SIVmac239 Env) has been previously described (39) and was used at 106 PFU/mouse. All animal experiments were performed in accordance with the Institutional Animal Care and Use Committee guidelines of Beth Israel Deaconess Medical Center.
mAb administration
The monoclonal anti-CD4 Ab (GK1.5; BioXCell) was administered by two i.p. injections of 500 μg 1 d prior to immunization and on the day of immunization. To maintain CD4 T cell depletion, this injection regimen was repeated every 21 d following immunization. Anti–PD-ligand 1 (PD-L1) (10F.9G2; BioXCell), anti–PD-1 (29F.1A12; BioLegend), or isotype IgG2b (clone LTF-2; BioXCell) were administered by 200 μg i.p. injections every third day, as described previously (40).
rIL-2 administration
Mouse rIL-2 (R&D Systems) resuspended in 0.1% normal mouse serum was given 1 μg i.p. twice daily from days 3 to 12, according to published dosing protocols (41, 42).
Tissue processing
Single-cell suspensions of splenocytes and lymph nodes were generated by grinding the tissue through a 70-μm filter (Fisher Scientific). Tissue RBCs were lysed by ammonium-chloride-potassium treatment for 3 min. Cellular debris was further clarified by passage through a 30-μm filter (Miltenyi Biotec). For purification of PBMCs, Ficoll–Histopaque density centrifugation was performed at 1900 rpm for 20 min.
Flow cytometery
Identification of Ag-specific cells by MHC class I tetramer staining was performed using either H-2Db tetramer loaded with the immunodominant AL11 peptide (AAVKNWMTQTL) of Gag (43) or H-2Kb tetramer loaded with the immunodominant OVA257–264 (SIINFEKL) epitope from chicken OVA (44). Biotinylated class I monomers were provided by the NIH Tetramer Core Facility (Emory University, Atlanta, GA). Background staining of cells from naive animals was ≤0.1%. Surface staining was performed for 30 min at 4°C. The following Abs were used for staining: anti-CD8α (53-6.7), -CD4 (RM4-5), -CD44 (IM7), -CD45.2 (104), -CD45.1 (A20), -TCRα V2 (B20.1), -CD127 (A7R34), -killer cell lectin–like receptor G1 (KLRG1) (2F1), -CD27 (LG.3A10), -CD43 (1B11), -PD-1 (RMP1-30), -Tim-3 (RMT3-23), -LAG-3 (C9B7W), -2B4 (eBio244F4), -CD25 (PC61), -CD28 (37.51), -OX-40 (OX-86), -4-1BB (17B5), -CD71 (RI7217), -CD98 (RL388), -CD11a (2D7), and -CD69 (H1.2F3). For detection of granzyme B, cells were surface stained and then permeabilized for 15 min at room temperature with Cytofix/Cytoperm (BD Biosciences). Permeabilized cells were subsequently stained with anti-granzyme B (GB11). Ki-67 detection was performed using BD Perm Buffer 2 (BD Biosciences) for 15 min at room temperature, followed by anti–Ki-67 (B56) staining. For intracellular cytokine staining, cells were incubated for 5 h at 37°C with 2 μg/ml OVA257–264 peptide (AnaSpec) or 1 μg/ml overlapping SIVmac239 Env peptide pool (NIH AIDS Reagent Program). At the time of peptide incubation, brefeldin A and monensin (BD Biosciences) and anti-CD107a (ID4B) were added. After peptide incubation, cells were surface stained, washed, permeabilized with Cytofix/Cytoperm (BD Biosciences) for 20 min at 4°C, and subsequently stained with anti–IFN-γ (XMG1.2) and –TNF-α (MP6-XT22). All Abs were purchased from BD Biosciences, eBioscience, BioLegend, or Life Technologies. Dead cells were excluded by use of vital exclusion dye (Life Technologies). Annexin V staining was performed using an Annexin V staining kit (BioLegend). Samples were acquired on an LSR II flow cytometer (BD Biosciences) and analyzed using FlowJo version 9.7.2 (Tree Star).
Ex vivo killing assay
Ex vivo cytotoxic potential of CD8+ T cells was measured as described previously (45). Briefly, EL-4 cells (American Type Culture Collection) were pulsed with 2 μM OVA257–264 peptide (AnaSpec) or no peptide for 1 h at 37°C. Peptide-pulsed cells were stained with CFSE (Life Technologies), and unpulsed cells were stained with CellTrace Violet (Life Technologies) according to the manufacturer’s instructions. CD8+ T cells were enriched by negative selection using the EasySep Mouse CD8+ T cell enrichment kit (StemCell Technologies). The number of effector cells was determined by quantification of Kb/OVA+CD8+ T cells, the appropriate numbers were added, and the mixtures were incubated for 5 h at 37°C. Following incubation, cells were stained with a vital exclusion dye (Life Technologies) to exclude dead cells. Fixed cells were acquired on an LSR II flow cytometer (BD Biosciences) and analyzed using FlowJo version 9.7.2 (Tree Star). Specific lysis was calculated as 100 − [100 × (percent survival/average percent survival in absence of effector cells)].
In vivo luciferase imaging
Quantification of luciferase transgene expression was performed as described previously (37, 46). Briefly, hair was removed from the hind legs and ventral posterior area of the mouse. Mice were injected i.p. with 150 μl of 30 mg/ml Luciferin (Caliper Life Sciences) according to the manufacturer’s instructions. Mice were anesthetized by 4% isofluorane inhalation and maintained under anesthesia by nose cone and 1.5% isofluorane. Luminescence was quantitated using an IVIS Lumina II charge-coupled device imaging system and Living Image software (Caliper Life Sciences). Image integration time was 240 s, f/stop was 1.2, and binning was large.
Microarray
Gene expression profiling was performed as described previously (47, 48). Briefly, CD8+ T cells were enriched by negative selection using the CD8+ T cell isolation kit II (Miltenyi Biotec). Db/AL11+CD44+CD8+ T cells were subsequently sorted to >95% purity on a FACSAria (BD Biosciences). Sorted cells were stored at −80°C in 1 ml of TRIzol (Life Sciences). RNA extraction was performed using the RNAdvance Tissue Isolation kit (Agencourt), according to the manufacturer’s instructions. The cDNA synthesis was performed using the Ovation Pico WTA v2 kit (NuGEN), according to the manufacturer’s instructions. Proper amplification of cDNA was confirmed using an Agilent 2100 Bioanalyzer (Agilent Technologies) and performed by the Harvard Biopolymers Facility. To confirm optimal amplification of mRNA and absence of contaminating gDNA (data not shown), quantitative RT-PCR for Rn18s (RefSeq: NR_003278.2) and mouse RT2-qPCR primer control (Qiagen) were performed using the SYBR Green Quantification System (Qiagen). Data were acquired on a StepOnePlus Real-Time PCR System (Applied Biosystems). Cycle conditions were as follows: 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. cDNA was subsequently fragmented and biotinylated using an Encore Biotin Module 4200 (NuGEN). cDNA was hybridized to Mouse Genome 430 version 2.0 chip (Affymetrix) at the Microarray Core of Dana Farber Cancer Institute. The Robust Multiarray Average method was used to process data image files using GenePattern (Broad Institute).
Functional enrichment and network analysis
Differential gene expression was determined using GENE-E version 3.0.163 (Broad Institute). Functional enrichment, pathway, and network analyses were performed using genes ranked by adjusted p values, fold change, and modified t-statistics from limma analysis (49). Gene set enrichment analysis (GSEA) was performed using GSEA software (http://www.broadinstitute.org/gsea). All 45,282 probes, ranked by limma-derived modified t statistics, were imported into the GSEA software. We tested for the enrichment of C2 and C7 and C3.tft Molecular Signatures Database. GSEA was run according to default parameters: probes for the same gene were collapsed into a single gene symbol (identified by its HUGO gene symbol), permutation number = 1000, and permutation type = gene sets. By convention, a nominal p value <0.05 was used as the cutoff value for significance. Enriched gene sets were grouped into modules according to shared leading edge genes using the cytoscape plugin EnrichmentMap (http://www.cytoscape.org/) by calculating the Jaccard index between pathways using a threshold of 0.50. The Jaccard index characterizes the overlap between two gene sets, relative to the size of their union and stratifies gene sets that correspond to similar biological processes.
Statistical analysis
Statistical analysis was performed using a two-tailed non-parametric Mann–Whitney U test in Prism version 6.0f (GraphPad Software).
Accession number
Microarray data have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE73001.
Results
Depletion of CD4+ T cells at the time of immunization results in aberrant CD8+ T cell differentiation with decreased effector function
To investigate the role of CD4+ T cells in controlling the early stages of the CD8+ T cell response following Ad vector immunization, we used an Ad5-based immunization regimen. This vaccination regimen generates small, but measurable, CD8+ T cell responses in the absence of CD4+ T cell help (13, 20, 21). Mice were depleted of CD4+ T cells by two sequential doses of the anti-CD4 Ab (clone GK1.5) beginning on the day prior to immunization, and CD4+ T cell depletion was maintained by readministration of the Ab every 21 d. This regimen led to complete and sustained depletion of CD4+ T cells (data not shown). Following i.m. immunization with Ad5-SIINFEKL-Luc, there was a 10-fold reduction in the frequency of Kb/OVA+CD8+ T cells in the anti-CD4–treated mice at the peak of the response compared with untreated control animals (Fig. 1A).
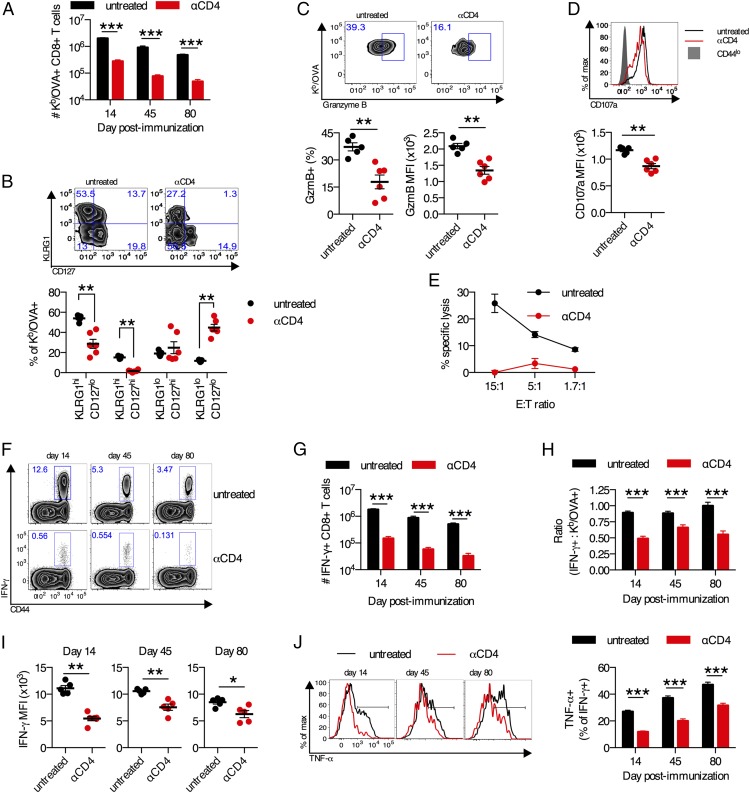
FIGURE 1.
Aberrant differentiation of CD8+ T cells primed in the absence of CD4+ T cells. (A–J) C57BL/6 mice were treated with anti-CD4 Ab or left untreated and immunized i.m. with 109 viral particles of Ad5-SIINFEKL-Luc. (A) Frequency of Kb/OVA+CD8+ T cells in the spleen. (B) KLRG1 and CD127 expression on Kb/OVA+ CD8+ T cells in the spleen on day 14 postimmunization. (C) Expression of granzyme B by percentage and mean fluorescence intensity (MFI) on Kb/OVA+CD8+ T cells in the spleen on day 14 postimmunization. (D) Expression of CD107a on IFN-γ+CD8+ T cells following OVA257–264 peptide stimulation of splenocytes from day 14 postimmunization. (E) Five-hour killing assay of OVA257–264 peptide–pulsed EL-4 cells by pooled splenic CD8+ T cells at day 14 postimmunization. Each E:T ratio was performed in duplicate. (F) Representative plots of IFN-γ+CD8+ T cells from the spleen. (G) Absolute frequency of IFN-γ+CD8+ T cells. (H) The fraction of OVA257–264-specific CD8+ T cells that produce IFN-γ. (I) MFI of IFN-γ+CD8+ T cells. (J) Representative plots of TNF-α production by IFN-γ+CD8+ T cells and group averages. Data are representative of three experiments (B–E and I) or pooled from three experiments (A, F–H, and J). n = 5–6 per group per experiment. Mean ± SEM are shown. ***p < 0.001, **p < 0.01, *p < 0.05, Mann–Whitney U test.
We first sought to determine whether the absence of CD4+ T cell help at the time of priming altered effector CD8+ T cell differentiation. Differentiation into memory precursor or terminal effector cells was determined by the expression of CD127 and KLRG1, respectively (50–52). On day 14 postimmunization, Kb/OVA+CD8+ T cells from anti-CD4–treated mice exhibited a significant impairment in KLRG1 upregulation (p < 0.01), and ∼50% of OVA257–264-specific CD8+ T cells were undifferentiated KLRG1loCD127lo cells (p < 0.01; Fig. 1B). A recent report has identified an alternative scheme for identifying effector phenotype cells as CD43loCD27lo (53). This CD43loCD27lo subset was also significantly reduced in anti-CD4–treated mice compared with controls (p < 0.01; data not shown). Consistent with these phenotypic data, we observed decreased expression of granzyme B (p < 0.01; Fig. 1C) and decreased CD107a expression (upon cognate peptide restimulation; p < 0.01; Fig. 1D) on OVA-specific CD8+ T cells from anti-CD4–treated mice. Furthermore, in a 5-h ex vivo cytotoxicity assay, Kb/OVA+ CD8+ T cells from anti-CD4–treated mice displayed a major defect in cytotoxicity (Fig. 1E). Collectively, these data demonstrate a major defect in CD8+ T cell differentiation in the absence CD4+ T cell help.
Given the impaired effector phenotype and cytotoxicity of CD8+ T cells primed without CD4+ T cell help, we sought to determine whether these cells displayed alterations in their ability to secrete cytokines. There was a significant reduction in the frequency and absolute number of CD8+ T cells that produced IFN-γ upon ex vivo restimulation in mice treated with anti-CD4 Ab compared with untreated control mice on day 14 (12-fold reduction), day 45 (15-fold reduction), and day 80 (15.5-fold reduction) postimmunization (p < 0.001; Fig. 1F, 1G). The frequency of Ag-specific CD8+ T cells that express IFN-γ was substantially reduced in anti-CD4–treated mice (p < 0.001; Fig. 1H), and these cells produced less per-cell IFN-γ than did cells from untreated control animals (p < 0.05; Fig. 1I). Finally, the fraction of IFN-γ+CD8+ T cells from anti-CD4–treated mice that were capable of coproducing TNF-α was significantly reduced compared with IFN-γ+CD8+ T cells from untreated control animals (p < 0.001; Fig. 1J). These results were confirmed using CD4 KO and MHC class II KO mice, which displayed similar defects in IFN-γ and TNF-α production (data not shown). These data show that, without CD4+ T cell help at priming, Ad vector–elicited CD8+ T cells undergo aberrant differentiation consisting of impaired effector phenotype, decreased cytotoxicity, and decreased expression of effector cytokines.
CD8+ T cells primed without CD4+ T cell help express multiple inhibitory receptors
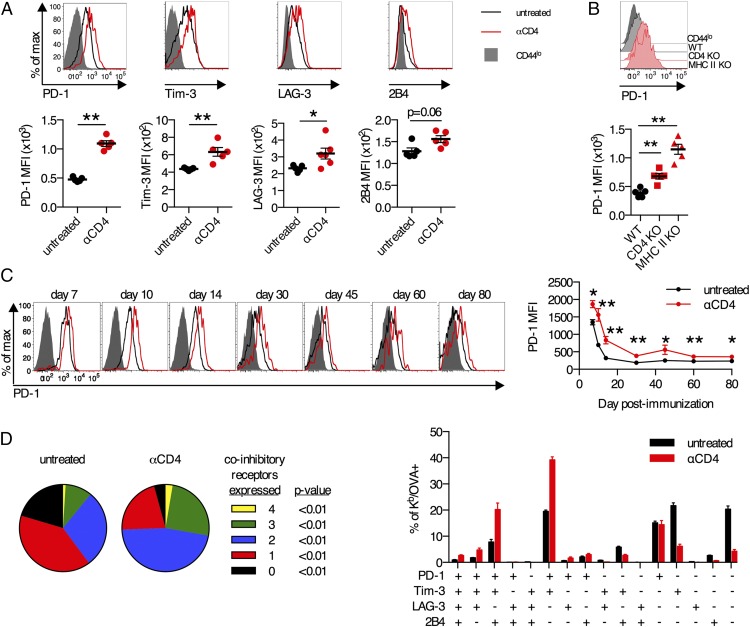
To better understand the atypical differentiation of unhelped CD8+ T cells, we also examined the expression of inhibitory receptors on Ag-specific CD8+ T cells following immunization of anti-CD4–treated mice. There was a substantial increase in the expression of PD-1 on Kb/OVA+CD8+ T cells from anti-CD4–treated mice compared with untreated controls (p < 0.01; Fig. 2A). Increased expression of Tim-3, LAG-3, and 2B4 was also observed in anti-CD4–treated mice (Fig. 2A). Elevated PD-1 expression was also observed on Kb/OVA+ CD8+ T cells from CD4 KO and MHC class II KO mice (p < 0.01; Fig. 2B). In both untreated controls and anti-CD4–treated mice, PD-1 expression on Kb/OVA+ CD8+ T cells in the blood declined from days 7 to 14 and then remained steady through day 80, and at all times, PD-1 expression was significantly higher in anti-CD4–treated mice (p < 0.05; Fig. 2C). Increased coexpression of multiple inhibitory receptors has been shown to correlate with reduced functionality of CD8+ T cells in the context of chronic viral infection (54). Kb/OVA+CD8+ T cells from anti-CD4–treated mice exhibited significant increases (p < 0.01) in the fraction of cells that coexpressed two, three, or all four of the inhibitory receptors PD-1, Tim-3, LAG-3, and 2B4 compared with untreated controls (Fig. 2C, 2D). Thus, CD8+ T cells primed in the absence of CD4+ T cell help exhibit increased and prolonged expression of inhibitory receptors.
FIGURE 2.
Elevated expression of inhibitory receptors on CD8+ T cells primed in the absence of CD4+ T cells. (A, C, and D) C57BL/6 mice were treated with anti-CD4 Ab or left untreated and immunized i.m. with 109 viral particles (vp) of Ad5-SIINFEKL-Luc. (A) Expression of inhibitory receptors PD-1, Tim-3, LAG-3, and 2B4 on Kb/OVA+CD8+ T cells in the spleen on day 14 postimmunization. (B) C57BL/6, CD4 KO, or MHC II KO mice were immunized i.m. with 109 vp of Ad5-SIINFEKL-Luc. PD-1 expression on Kb/OVA+CD8+ T cells in the blood on day 14 postimmunization. (C) Longitudinal expression of PD-1 on Kb/OVA+CD8+ T cells in the blood. (D) Coexpression of inhibitory receptors PD-1, Tim-3, LAG-3, and 2B4 on Kb/OVA+CD8+ T cells in the spleen on day 14 postimmunization, either as number of inhibitory receptors expressed (left) or specific combinations (right). Data are representative of three experiments (n = 5–6 per group per experiment) (A, C, and D) or two experiments (n = 5 per group per experiment) (B). Mean ± SEM are shown. **p < 0.01, *p < 0.05, Mann–Whitney U test.
CD8+ T cells primed in the absence of CD4+ T cells by poxvirus vector immunization have aberrant effector characteristics
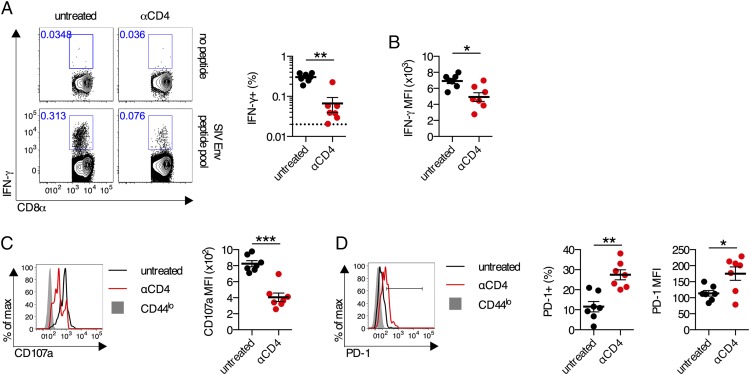
To determine whether aberrant CD8+ T cell differentiation in the absence of CD4+ T cells is generalizable to multiple viral vector–based immunization regimens, we used a poxvirus vector-based immunization regimen. Following immunization with a replication-incompetent recombinant poxvirus vector expressing an SIV Env transgene (NYVAC-Env), Env-specific CD8+ T cell responses from anti-CD4–treated mice exhibited a significantly reduced peak magnitude compared with untreated controls (p < 0.01; Fig. 3A). The responding IFN-γ+ cells produced less IFN-γ on a per-cell basis when CD4+ T cells were absent (p < 0.05; Fig. 3B). In addition, in the absence of CD4+ T cells, these responding cells displayed a significantly less cytotoxic phenotype, as measured by CD107a expression (p < 0.001; Fig. 3C). Unhelped Env-specific CD8+ T cells also exhibited elevated PD-1 expression (p < 0.05; Fig. 3D). These data demonstrate that CD8+ T cell effector dysfunction following viral vector immunization in the absence of CD4+ T cells is generalizable to multiple replication-incompetent viral vector platforms.
FIGURE 3.
Aberrant differentiation of CD8+ T cells in the absence of CD4 T cells following poxvirus vector immunization. (A–D) C57BL/6 mice were treated with anti-CD4 Ab or left untreated and immunized i.m. with 106 PFU of NYVAC-Env (n = 7 per group). (A) Representative flow plots and frequency of IFN-γ+ Env-specific CD8+ T cells in the spleen on day 14. (B) Per-cell production of IFN-γ by IFN-γ+CD8+ T cells as measured by mean fluorescence intensity (MFI). (C) Representative flow plots and group averages of CD107a expression by IFN-γ+CD8+ T cells upon Env peptide pool stimulation. (D) Representative flow plots and group averages of PD-1 expression on IFN-γ+CD8+ T cells as measured by percentage and MFI. Mean ± SEM are shown. ***p < 0.001, **p < 0.01, *p < 0.05, Mann–Whitney U test.
Absence of CD4+ T cells induces early transcriptional signatures of T cell exhaustion and anergy in CD8+ T cells
Given the multifaceted abnormalities of CD8+ T cells primed without CD4+ T cell help, we sought to understand the transcriptional state of these cells. To examine this, mice were treated with anti-CD4 or left untreated and immunized with Ad5HVR48-Gag, which also induces dysfunctional CD8+ T cells in the absence of CD4+ T cell help (data not shown). On day 14 postimmunization, Db/AL11+ CD8+ T cells from the spleen of untreated control or anti-CD4–treated mice were sorted, and gene expression profiling was performed. Highly upregulated genes in Db/AL11+CD8+ T cells from anti-CD4–treated mice compared with untreated controls were Havcr2 (encodes Tim-3), Pdcd1 (encodes PD-1), Cd244 (encodes 2B4), Lag3 (encodes LAG-3), and Ctla4 (encodes CTLA4) (Fig. 4A, Supplemental Table I). In contrast, highly upregulated genes in Db/AL11+CD8+ T cells from untreated control mice compared with anti-CD4–treated were Klrg1 (encodes KLRG1), Gzmm (encodes granzyme M), and Gzma (encodes granzyme A) (Fig. 4A, Supplemental Table I). These results highlight the overexpression of inhibitory markers by CD8+ T cells as a key difference in the absence of CD4+ T cell help.
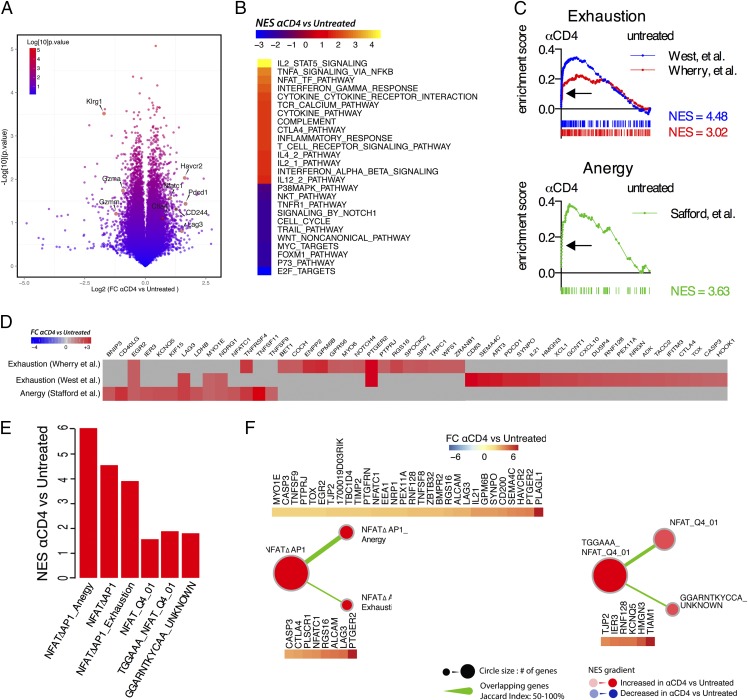
FIGURE 4.
Transcriptional signatures of exhaustion and anergy in CD8+ T cells primed in the absence of CD4+ T cells. (A–F) C57BL/6 mice (n = 4/group) were treated with anti-CD4 Ab or left untreated and immunized i.m. with 109 viral particles of Ad5HVR48-Gag. Gene expression profiling of cDNA from sorted Db/AL11+ CD8+ T cells from day 14 postimmunization was performed. (A) Volcano plot of differentially expressed genes between Db/AL11+ CD8+ T cells from untreated or anti-CD4 Ab–treated animals. Select genes are highlighted. (B) GSEA was performed on the C2 dataset from the MSigDB (Broad Institute) and select gene signatures of interest with a false discovery rate [FDR] < 0.05 are shown). (C) GSEA was performed of two different molecular signatures of exhaustion (GSE9650 (57) and GSE30962 (56)) and one signature of anergy (GSE2323 (86)). FDR < 0.05 for all gene signatures. (D) Genes within the leading edge of the gene signatures from (C). (E and F) GSEA using gene sets composed of NFAT target genes and overlap and shared enriched genes in gene signatures. NFATΔAP-1 denotes AP-1–independent NFAT target genes as described in Ref. 58.
To further probe the concerted transcriptional differences between CD8+ T cells primed with and without CD4+ T cell help, we used GSEA (55). GSEA analysis identified multiple cell cycle pathways that were downregulated in Db/AL11+CD8+ T cells from anti-CD4–treated mice (Fig. 4B, Supplemental Table II). Consistent with this, reduced proliferation of Db/AL11+CD8+ T cells from anti-CD4–treated mice was confirmed experimentally (data not shown). Conversely, multiple TCR signaling, cytokine signaling, and cell death pathways were enriched in Db/AL11+CD8+ T cells from anti-CD4–treated mice (Fig. 4B, Supplemental Table II). Increased apoptosis of Db/AL11+CD8+ T cells from anti-CD4–treated mice was also confirmed experimentally (data not shown). Taken together, the decreased proliferation and increased apoptosis of unhelped Db/AL11+CD8+ T cells explains their reduced frequencies.
The molecular signatures of CD8+ T cell exhaustion from two previous studies (56, 57) were significantly enriched in Db/AL11+CD8+ T cells from anti-CD4–treated mice (Fig. 4C, Supplemental Table III). The converse effector gene signature from each study was significantly enriched in Db/AL11+CD8+ T cells from untreated control animals (Supplemental Table III). A molecular signature of T cell anergy was also significantly enriched in Db/AL11+CD8+ T cells from anti-CD4–treated mice (Fig. 4C). Leading edge analysis of enriched genes from the exhaustion and anergy signatures identified that these two signatures were largely nonoverlapping (Fig. 4D), which suggests the simultaneous expression of two distinct transcriptional programs in these cells. The transcriptional signature of exhaustion in CD8+ T cells primed without CD4+ T cell help is consistent with the elevated expression of inhibitory receptors and hypofunctionality, and the transcriptional signature of anergy is consistent with the apparent rapidity of this dysfunction.
A recent report (58) has demonstrated that the transcription factor NFAT when acting independently of AP-1 can induce expression of exhaustion- and anergy-associated genes. NFATc1 was substantially overexpressed in Db/AL11+CD8+ T cells from anti-CD4–treated mice (Fig. 4A), and these cells were enriched for signatures of NFAT signaling (Fig. 4B, Supplemental Table II). We thus hypothesized that AP-1–independent NFAT signaling (in this article termed NFATΔAP-1) was driving the simultaneous expression of exhaustion and anergy transcriptional signatures. Consistent with this hypothesis, the NFATΔAP-1 gene signature was enriched in cells from anti-CD4–treated mice, as were the specific subsets of genes previously shown to be regulated by NFATΔAP-1 and involved in anergy or exhaustion (Fig. 4E, 4F, Supplemental Table IV). An additional signature for non-canonical NFAT signaling was also significantly enriched (Fig. 4E, 4F). Of note, no AP-1 target pathways were enriched in cells from anti-CD4–treated mice compared with untreated controls (data not shown). These data are consistent with a model where atypical NFAT signaling in Ag-specific CD8+ T cells from anti-CD4–treated mice appears to drive, in part, the simultaneous expression of exhaustion and anergy gene signatures in these cells, which transcriptionally underpins the dysfunctional effector differentiation of these cells.
Blockade of PD-1 signaling during priming partially rescues the CD8+ T cell response generated without CD4+ T cell help
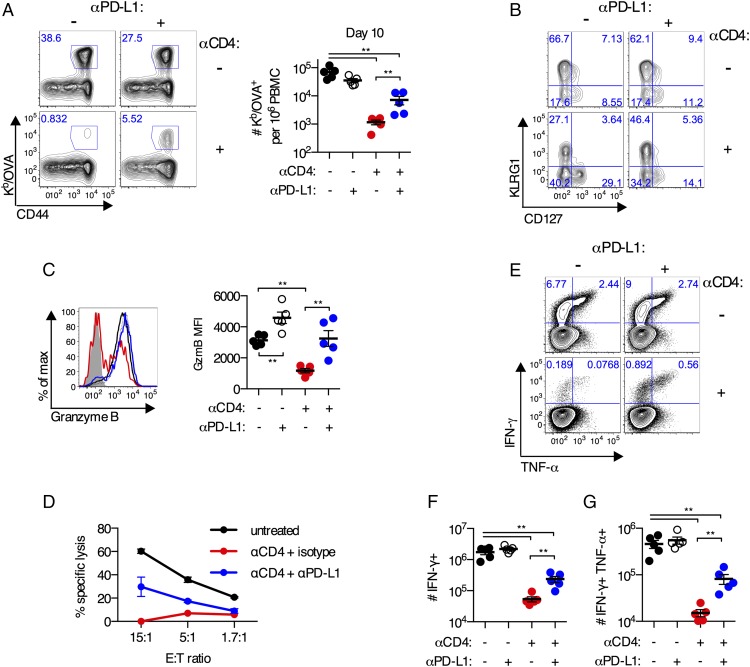
Given the elevated expression of PD-1 on CD8+ T cells primed without CD4+ T cell help and the expression of an exhaustion transcriptional signature, we hypothesized that blockade of PD-1 might rescue the differentiation of these cells, as has been reported for chronic infections or cancer (40, 59–62). Indeed, blockade of PD-1 signaling (via anti–PD-L1 Ab) led to a partial rescue (6-fold; p < 0.01) in the accumulation of Kb/OVA+CD8+ T cells primed in the absence of CD4+ T cell help (Fig. 5A). Administration of anti–PD-L1 Ab significantly increased the fraction of unhelped Kb/OVA+CD8+ T cells that expressed a KLRG1hiCD127lo phenotype (isotype Ab: 24 ± 4% versus anti–PD-L1 Ab: 48 ± 3%; p < 0.01; Fig. 5B) and decreased the fraction of cells that were in an undifferentiated KLRG1loCD127lo state (isotype Ab: 40 ± 3% versus anti–PD-L1 Ab: 32 ± 2%; p = 0.06; Fig. 5B). Blockade of PD-1 signaling at the time of priming improved the accumulation and phenotype of Ag-specific CD8+ T cells primed without CD4+ T cell help.
FIGURE 5.
Anti–PD-L1 Ab treatment partially rescues differentiation of CD8+ T cells primed in the absence of CD4+ T cells. (A–G) C57BL/6 mice were treated with anti-CD4 Ab or left untreated and immunized i.m. with 109 viral particles of Ad5-SIINFEKL-Luc. Mice were administered anti–PD-L1 or isotype control Ab every 3 d beginning 1 d prior to immunization. (A) Representative plots and absolute number of Kb/OVA+CD8+ T cells in the blood on day 10 postimmunization. (B) KLRG1 and CD127 expression on Kb/OVA+CD8+ T cells in the blood on day 10 postimmunization. (C) Expression of granzyme B on Kb/OVA+ CD8+ T cells in the blood on day 10 postimmunization. (D) Five-hour killing assay of OVA257–264 peptide–pulsed EL-4 cells by pooled CD8+ T cells from day 10 postimmunization. E:T ratio was performed in duplicate for untreated and anti-CD4+ anti–PD-L1 and singlet for anti-CD4+isotype. (E) Representative plots of IFN-γ and TNF-α by CD8+ T cells from the spleen upon ex vivo stimulation with OVA257–264 peptide. (F) Absolute number of OVA257–264-specific CD8+ T cells that produce IFN-γ. (G) Absolute number of OVA257–264-specific CD8+ T cells that coproduce IFN-γ and TNF-α. Data are representative of two to four experiments per group (n = 5–8 per group per experiment). Mean ± SEM are shown. **p < 0.01, Mann–Whitney U test.
Blockade of PD-1 signaling also dramatically improved the functionality of CD8+ T cells primed in the absence of CD4+ T cell help. Anti–PD-L1 Ab treatment rescued the defect in granzyme B expression observed in unhelped Kb/OVA+CD8+ T cells (p < 0.01; Fig. 5C). This coincided with a substantial increase in per-cell cytotoxicity as measured using an ex vivo killing assay (Fig. 5D). The fraction and number of IFN-γ+CD8+ T cells was significantly increased in anti–PD-L1 Ab–treated mice compared with isotype controls (p < 0.01; Fig. 5E, 5F). Finally, anti–PD-L1 treatment lead to a partial rescue in the ability of IFN-γ+CD8+ T cells to coproduce TNF-α compared with mice that received an isotype control Ab (p < 0.01; Fig. 5E, 5G). Treatment with an anti–PD-1 Ab recapitulated the effect of treatment with an anti–PD-L1 Ab for all parameters analyzed (data not shown). These data demonstrate that in the absence of CD4+ T cells increased PD-1 signaling impairs the functionality of Ad vector–elicited CD8+ T cell responses.
Provision of CD4+ T cell help within days of immunization does not prevent dysfunction of CD8+ T cells
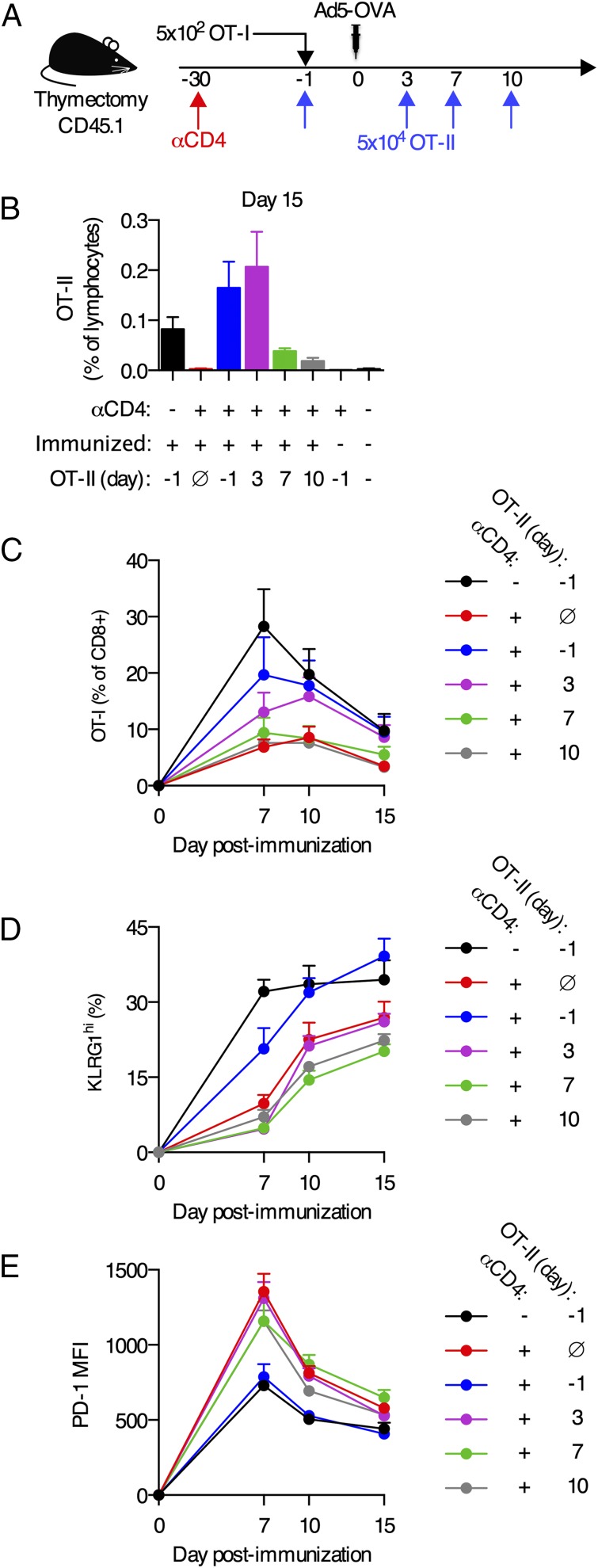
To better understand how CD4+ T cells prevented CD8+ T cell dysfunction, we sought to determine whether the defects of CD8+ T cells primed without CD4+ T cell help were imprinted at priming or whether the atypical differentiation was a progressive process. We experimentally reconstituted CD4+ T cell help after immunization to explore whether this could rescue the dysfunctional state of CD8+ T cells primed in the absence of CD4 T cells, using a recently described experimental system (63, 64). CD45.1+ congenic mice underwent adult thymectomy and were treated with anti-CD4 Ab or left untreated as a control (Fig. 6A). One month after anti-CD4 Ab treatment, no anti-CD4 Ab was detectable in the serum (data not shown). A total of 5 × 102 naive OT-I CD8+ T cells (specific for the Kb/OVA257–264 epitope) were adoptively transferred into all animals, and mice either received no adoptively transferred CD4+ T cells or 5 × 104 naive OT-II CD4+ T cells (specific for the I-Ab/OVA323–339 epitope) on either day −1, 3, 7, or 10 postimmunization (Fig. 6A). We confirmed that transferred OT-II CD4+ T cells were primed and expanded in all experimental groups because the frequency of OT-II CD4+ T cells was significantly greater than mice that received OT-II CD4+ T cells and were left unimmunized (p < 0.05; Fig. 6B), and all OT-II CD4+ T cells were CD44hi (data not shown).
FIGURE 6.
Provision of CD4+ T cell help after immunization does not rescue the dysfunctional phenotype of CD8+ T cells primed without CD4+ T cell help. (A) Experimental schematic. CD45.1+ C57BL/6 mice underwent adult thymectomy and were treated with anti-CD4 Ab or left untreated 30 d prior to immunization. A total of 5 × 102 naive CD45.2+ OT-I CD8+ T cells were adoptively transferred 1 d prior to immunization. Mice were immunized i.m. with 109 viral particles of Ad5-OVA. A total of 5 × 104 naive CD45.2+OT-II CD4+ T cells were adoptively transferred on day −1, 3, 7, or 10 postimmunization. For one control group, no OT-II CD4+ T cells were transferred. (B) Frequency of CD45.2+OT-II CD4+ T cells in the blood on day 15 postimmunization. (C) Frequency of OT-I CD8+ T cells in the blood. (D) Fraction of blood OT-I CD8+ T cells expressing KLRG1. (E) PD-1 expression on blood OT-I CD8+ T cells. Data are pooled from two experiments (B–D), expect for (B) where unimmunized mice are from a single experiment or representative of two experiments (E). n = 4–5 per group per experiment. Mean + SEM are shown.
Mice that received anti-CD4 Ab and did not receive adoptive transfer of CD4+ T cells had OT-I CD8+ T cell responses of a reduced magnitude, and the responding cells expressed less KLRG1 and more PD-1 than OT-I cells from control mice that did not receive anti-CD4 Ab (black versus red lines; Fig. 6C–E), which recapitulated the responses seen when endogenous Kb/OVA-specific CD8+ T cells were measured (Figs. 1, 2). Adoptive transfer of naive OT-II CD4+ T cells prior to immunization resulted in expansion and differentiation of OT-I CD8+ T cells that were largely equivalent to the responses seen in mice that had not been treated with anti-CD4 Ab (black versus blue lines; Fig. 6C–E). Thus, this experimental system recapitulates the CD4+ T cell help provided by endogenous CD4+ T cells.
When naive OT-II CD4+ T cells were transferred into mice on day 3 postimmunization, the expansion of the OT-I CD8+ T cell responses was only partially rescued, and no rescue in impaired expression of KLRG1 or overexpression of PD-1 was observed (purple line; Fig. 6C–E). Adoptive transfer of naive OT-II CD4+ T cells on day 7 or 10 postimmunization had no detectable impact on the magnitude of the OT-I CD8+ T cell responses nor did it rescue the dysfunctional phenotype (green and gray lines; Fig. 6C–E). These data suggest that CD4+ T cells are essential during priming to provide help to responding CD8+ T cells, and the absence of help during the first 72 h postimmunization is sufficient for dysfunctional differentiation to occur.
Phenotypic alterations in unhelped CD8+ T cells are apparent from the earliest time points
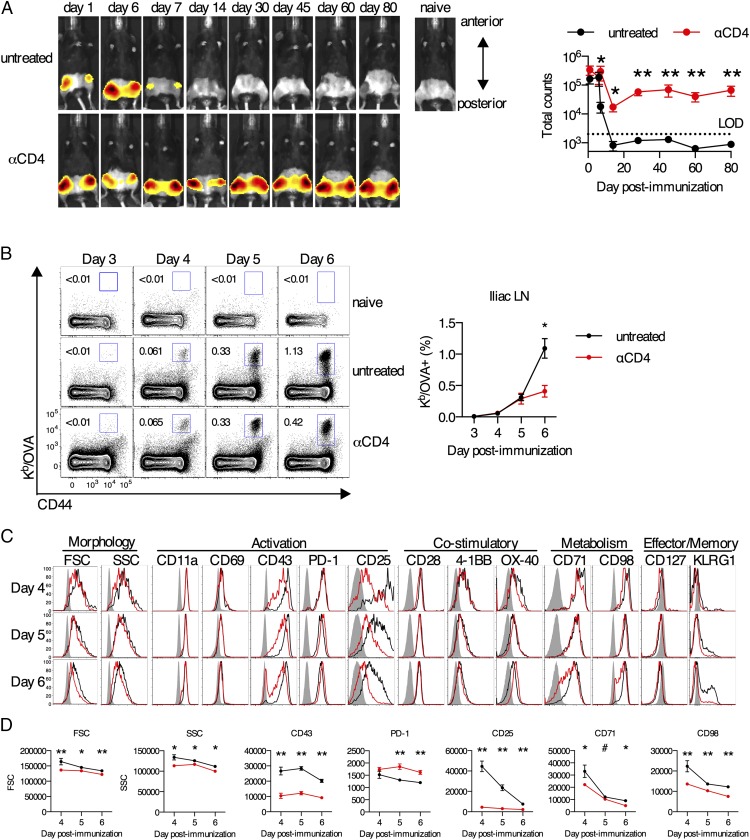
Persistent Ag is a critical driving factor in chronic infection–induced exhaustion (65–67). Thus, we sought to determine whether differences in Ag levels in the presence or absence of CD4+ T cells might be the reason for the dysfunction of unhelped CD8+ T cells. Assessing Ag levels by quantification of luciferase production using live animal imaging, we observed no differences in the amount of Ag in the presence or absence of CD4+ T cells through the first 6 d postimmunization (Fig. 7A). However, from days 6 to 7, there was a sharp decline in the levels of detectable Ag in the presence of CD4+ T cells that was not observed when CD4+ T cells were absent, which resulted in sustained Ag expression for at least 80 d (p < 0.05; Fig. 7A). These observations are consistent with a previous report that found immunization of CD4+ T cell–deficient animals with Ad vectors resulted in normal Ag levels early postimmunization but impaired long-term transgene clearance (13). Thus, alterations in transgene expression kinetics in the absence of CD4+ T cells did not occur until 7 d postimmunization.
FIGURE 7.
Early phenotype of Ag-specific CD8+ T cells primed with or without CD4+ T cell help. (A–D) C57BL/6 mice were treated with anti-CD4 Ab or left untreated and immunized i.m. with 109 viral particles of Ad5-SIINFEKL-Luc. (A) In vivo luciferase transgene expression in a single representative animal per group over time or group average as quantified by IVIS. (B) Representative flow plots and group averages of Kb/OVA+ CD8+ T cells in the iliac lymph nodes (LNs). (C) Representative flow plots of cellular morphology, activation markers, costimulatory receptors, metabolic receptors, and effector/memory markers on Kb/OVA+CD8+ T cells in the iliac LNs. (D) Group average expression of markers with significant differences from (B). Data are representative of two independent experiments (n = 5 mice per group per experiment). Mean ± SEM are shown. **p < 0.01, *p < 0.05, #p = 0.06, Mann–Whitney U test.
Given that differences in Ag levels were not apparent until 7 d postimmunization, we performed detailed phenotypic analysis of the OVA-specific CD8+ T cell responses at the earliest time points to determine when abnormalities were first observed. Endogenous OVA-specific CD8+ T cells were first detectable in the draining (iliac) lymph nodes on day 4 postimmunization (Fig. 7B), consistent with a previous report (68). Unhelped CD8+ T cells already displayed phenotypic abnormalities by day 4 postimmunization (Fig. 7C, 7D). Impaired upregulation of CD25 (high-affinity IL-2R) on Kb/OVA+CD8+ T cells from anti-CD4–treated mice was the most striking defect (p < 0.01; Fig. 7C, 7D). Intriguingly, several other genes that are known to be regulated by IL-2 signaling, including upregulation of CD71, CD98, and granzyme B as well as increased cell size and granularity during proliferation (69–71), were all perturbed in the absence of CD4+ T cells (Figs. 1, 7C, 7D). These data demonstrate that in the absence of CD4+ T cell help, phenotypic defects (day 4 or earlier) and defects in frequency (day 6) occur before differences in Ag load are observed. Furthermore, these data suggest that in the absence of CD4+ T cell help there are alterations in IL-2 signaling to CD8+ T cells.
Perturbed IL-2 signaling in the absence of CD4+ T cells is involved in immediate CD8+ T cell dysfunction
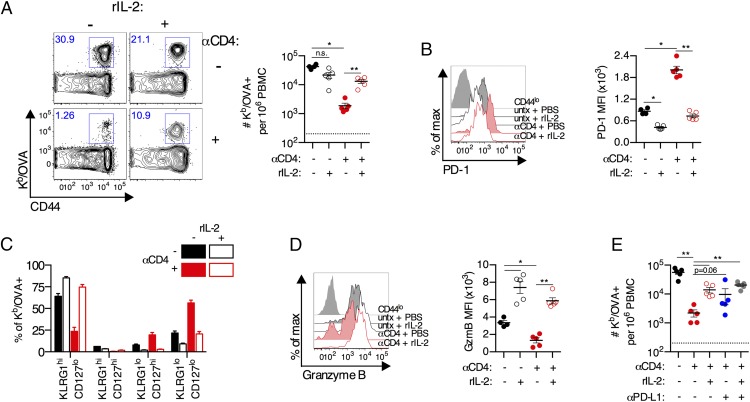
Given the observed perturbation in IL-2 signaling, we hypothesized that administration of rIL-2 might correct the dysfunctional differentiation of CD8+ T cells in the absence of CD4+ T cell help. Administration of rIL-2 to anti-CD4 Ab–treated mice, but not untreated controls, increased the frequency of Kb/OVA+ CD8+ T cells by 8.6-fold and the absolute number by 7-fold on day 10 postimmunization (p < 0.01; Fig. 8A). Administration of rIL-2 decreased PD-1 expression on Kb/OVA+ CD8+ T cells from anti-CD4–treated mice to levels seen on Kb/OVA+ CD8+ T cells from untreated control animals (p < 0.01; Fig. 8B). Administration of rIL-2 corrected the defect in differentiation to a KLRG1hiCD127lo effector phenotype (Fig. 8C) and also significantly increased granzyme B expression (p < 0.01; Fig. 8D). Thus, administration of rIL-2 can partially rescue the normal differentiation of unhelped CD8+ T cells. Taken together, these data suggest a major mechanism of CD4+ T cell help to drive proper CD8+ T cell expansion and differentiation following viral vector immunization is IL-2.
FIGURE 8.
Early IL-2 treatment rescues proliferation and effector differentiation of CD8+ T cells primed in the absence of CD4+ T cells. (A–D) C57BL/6 mice were treated with anti-CD4 Ab or left untreated and immunized i.m. with 109 viral particles of Ad5-SIINFEKL-Luc. Mice were administered with recombinant mouse IL-2 (rIL-2) or PBS control twice daily from days 3 to 10 postimmunization. (A) Frequency of Kb/OVA+CD8+ T cells in blood on day 10 postimmunization. (B) Expression of PD-1 on Kb/OVA+CD8+ T cells in the blood. (C) KLRG1 and CD127 expression on Kb/OVA+CD8+ T cells in the blood. (D) Expression of granzyme B on Kb/OVA+CD8+ T cells in the blood. (E) Mice were administered with recombinant mouse IL-2 (rIL-2) or PBS control twice daily from days 3 to 10 postimmunization and anti–PD-L1 Ab or isotype control once every 3 d beginning on day 0. Frequency of Kb/OVA+CD8+ T cells in blood on day 10 postimmunization. Data are representative of two experiments (n = 5 per group per experiment). Mean ± SEM are shown. **p < 0.01, *p < 0.05, Mann–Whitney U test.
Because administration of rIL-2 decreased PD-1 expression on unhelped CD8+ T cells and largely rescued differentiation of these cells (Fig. 5), we sought to determine whether this reflected a single mechanistic pathway of CD4+ T cell help. Coadministration of rIL-2 and anti–PD-L1 Ab did not significantly increase the frequency of Kb/OVA+CD8+ T cells in the blood compared with either treatment alone (Fig. 8E), and it also did not impact on PD-1, KLRG1, or granzyme B expression any more than one treatment alone (data not shown). Thus, IL-2 signaling and PD-1 signaling appear to represent a single signaling axis in this system. These data suggest that following Ad vector immunization CD4+ T cells provide IL-2 early postimmunization to, in part, suppress PD-1 signaling and thereby allow expansion and differentiation of functional CD8+ T cells.
Discussion
This study sought to provide further insights into the functionality and transcriptional state of primary CD8+ T cell responses elicited in the absence of CD4+ T cell help. Toward that end, our data demonstrate an immediate need for CD4+ T cell help following Ad vector and poxvirus vector immunization to promote expansion and acquisition of effector functions, and prevent early exhaustion-like dysfunction of vaccine-elicited CD8 T cells. Following viral vector immunization of CD4+ T cell–deficient mice, the CD8+ T cell responses are of reduced frequency, have reduced cytotoxicity and cytokine production, and exhibit an aberrant phenotype characterized by elevated expression of inhibitory receptors and inhibitory exhaustion and anergy transcriptional programs. Taken together, this study demonstrates that viral vector–elicited CD8+ T cells are immediately dysfunctional when primed without CD4+ T cell help.
Although CD4+ T cells are critical for driving optimal primary CD8+ T cell expansion in several experimental systems (4–7, 11–15), previous studies that have directly assessed the per-cell functionality of unhelped CD8+ T cells identified no major defects in effector function when primed without CD4+ T cell help (12, 16–19). These data would suggest that prior reports of reduced Ag clearance CD4+ T cell–deficient animals might instead reflect insufficient expansion of an otherwise functional CD8+ T cell response (3–10). In this study, we demonstrate that CD4+ T cells are required at priming for optimal expansion of CD8+ T cell responses by Ad vector and poxvirus vector immunization, consistent with previous reports (13, 20–22). In addition, we identify a critical role for CD4+ T cells in driving acquisition of CD8+ T cell effector functions and preventing dysfunctional differentiation, in contrast to previous reports showing largely functional primary CD8+ T cell effector responses in the absence of CD4+ T cells (12, 16–19). A potential difference is that these prior studies used infection models or cellular immunization models rather than viral vectors. Thus, at least in the context of viral vectors, reduced Ag clearance in the absence of CD4+ T cells (Fig. 7) (3, 72) appears to reflect both reduced frequency and reduced functionality of CD8+ T cells. Given the widespread use of viral vectors as candidate vaccine platforms, understanding how these responses are regulated is critical for rational vaccine design.
The magnitude of the CD8+ T cell response induced by Ad vectors appears to be regulated, in part, by IL-2 produced by CD4+ T cells, as has been described for vaccinia virus and tumor models (11, 73). Interestingly, although defects in phenotype were immediately apparent from the earliest detectable time point when CD4+ T cells were absent, the expansion of CD8+ T cells initially occurred unimpaired (days 4 and 5) but were impaired by day 6 (Fig. 7). With regard to CD8+ T cell expansion, IL-2 is not needed initially but is required to maintain proliferation (74). By contrast, IL-2 signaling appears to act very early postactivation to drive terminal effector differentiation (69, 70). Given the apparent perturbation in IL-2 signaling in the absence of CD4+ T cell help, these findings may explain the different time at which different defects in CD8+ T cell behavior are observed.
The most intriguing characteristic of CD8+ T cells primed without CD4+ T cell help is the simultaneous expression of exhaustion and anergy transcriptional signatures, which are largely distinct differentiation states (32), and thus the underlying dysfunction of unhelped CD8+ T cells appears to be caused by simultaneous expression of multiple inhibitory transcriptional programs. CD8+ T cell exhaustion is characterized by elevated expression of inhibitory receptors, decreased ex vivo cytotoxicity, decreased ability to produce cytokines, a unique transcriptional signature, and the ability to acquire effector functionality in response to blockade of PD-1 signaling (40, 54, 56, 57, 75–78). Unhelped CD8+ T cells share all of these characteristics (Figs. 1–5). However, CD8+ T cell exhaustion involves a strong temporal component where CD8+ T cells become progressively more impaired the longer the cells remain in an environment of persistent viremia (75, 79–82). By contrast, in our model, CD8+ T cell dysfunction occurs within days of immunization (Figs. 6, 7), which is more consistent with the process of anergy.
It has recently been demonstrated that expression of an engineered NFAT incapable of interacting with AP-1 is sufficient to drive expression of anergy and exhaustion-related genes (58). Elevated NFAT, but not AP-1 family proteins, was enriched in CD8+ T cells from CD4+ T cell–deficient mice, and the gene signature of AP-1–independent NFAT signaling was enriched (Fig. 4). Thus, we propose a model whereby a major driving factor of the observed CD8+ T cell dysfunction is due to elevated noncanonical NFAT signaling. The partial prevention of this phenotype by rIL-2 administration suggests that, in the context of Ad vector immunization, CD4+ T cell–derived IL-2 is responsible for inducing AP-1 signaling in CD8+ T cells, as has been reported in an in vitro model of CD4+ T cell anergy (83). Thus, a shared characteristic of the dysfunctional CD8+ T cells described in this study, anergic T cells, and exhausted CD8+ T cells is responsiveness to reinvigoration by restoration of IL-2 signaling (Fig. 8) (42, 83, 84). Interestingly, impaired SLAT-induced NFAT signaling has been shown to reduce primary CD8+ T cell proliferation without altering effector functionality (85). By contrast, our data suggest that increased (noncanonical) NFAT signaling can also be involved in impaired CD8+ T cell proliferation while also impacting effector functionality. Thus, it appears that in the context of Ad vector immunization in the absence of CD4+ T cells, the lack of sufficient IL-2 signaling leads to noncanonical NFAT signaling in CD8+ T cells, which serves to impair proliferation and drive dysfunctional differentiation of these cells.
In conclusion, we demonstrate that CD4+ T cells are required immediately upon Ag priming for the optimal expansion and appropriate differentiation of viral vector vaccine–induced CD8+ T cells. The absence of CD4+ T cell help results in a multifaceted and immediate dysfunction of responding CD8+ T cells, which transcriptionally appears regulated by AP-1–independent NFAT-driven expression of exhaustion and anergy programs. This dysfunction is imprinted immediately postimmunization, and IL-2 is a critical factor in promoting functionality and suppressing expression of inhibitory receptors. Thus, CD4+ T cells play a critical role in preventing the immediate dysfunction of CD8+ T cells following immunization with viral vectors. These findings suggest potential pathways to target to improve the immunogenicity of viral vector immunization in the context of reduced CD4+ T cell frequency and/or functionality.
Supplementary Material
Acknowledgments
We thank Lily Parenteau and Stephen Blackmore for technical assistance, the CVVR Flow Cytometry core for assistance with cell sorting, David Knipe (Harvard Medical School) and Arlene Sharpe (Harvard Medical School) for advice and feedback, and the National Institutes of Health Tetramer Core Facility (Emory University) for MHC class I monomers.
This work was supported by National Institutes of Health Grants AI078526 and AI096040 and Bill and Melinda Gates Foundation Grant OPP1033091 (to D.H.B.), a Herchel Smith Graduate Fellowship from Harvard University (to N.M.P.), and National Institutes of Health Grants AI007245 and AI07387 (to P. P.-M.).
The microarray data presented in this article have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE73001.
The online version of this article contains supplemental material.
- Ad
- adenovirus
- Ad5
- adenovirus serotype 5
- Ad5HVR48
- chimeric Ad5 with hypervariable regions 1–7 of Ad48
- Env
- SIVmac239 Env
- Gag
- SIVmac239 Gag
- GSEA
- gene set enrichment analysis
- KLRG1
- killer cell lectin–like receptor G1
- KO
- knockout
- NYVAC
- attenuated Copenhagen vaccinia strain
- PD-1
- programmed death-1
- PD-L1
- PD-ligand 1
- SIINFEKL-luc
- fusion protein of luciferase and the OVA257–264 peptide.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Laidlaw B. J., Craft J. E., Kaech S. M. 2016. The multifaceted role of CD4+ T cells in CD8+ T cell memory. Nat. Rev. Immunol. 16: 102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiesel M., Oxenius A. 2012. From crucial to negligible: functional CD8+ T-cell responses and their dependence on CD4+ T-cell help. Eur. J. Immunol. 42: 1080–1088. [DOI] [PubMed] [Google Scholar]
- 3.Yang Y., Xiang Z., Ertl H. C., Wilson J. M. 1995. Upregulation of class I major histocompatibility complex antigens by interferon γ is necessary for T-cell–mediated elimination of recombinant adenovirus-infected hepatocytes in vivo. Proc. Natl. Acad. Sci. USA 92: 7257–7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed K. A., Wang L., Munegowda M. A., Mulligan S. J., Gordon J. R., Griebel P., Xiang J. 2012. Direct in vivo evidence of CD4+ T cell requirement for CTL response and memory via pMHC-I targeting and CD40L signaling. [Published erratum appears in 2012 J. Leukoc. Biol. 92: 1123.] J. Leukoc. Biol. 92: 289–300. [DOI] [PubMed] [Google Scholar]
- 5.Sokke Umeshappa C., Hebbandi Nanjundappa R., Xie Y., Freywald A., Deng Y., Ma H., Xiang J. 2012. CD154 and IL-2 signaling of CD4+ T cells play a critical role in multiple phases of CD8+ CTL responses following adenovirus vaccination. PLoS One 7: e47004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith C. M., Wilson N. S., Waithman J., Villadangos J. A., Carbone F. R., Heath W. R., Belz G. T. 2004. Cognate CD4+ T cell licensing of dendritic cells in CD8+ T cell immunity. Nat. Immunol. 5: 1143–1148. [DOI] [PubMed] [Google Scholar]
- 7.Behrens G. M. N., Li M., Davey G. M., Allison J., Flavell R. A., Carbone F. R., Heath W. R. 2004. Helper requirements for generation of effector CTL to islet β cell antigens. J. Immunol. 172: 5420–5426. [DOI] [PubMed] [Google Scholar]
- 8.Guerder S., Matzinger P. 1992. A fail-safe mechanism for maintaining self-tolerance. J. Exp. Med. 176: 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toka F. N., Gieryńska M., Suvas S., Schoenberger S. P., Rouse B. T. 2005. Rescue of memory CD8+ T cell reactivity in peptide/TLR9 ligand immunization by codelivery of cytokines or CD40 ligation. Virology 331: 151–158. [DOI] [PubMed] [Google Scholar]
- 10.Fernando G. J. P., Khammanivong V., Leggatt G. R., Liu W. J., Frazer I. H. 2002. The number of long-lasting functional memory CD8+ T cells generated depends on the nature of the initial nonspecific stimulation. Eur. J. Immunol. 32: 1541–1549. [DOI] [PubMed] [Google Scholar]
- 11.Wiesel M., Joller N., Ehlert A.-K., Crouse J., Spörri R., Bachmann M. F., Oxenius A. 2010. Th cells act via two synergistic pathways to promote antiviral CD8+ T cell responses. J. Immunol. 185: 5188–5197. [DOI] [PubMed] [Google Scholar]
- 12.Novy P., Quigley M., Huang X., Yang Y. 2007. CD4 T cells are required for CD8 T cell survival during both primary and memory recall responses. J. Immunol. 179: 8243–8251. [DOI] [PubMed] [Google Scholar]
- 13.Yang T. C., Millar J., Groves T., Zhou W., Grinshtein N., Parsons R., Evelegh C., Xing Z., Wan Y., Bramson J. 2007. On the role of CD4+ T cells in the CD8+ T-cell response elicited by recombinant adenovirus vaccines. Mol. Ther. 15: 997–1006. [DOI] [PubMed] [Google Scholar]
- 14.Wang J.-C. E., Livingstone A. M. 2003. Cutting edge: CD4+ T cell help can be essential for primary CD8+ T cell responses in vivo. J. Immunol. 171: 6339–6343. [DOI] [PubMed] [Google Scholar]
- 15.Le Bon A., Etchart N., Rossmann C., Ashton M., Hou S., Gewert D., Borrow P., Tough D. F. 2003. Cross-priming of CD8+ T cells stimulated by virus-induced type I interferon. Nat. Immunol. 4: 1009–1015. [DOI] [PubMed] [Google Scholar]
- 16.Sun J. C., Williams M. A., Bevan M. J. 2004. CD4+ T cells are required for the maintenance, not programming, of memory CD8+ T cells after acute infection. Nat. Immunol. 5: 927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun J. C., Bevan M. J. 2003. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science 300: 339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janssen E. M., Lemmens E. E., Wolfe T., Christen U., von Herrath M. G., Schoenberger S. P. 2003. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature 421: 852–856. [DOI] [PubMed] [Google Scholar]
- 19.Northrop J. K., Thomas R. M., Wells A. D., Shen H. 2006. Epigenetic remodeling of the IL-2 and IFN-γ loci in memory CD8 T cells is influenced by CD4 T cells. J. Immunol. 177: 1062–1069. [DOI] [PubMed] [Google Scholar]
- 20.Provine N. M., Larocca R. A., Penaloza-MacMaster P., Borducchi E. N., McNally A., Parenteau L. R., Kaufman D. R., Barouch D. H. 2014. Longitudinal requirement for CD4+ T cell help for adenovirus vector-elicited CD8+ T cell responses. J. Immunol. 192: 5214–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holst P. J., Bartholdy C., Stryhn A., Thomsen A. R., Christensen J. P. 2007. Rapid and sustained CD4+ T-cell‑independent immunity from adenovirus-encoded vaccine antigens. J. Gen. Virol. 88: 1708–1716. [DOI] [PubMed] [Google Scholar]
- 22.Holst P. J., Christensen J. P., Thomsen A. R. 2011. Vaccination against lymphocytic choriomeningitis virus infection in MHC class II‑deficient mice. J. Immunol. 186: 3997–4007. [DOI] [PubMed] [Google Scholar]
- 23.Baden L. R., Walsh S. R., Seaman M. S., Tucker R. P., Krause K. H., Patel A., Johnson J. A., Kleinjan J., Yanosick K. E., Perry J., et al. 2013. First-in-human evaluation of the safety and immunogenicity of a recombinant adenovirus serotype 26 HIV-1 Env vaccine (IPCAVD 001). J. Infect. Dis. 207: 240–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barouch D. H., Liu J., Peter L., Abbink P., Iampietro M. J., Cheung A., Alter G., Chung A., Dugast A.-S., Frahm N., et al. 2013. Characterization of humoral and cellular immune responses elicited by a recombinant adenovirus serotype 26 HIV-1 Env vaccine in healthy adults (IPCAVD 001). J. Infect. Dis. 207: 248–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barnes E., Folgori A., Capone S., Swadling L., Aston S., Kurioka A., Meyer J., Huddart R., Smith K., Townsend R., et al. 2012. Novel adenovirus-based vaccines induce broad and sustained T cell responses to HCV in man. Sci. Transl. Med. 4: 115ra1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swadling L., Capone S., Antrobus R. D., Brown A., Richardson R., Newell E. W., Halliday J., Kelly C., Bowen D., Fergusson J., et al. 2014. A human vaccine strategy based on chimpanzee adenoviral and MVA vectors that primes, boosts, and sustains functional HCV-specific T cell memory. Sci. Transl. Med. 6: 261ra153 doi:10.1126/scitranslmed.3009185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smaill F., Jeyanathan M., Smieja M., Medina M. F., Thanthrige-Don N., Zganiacz A., Yin C., Heriazon A., Damjanovic D., Puri L., et al. 2013. A human type 5 adenovirus-based tuberculosis vaccine induces robust T cell responses in humans despite preexisting anti-adenovirus immunity. Sci. Transl. Med. 5: 205ra134. [DOI] [PubMed] [Google Scholar]
- 28.Ewer K. J., O’Hara G. A., Duncan C. J. A., Collins K. A., Sheehy S. H., Reyes-Sandoval A., Goodman A. L., Edwards N. J., Elias S. C., Halstead F. D., et al. 2013. Protective CD8+ T-cell immunity to human malaria induced by chimpanzee adenovirus-MVA immunisation. Nat. Commun. 4: 2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sullivan N. J., Hensley L., Asiedu C., Geisbert T. W., Stanley D., Johnson J., Honko A., Olinger G., Bailey M., Geisbert J. B., et al. 2011. CD8+ cellular immunity mediates rAd5 vaccine protection against Ebola virus infection of nonhuman primates. Nat. Med. 17: 1128–1131. [DOI] [PubMed] [Google Scholar]
- 30.Stanley D. A., Honko A. N., Asiedu C., Trefry J. C., Lau-Kilby A. W., Johnson J. C., Hensley L., Ammendola V., Abbate A., Grazioli F., et al. 2014. Chimpanzee adenovirus vaccine generates acute and durable protective immunity against ebolavirus challenge. Nat. Med. 20: 1126–1129. [DOI] [PubMed] [Google Scholar]
- 31.Kaech S. M., Cui W. 2012. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12: 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wherry E. J., Kurachi M. 2015. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 15: 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mescher M. F., Curtsinger J. M., Agarwal P., Casey K. A., Gerner M., Hammerbeck C. D., Popescu F., Xiao Z. 2006. Signals required for programming effector and memory development by CD8+ T cells. Immunol. Rev. 211: 81–92. [DOI] [PubMed] [Google Scholar]
- 34.Wherry E. J., Teichgräber V., Becker T. C., Masopust D., Kaech S. M., Antia R., von Andrian U. H., Ahmed R. 2003. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol. 4: 225–234. [DOI] [PubMed] [Google Scholar]
- 35.Kaufman D. R., De Calisto J., Simmons N. L., Cruz A. N., Villablanca E. J., Mora J. R., Barouch D. H. 2011. Vitamin A deficiency impairs vaccine-elicited gastrointestinal immunity. J. Immunol. 187: 1877–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts D. M., Nanda A., Havenga M. J. E., Abbink P., Lynch D. M., Ewald B. A., Liu J., Thorner A. R., Swanson P. E., Gorgone D. A., et al. 2006. Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. Nature 441: 239–243. [DOI] [PubMed] [Google Scholar]
- 37.Kaufman D. R., Bivas-Benita M., Simmons N. L., Miller D., Barouch D. H. 2010. Route of adenovirus-based HIV-1 vaccine delivery impacts the phenotype and trafficking of vaccine-elicited CD8+ T lymphocytes. J. Virol. 84: 5986–5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finn J. D., Bassett J., Millar J. B., Grinshtein N., Yang T. C., Parsons R., Evelegh C., Wan Y., Parks R. J., Bramson J. L. 2009. Persistence of transgene expression influences CD8+ T-cell expansion and maintenance following immunization with recombinant adenovirus. J. Virol. 83: 12027–12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teigler J. E., Phogat S., Franchini G., Hirsch V. M., Michael N. L., Barouch D. H. 2014. The canarypox virus vector ALVAC induces distinct cytokine responses compared to the vaccinia virus‑based vectors MVA and NYVAC in rhesus monkeys. J. Virol. 88: 1809–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barber D. L., Wherry E. J., Masopust D., Zhu B., Allison J. P., Sharpe A. H., Freeman G. J., Ahmed R. 2006. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439: 682–687. [DOI] [PubMed] [Google Scholar]
- 41.Blattman J. N., Grayson J. M., Wherry E. J., Kaech S. M., Smith K. A., Ahmed R. 2003. Therapeutic use of IL-2 to enhance antiviral T-cell responses in vivo. Nat. Med. 9: 540–547. [DOI] [PubMed] [Google Scholar]
- 42.West E. E., Jin H.-T., Rasheed A. U., Penaloza-Macmaster P., Ha S.-J., Tan W. G., Youngblood B., Freeman G. J., Smith K. A., Ahmed R. 2013. PD-L1 blockade synergizes with IL-2 therapy in reinvigorating exhausted T cells. J. Clin. Invest. 123: 2604–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barouch D. H., Pau M. G., Custers J. H. H. V., Koudstaal W., Kostense S., Havenga M. J. E., Truitt D. M., Sumida S. M., Kishko M. G., Arthur J. C., et al. 2004. Immunogenicity of recombinant adenovirus serotype 35 vaccine in the presence of pre-existing anti-Ad5 immunity. J. Immunol. 172: 6290–6297. [DOI] [PubMed] [Google Scholar]
- 44.Rötzschke O., Falk K., Stevanović S., Jung G., Walden P., Rammensee H. G. 1991. Exact prediction of a natural T cell epitope. Eur. J. Immunol. 21: 2891–2894. [DOI] [PubMed] [Google Scholar]
- 45.Hermans I. F., Silk J. D., Yang J., Palmowski M. J., Gileadi U., McCarthy C., Salio M., Ronchese F., Cerundolo V. 2004. The VITAL assay: a versatile fluorometric technique for assessing CTL- and NKT-mediated cytotoxicity against multiple targets in vitro and in vivo. J. Immunol. Methods 285: 25–40. [DOI] [PubMed] [Google Scholar]
- 46.Contag C. H., Contag P. R., Mullins J. I., Spilman S. D., Stevenson D. K., Benaron D. A. 1995. Photonic detection of bacterial pathogens in living hosts. Mol. Microbiol. 18: 593–603. [DOI] [PubMed] [Google Scholar]
- 47.Quigley M., Pereyra F., Nilsson B., Porichis F., Fonseca C., Eichbaum Q., Julg B., Jesneck J. L., Brosnahan K., Imam S., et al. 2010. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat. Med. 16: 1147–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barnitz R. A., Imam S., Yates K., Haining W. N. 2013. Isolation of RNA and the synthesis and amplification of cDNA from antigen-specific T cells for genome-wide expression analysis. Methods Mol. Biol. 979: 161–173. [DOI] [PubMed] [Google Scholar]
- 49.Gentleman R., Carey V., Huber W., Irizarry R., Dudoit S. 2006. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer Science & Business Media, New York. [Google Scholar]
- 50.Joshi N. S., Cui W., Chandele A., Lee H. K., Urso D. R., Hagman J., Gapin L., Kaech S. M. 2007. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity 27: 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaech S. M., Tan J. T., Wherry E. J., Konieczny B. T., Surh C. D., Ahmed R. 2003. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4: 1191–1198. [DOI] [PubMed] [Google Scholar]
- 52.Sarkar S., Kalia V., Haining W. N., Konieczny B. T., Subramaniam S., Ahmed R. 2008. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 205: 625–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olson J. A., McDonald-Hyman C., Jameson S. C., Hamilton S. E. 2013. Effector-like CD8+ T cells in the memory population mediate potent protective immunity. Immunity 38: 1250–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blackburn S. D., Shin H., Haining W. N., Zou T., Workman C. J., Polley A., Betts M. R., Freeman G. J., Vignali D. A. A., Wherry E. J. 2009. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol. 10: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.West E. E., Youngblood B., Tan W. G., Jin H.-T., Araki K., Alexe G., Konieczny B. T., Calpe S., Freeman G. J., Terhorst C., et al. 2011. Tight regulation of memory CD8+ T cells limits their effectiveness during sustained high viral load. Immunity 35: 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wherry E. J., Ha S.-J., Kaech S. M., Haining W. N., Sarkar S., Kalia V., Subramaniam S., Blattman J. N., Barber D. L., Ahmed R. 2007. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27: 670–684. [DOI] [PubMed] [Google Scholar]
- 58.Martinez G. J., Pereira R. M., Äijö T., Kim E. Y., Marangoni F., Pipkin M. E., Togher S., Heissmeyer V., Zhang Y. C., Crotty S., et al. 2015. The transcription factor NFAT promotes exhaustion of activated CD8+ T cells. Immunity 42: 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakuishi K., Apetoh L., Sullivan J. M., Blazar B. R., Kuchroo V. K., Anderson A. C. 2010. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. [Published erratum appears in 2011 J. Exp. Med. 208: 1331.] J. Exp. Med. 207: 2187–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Velu V., Titanji K., Zhu B., Husain S., Pladevega A., Lai L., Vanderford T. H., Chennareddi L., Silvestri G., Freeman G. J., et al. 2009. Enhancing SIV-specific immunity in vivo by PD-1 blockade. Nature 458: 206–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fuller M. J., Callendret B., Zhu B., Freeman G. J., Hasselschwert D. L., Satterfield W., Sharpe A. H., Dustin L. B., Rice C. M., Grakoui A., et al. 2013. Immunotherapy of chronic hepatitis C virus infection with antibodies against programmed cell death-1 (PD-1). Proc. Natl. Acad. Sci. USA 110: 15001–15006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horne-Debets J. M., Faleiro R., Karunarathne D. S., Liu X. Q., Lineburg K. E., Poh C. M., Grotenbreg G. M., Hill G. R., MacDonald K. P. A., Good M. F., et al. 2013. PD-1 dependent exhaustion of CD8+ T cells drives chronic malaria. Cell Rep. 5: 1204–1213. [DOI] [PubMed] [Google Scholar]
- 63.Feau S., Arens R., Togher S., Schoenberger S. P. 2011. Autocrine IL-2 is required for secondary population expansion of CD8+ memory T cells. Nat. Immunol. 12: 908–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feau S., Garcia Z., Arens R., Yagita H., Borst J., Schoenberger S. P. 2012. The CD4+ T-cell help signal is transmitted from APC to CD8+ T-cells via CD27-CD70 interactions. Nat. Commun. 3: 948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mueller S. N., Ahmed R. 2009. High antigen levels are the cause of T cell exhaustion during chronic viral infection. Proc. Natl. Acad. Sci. USA 106: 8623–8628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blattman J. N., Wherry E. J., Ha S.-J., van der Most R. G., Ahmed R. 2009. Impact of epitope escape on PD-1 expression and CD8 T-cell exhaustion during chronic infection. J. Virol. 83: 4386–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.D’Souza M., Fontenot A. P., Mack D. G., Lozupone C., Dillon S., Meditz A., Wilson C. C., Connick E., Palmer B. E. 2007. Programmed death 1 expression on HIV-specific CD4+ T cells is driven by viral replication and associated with T cell dysfunction. J. Immunol. 179: 1979–1987. [DOI] [PubMed] [Google Scholar]
- 68.Yang T. C., Dayball K., Wan Y. H., Bramson J. 2003. Detailed analysis of the CD8+ T-cell response following adenovirus vaccination. J. Virol. 77: 13407–13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pipkin M. E., Sacks J. A., Cruz-Guilloty F., Lichtenheld M. G., Bevan M. J., Rao A. 2010. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity 32: 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kalia V., Sarkar S., Subramaniam S., Haining W. N., Smith K. A., Ahmed R. 2010. Prolonged interleukin-2Rα expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity 32: 91–103. [DOI] [PubMed] [Google Scholar]
- 71.Cornish G. H., Sinclair L. V., Cantrell D. A. 2006. Differential regulation of T-cell growth by IL-2 and IL-15. Blood 108: 600–608. [DOI] [PubMed] [Google Scholar]
- 72.Yang Y., Ertl H. C., Wilson J. M. 1994. MHC class I‑restricted cytotoxic T lymphocytes to viral antigens destroy hepatocytes in mice infected with E1-deleted recombinant adenoviruses. Immunity 1: 433–442. [DOI] [PubMed] [Google Scholar]
- 73.Bos R., Sherman L. A. 2010. CD4+ T-cell help in the tumor milieu is required for recruitment and cytolytic function of CD8+ T lymphocytes. Cancer Res. 70: 8368–8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.D’Souza W. N., Lefrançois L. 2003. IL-2 is not required for the initiation of CD8 T cell cycling but sustains expansion. J. Immunol. 171: 5727–5735. [DOI] [PubMed] [Google Scholar]
- 75.Wherry E. J., Blattman J. N., Murali-Krishna K., van der Most R., Ahmed R. 2003. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J. Virol. 77: 4911–4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ahmed R., Salmi A., Butler L. D., Chiller J. M., Oldstone M. B. 1984. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice: role in suppression of cytotoxic T lymphocyte response and viral persistence. J. Exp. Med. 160: 521–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Doering T. A., Crawford A., Angelosanto J. M., Paley M. A., Ziegler C. G., Wherry E. J. 2012. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity 37: 1130–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jin H.-T., Anderson A. C., Tan W. G., West E. E., Ha S.-J., Araki K., Freeman G. J., Kuchroo V. K., Ahmed R. 2010. Cooperation of Tim-3 and PD-1 in CD8 T-cell exhaustion during chronic viral infection. Proc. Natl. Acad. Sci. USA 107: 14733–14738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fuller M. J., Zajac A. J. 2003. Ablation of CD8 and CD4 T cell responses by high viral loads. J. Immunol. 170: 477–486. [DOI] [PubMed] [Google Scholar]
- 80.Zajac A. J., Blattman J. N., Murali-Krishna K., Sourdive D. J., Suresh M., Altman J. D., Ahmed R. 1998. Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med. 188: 2205–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gallimore A., Glithero A., Godkin A., Tissot A. C., Plückthun A., Elliott T., Hengartner H., Zinkernagel R. 1998. Induction and exhaustion of lymphocytic choriomeningitis virus-specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I-peptide complexes. J. Exp. Med. 187: 1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Angelosanto J. M., Blackburn S. D., Crawford A., Wherry E. J. 2012. Progressive loss of memory T cell potential and commitment to exhaustion during chronic viral infection. J. Virol. 86: 8161–8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Duré M., Macián F. 2009. IL-2 signaling prevents T cell anergy by inhibiting the expression of anergy-inducing genes. Mol. Immunol. 46: 999–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tham E. L., Shrikant P., Mescher M. F. 2002. Activation-induced nonresponsiveness: a Th-dependent regulatory checkpoint in the CTL response. J. Immunol. 168: 1190–1197. [DOI] [PubMed] [Google Scholar]
- 85.Feau S., Schoenberger S. P., Altman A., Bécart S. 2013. SLAT regulates CD8+ T cell clonal expansion in a Cdc42- and NFAT1-dependent manner. J. Immunol. 190: 174–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Safford M., Collins S., Lutz M. A. M., Allen A., Huang C.-T. C., Kowalski J., Blackford A., Horton M. R. M., Drake C., Schwartz R. H. R., Powell J. D. J. 2005. Egr-2 and Egr-3 are negative regulators of T cell activation. Nat. Immunol. 6: 472–480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.