ABSTRACT
Sterols are the basal components of the membranes of the fungal pathogen Candida albicans, and these membranes determine the susceptibility of C. albicans cells to a variety of stresses, such as ionic, osmotic and oxidative pressures, and treatment with antifungal drugs. The common antifungal azoles in clinical use are targeted to the biosynthesis of ergosterol. In the past years, the synthesis, storage and metabolism of ergosterol in Saccharomyces cerevisiae has been characterized in some detail; however, these processes has not been as well investigated in the human opportunistic pathogen C. albicans. In this review, we summarize the genes involved in ergosterol synthesis and regulation in C. albicans. As well, genes in S. cerevisiae implicated in ergosterol storage and conversions with other lipids are noted, as these provide us clues and directions for the study of the homologous genes in C. albicans. In this report we have particularly focused on the essential roles of ergosterol in the dynamic process of cell biology and its fundamental status in the biological membrane system that includes lipid rafts, lipid droplets, vacuoles and mitochondria. We believe that a thorough understanding of this classic and essential pathway will give us new ideas about drug resistance and morphological switching in C. albicans.
KEYWORDS: biosynthesis, Candida albicans, function, regulation, sterols
The relative evolutionary relationship between humans and their fungal pathogens has made the treatment of fungal infections with chemotherapeutic agents encounter great challenges. The majority of life-threatening fungal infections are caused by C. albicans, Cryptococcus neoformans and Aspergillus fumigatus, with C. albicans predominating with a death toll of over 400,000 each year (the mortality rate can approach 40% for systemic infections).1 Worryingly, in the past years C. albicans infections have been extended from life-threatening systemic infections in immunocompromised patients to nosocomial fungus infections in healthy people.2 Currently the treatment of candidiasis in the clinic is limited by the relatively small number of traditional types of antifungal drugs and by drug resistance, which has increased through antifungal drug abuse and the need for long-term treatment options in response to recurrent attacks.3
At present, the azoles, polyenes, and echinocandins have been widely used for the treatment of fungal infections.4 Although there are limited reports of resistance to polyenes or echinocadins, azole resistance is still the most clinically relevant issue. Recently, fluconazole-(diflucan-)resistant C. albicans, which leads to about 46,000 hospitalized patients infected each year, has been classified as a serious pathogen by the Centers for Disease Control and Prevention.5,6 There are 4 major mechanisms that generate resistance to azoles in C. albicans: transcriptional regulation or mutations in the ergosterol biosynthetic pathway, including the up-regulation or mutations of the ERG11 gene whose product is the direct target for azoles;7,8 reduction in intracellular drug concentrations by the overexpression of transmembrane efflux pump proteins, such as major facilitator superfamily (MFS) and ATP-binding cassette (ABC) transporter proteins;9-11 cellular stress responses through changes in metabolic pathways, which depend on the regulation of transcription factors (Mdr1p, Tac1p, Crz1p) or protein kinase A;12-14 and the formation of biofilms consisting of an extracellular exopolymeric matrix, yeast cells, hyphae and pseudohyphae.15 Most of the drug-resistant isolates are related in some way to alterations in the content and composition of membrane sterols. To address the problem of drug resistance in C. albicans, we need a clear understanding of the synthesis of sterols and the transcriptional or post-transcriptional regulation of the process.
In yeast, the major sterol is ergosterol, which is mainly synthesized in the endoplasmic reticulum (ER) and in lipid droplets.16 Sterols in yeast are indispensable factors that coordinate membrane heterogeneity, prevent water penetration, and maintain the integrity, rigidity and fluidity of the plasma membrane.17 In a spin probe electron paramagnetic resonance (EPR) study, Sgherri et al. detected that azole treatment disrupted the formation of the ordered lipid bilayer and/or decreased the content of integral membrane proteins.18 In addition, the membranes of organelles and the fusion of transport vesicles are dependent on the correct assemblies of sterols and other lipids. In this review, we present the synthesis of ergosterol in the opportunistic pathogenic fungus C. albicans and summarize key genetic mutations in ergosterol biosynthesis and transcriptional factors, focusing on their effects on drug sensitivity. As a basic component of all kinds of membranes, ergosterol depletion changes many cellular biological processes and destroys the normal membrane properties. The impact of ergosterol depletion on lipid rafts, vacuoles and mitochondria in C. albicans are also covered. Finally, we identify some issues that are worthy of study in C. albicans through reference to the formation and function of ergosterol in S. cerevisiae.
The biosynthesis and storage of sterols
The sterol biosynthetic pathway in eukaryotes is conserved. Both ergosterol as the final product in yeasts, and cholesterol as the final product in mammals, are synthesized from the same intermediate lanosterol, albeit using different enzymes and requiring different levels of oxygen consumption.19 Previous work had not identified direct evidence for the uptake of sterols from the external environment in C. albicans until Zavrel et al. found that C. albicans takes up sterols only under aerobic conditions during the post-exponential-growth phase, a process independent of UPC2.20,21 As a result, the pathway of steroid biosynthesis plays an important part in dealing with the external environmental stress. This pathway and the linked antifungal inhibitors are shown in Fig. 1, which identifies the major intermediates and the alternative biosynthesis routes bypassing ERG11 or ERG3.22,23 This pathway includes 2 rate-limiting enzymes. One is 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase, which is necessary in regulation of cholesterol synthesis in mammals, and the other one is the lanosterol 14α-demethylase, which is essential in the ergosterol synthesis in fungi. By using the technology of gene disruption and controlled gene expression, ERG1, ERG7, ERG9, ERG25, ERG26, and ERG27 have been proved to be essential, while ERG2, ERG3, ERG5, ERG6, and ERG24 are considered to be non-essential in ergosterol biosynthesis.24-31 Interestingly, the direct target gene of azoles, ERG11, can only be disrupted in the ERG3-deficient strains or when strains are cultured in media with nystatin or amphotericin B, as the sterols with a C14-methyl group or diols especially for 14-methylergosta-8,24(28)-dien-3β,6α-diol accumulated by ERG3 is lethal in C. albicans.32-34 NCP1 encodes a NADPH-cytochrome P450 reductase that, in conjunction with Erg11p, is required for sterol C14-demethylation. NCP1 is an essential gene and highly expressed in cells treated with ketoconazole.35,36 The ERG gene disruptions change the components and distribution of sterols. These changes can be measured by gas chromatography-mass spectrometer (GC-MS), which provides a solid evidence for the function of the ERG enzymes.37 For instance, the clinical isolates displayed cross-resistant to azoles and amphotericin B have been detected to be the ERG3 mutated strain. And the direct evidence is that the upstream products of Erg3p, episterol and fecosterol, have accumulated about 10%. In consistent with this, ergosta-7,22-dienol, which is the product of the alternative route bypassing the Erg3p accounts for about 50% of all the sterols. These sterol composition data has provided a powerful evidence that the cross-resistance of clinical isolates depends on the mutation in ERG3.38
Figure 1.

The conserved pathway of sterol synthesis, storage and transformation in C. albicans and S. cerevisiae. A. Schematic representation of the common ergosterol synthesis process including the enzymes and intermediate products in C. albicans. The storage and transformation between steryl esters and fatty acids or sterols is uncharacterized in C. albicans and labeled with dotted boxes, based on the homologous genes and their roles in S. cerevisiae. B. Accumulated intermediates when ERG3 function is lost. C. Bypass pathway when C. albicans is treated with Erg11p inhibitors. The resulting aberrant sterol 14-methylergosta-8,24(28)-dien-3β,6α-diol is produced by ERG3. Solid arrows, single enzymatic process; dashed arrows, multiple enzymatic processes; underlined genes, S. cerevisiae genes and their homologous genes in C. albicans are shown in parentheses.
Although C. albicans can survive the inhibition of ergosterol synthesis,39 antifungals such as azoles, polyenes, and allylamines inhibit the growth of C. albicans severely. To be specific, azoles target to lanosterol 14-α demethylase (Cyp51p); polyenes disrupt the ergosterols distributed on the membrane of fungi; and allylamines are squalene synthase (Erg1p) inhibitors. Most of antifungal drugs in the clinic targeting the biosynthesis of ergosterol inhibit the activities of ERG enzymes and direct the biosynthesis of the ergosterol pathway to an alternative route (Fig. 1, B or C).40 Their fungistatic properties are caused not only the reduction of ergosterol in the membranes but also by accumulation of aberrantly formed sterols with a C14-methyl group changing the properties of membranes.17,41 The mutations or disruptions in ERG genes lead to increased or reduced sensitivity to the antifungal agents (Table 1). These sensitivity phenotypes provide clues for studying the uncharacterized genes or networks within the ergosterol biosynthetic pathway. Meanwhile, mutations in this pathway, causing alterations in enzymes activity or drug binding sites, are major factors which are related with azole resistance. Mutations in the ERG11 and ERG3 genes or the UPC2 gene which encodes a transcription factor contribute to azole resistance.22,42-46 Mutations in ERG11 are not limited to the point mutations, as the clinical isolates with 2 extra copies of Chromosome 5L, as an isochromosome, conferred increased fluconazole resistance. Recently, a clinical isolate of C. albicans with mutations in ERG11 and ERG5 was reported to be cross-resistant to azoles and amphotericin B.29
Table 1.
Sensitivities of the mutant strains with the mutations or disruptions in ERG genes referred in this article.
| Gene Description | Increased sensitivity | Reduced sensitivity |
|---|---|---|
| Met3p-ERG1 | fluconazole, terbinafine, amphotericin B, cycloheximide, nystatin | |
| erg2Δ/erg2Δ | fluconazole, 4-nitroquinoline oxide, terbinafine, o-phenanthroline | |
| erg6Δ/erg6Δ | terbinafine, tridemorph, fenpropiomorph, fluphenazine, cycloheximide, cerulenin, brefeldin A | |
| erg24Δ/erg24Δ | terbinafine, cycloheximide, cerulenin, fluphenazine, brefeldin A | fluconazole (slightly) |
| erg3Δ/erg3Δ | amphotericin B, cycloheximide, fluphenazine, brefeldin A, | fluconazole |
| erg3Δ/erg3Δ-erg11Δ/erg11Δ | terbinafine, cycloheximide, fluphenazine, brefeldin A, hygromycin B | fluconazole, amphotericin B |
| ncp1Δ/ncp1 | clotrimazole, fluconazole | |
| upc2Δ/upc2Δ | fluconazoles, terbinafine, fenpropimorph, lovastatin, nikkomycin Z, calcofluor white, SDS | |
| ndt80Δ/ndt80Δ | fluconazoles, caffeine, flucytosine, rapamycin, SDS | |
| efg1Δ/efg1Δ | fluconazoles, calcofluor white, congo red, amphotericin B, antimycin A, caspofungin, nystatin |
The synthesis of ergosterol in C. albicans has been demonstrated clearly. However, the storage of ergosterol, the transformation between sterols and fatty acids and the secretion of sterols have not been extensively investigated. In S. cerevisiae, sterol and steryl ester metabolism and the involved enzymes have been researched thoroughly (Fig. 1). The genes used in the metabolism of steryl esters, such as ARE2, ATF1, SAY1, TGL1, and YEH1, have corresponding homologs in C. albicans. All these genes are uncharacterized in C. albicans except ARE2, which has been verified as a sterol acyltransferase that uses cholesterol and oleoyl-CoA as substrates and is induced by ketoconazole.47 These genes control the storage and decomposition of sterols in lipid droplets, which in S. cerevisiae contain a neutral lipid core consisting of triacylglycerols (TG) and sterol esters (SE) surrounded by phospholipid.48 Recently, it has been noted for the first time in C. albicans that cells treated with arylquinuclidine derivatives (squalene synthase inhibitors) showed a cytoplasmic accumulation of lipid droplets that could be labeled with Nile Red.49 Lipid droplets not only play an important role in overcoming the potential toxic effect of abnormal sterols and free fatty acids (FA), but also affect cellular energy homeostasis and lipid metabolism.50,51 There are still few reports about the connections between lipid droplets and drug sensitivity in C. albicans, but only one literature refers that the loss-of-function of Pdr19p located in the lipid droplets in Candida glabrata renders yeast cells more sensitive to fluconazole.52 In our opinion, paying more attention to the features of lipid droplets and improving the understanding of the storage and decomposition mechanisms of the sterols in C. albicans may provide shortcuts to solving the problem of drug resistance and provide new strategies to identify antifungal targets.
Regulation of sterol biosynthesis in C. albicans
The regulatory mechanism for the sterol biosynthesis pathway is also conserved between animals and fungi. Sterol regulatory-element binding protein (SREBP), which consists of a sterol-sensing complex, is a key transcription activator of both cellular sterol import and de novo biosynthesis.19,53 Since the first fungal ortholog of SREBP, Sre1p, was identified in Schizosaccharomyces pombe, several SREBP-like proteins have been identified in S. cerevisiae and the pathogenic fungal species Cryptococcus neoformans and Aspergillus fumigates but no clear orthologs have been identified in C. albicans (Fig. 2).54 Maguire et al. illustrated that SREBP-like proteins in S. cerevisiae (Hms1p) and C. albicans (Cph2p) have lost a DUF2014 domain (Fig. 2). As a consequence, the proteins lack the ability to regulate sterol synthesis, and instead they are implicated in regulation of filamentous growth. At the same time, SRE1 appears to have an ancestral role in regulating hyphae growth, and has been replaced in its role in sterol gene regulation by the emergence of UPC2 in the Saccharomycotina. Similar to the situation in S. cerevisiae, Upc2p is a global transcriptional activator of the ERG genes in C. albicans and has been shown to up-regulate gene expression as a compensatory mechanism in response to sterol stress.55 Disruption of UPC2 impaired cell growth in anaerobic environments and enhanced azole sensitivity. The high expression of ERG genes (ERG2, ERG6, ERG7, ERG11, ERG24, ERG25, and ERG27) and genes encoding efflux pumps (CDR1, CDR2, and MDR1) induced by fluconazole disappeared in the upc2Δ/upc2Δ mutant as measured by Northern blot and microarray analysis.56 In contrast, gain-of-function mutations in UPC2 often lead to high expression of ERG11 and make a significant contribution to azole resistance.46 These results suggest that UPC2 is essential for azole resistance in C. albicans. In response to azoles, Upc2p has the ability of transcriptional self-regulation. Upc2p can always bind to a bipartite element within its own promoter, which is a domain including the sterol response element (SRE) and a partial homology to the SRE, named the short direct repeat (SDR).57 Furthermore, the data suggest that an additional transcription factor controls sterol biosynthesis by regulating UPC2 expression, but this gene has yet to be identified. In agreement with this, Gallo-Ebert et al. found that 3 inhibitors (VB00075177, VB00075853, and VB00074845) reduced the expression of both mRNA and protein levels of UPC2 and demonstrated that these compounds’ inhibiting effect of DNA-binding is irrelevant to the direct interaction with Upc2p.58 Given these points, there likely is another transcription factor that positively regulates the expression of UPC2 and the ERG genes.
Figure 2.
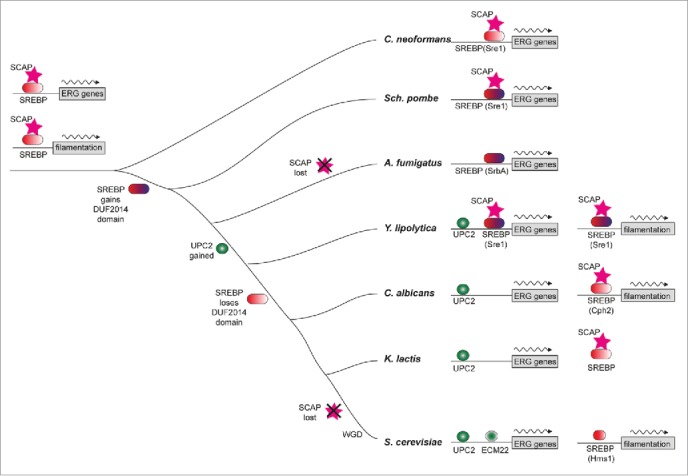
Model of sterol regulon evolution in Saccharomycotina illustrated by Maguire et al. in Plos Genetics (50). The ERG genes and filamentation genes are both regulated by SREBP and SCAP in the fungal progenitor. Upc2p gains binding sites in the promoters of ERG genes and the DUF2014 domain of SREBP, which may be important for interaction with Scap and retention of SREBP in the membrane, is lost but the protein still plays a role in regulation of filamentation in C. albicans.
As well as Upc2p, transcription factor Ndt80p can also bind to the promoters of ERG genes. By comparing transcriptional profiles of the ndt80Δ/ndt80Δ cells exposed to fluconazole or not, 7 of 10 ERG gene promoters have been identified as being transcriptionally dependent on Ndt80p, namely ERG3, ERG4, ERG6, ERG7, ERG11, ERG24, and ERG25, all of which are bound by Ndt80p directly.59 Ndt80p is required for the transcriptional activation of the sterol synthesis pathway in response to fluconazole, but the upregulation of ERG genes is not completely abolished in the ndt80Δ/ndt80Δ strains. In agreement with the ERG gene down-regulation, the absence of Ndt80p enhances antifungal drug susceptibility. However, blocking NDT80 plays little role in attempts to eliminate azole resistance in clinical isolates. This result is attributed to hyperactive forms of Mrr1p, Tac1p, and Upc2p, which up-regulate their target genes independent of Ndt80p and thereby mediate drug resistance in clinical strains.60
As well, there are other transcriptional factors that can regulate ERG gene expression and modify the sterol composition in C. albicans. The efg1Δ/efg1Δ mutant grown on solid media exhibited negative regulation of ERG11 and a simultaneous positive regulation of ERG3, and became more sensitive to azoles.61 All these transcription factors mentioned above regulate the sterol composition to promote cell growth and consequently control drug resistance and morphogenesis.
Ergosterol: The basis of normal biological membranes in C. albicans
Like cholesterol in mammals, ergosterol is an important component of membrane lipids and modulates the fluidity, permeability and integrity of the membrane in fungi. Through quantitative nano-electrospray ionization tandem mass spectrometry (nano-ESI-MS/MS) analysis of lipid extracts in S. cerevisiae, significant amounts of ergosterol have been identified in both the plasma membrane and the endomembrane system including peroxisomes, mitochondria, vacuoles and the ER.62 We have summarized the distribution and the important role of ergosterol in S.cerevisiae or C. albicans (Fig. 363).
Figure 3.
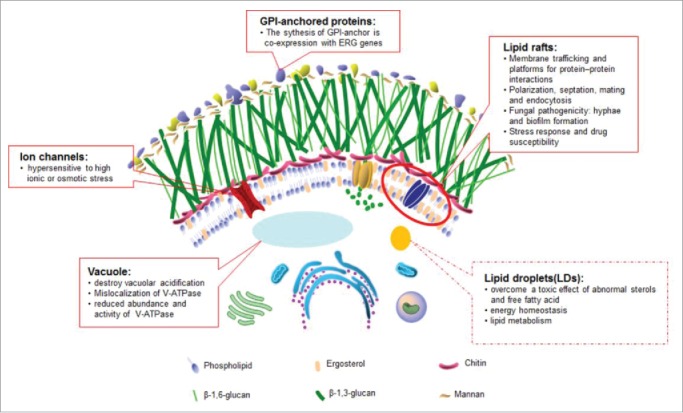
The effects of ergosterol are labeled on the C. albicans cell pattern diagram based on the picture from the Atlas of Fungal Infection.63 The functions and cellular properties of ergosterol in C. albicans are described in solid boxes, while the functions verified in S. cerevisiae but yet uncharacterized in C. albicans are noted in dotted boxes. Candida albicans Mutations in the Ergosterol Biosynthetic Pathway and Resistance to Several Antifungal Agents.
Within the plasma membrane or the membranes of intracellular organelles, microdomains with higher amounts of sterols and saturated fatty acids than the rest of the membrane are called lipid rafts; these rafts are resistant to the action of non-anionic detergents.64 The protein samples obtained from C. albicans lipid rafts were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and matrix assisted laser desorption ionization/time of flight (MALDI-TOF) mass spectroscopy. A total of 29 proteins were identified, including PMA1 (the marker for lipid rafts in S. cerevisiae), ECM33 (glycosylphosphatidylinositol (GPI)-anchored protein involved in cell wall biogenesis), MNN7, PMT2, MNT1 (mannosyltransferases), ERG11, SCS7 (proteins involved in lipid metabolism), SSA1, HSP90 (heat shock proteins) and, CDR1 (the ATP-binding cassette (ABC) multidrug transporter). The function of these proteins is consistent with the role that lipid rafts play in many biological processes, such as lipid metabolism, cell wall biogenesis, protein metabolism, electron transport, ATP synthesis and cellular stress.65-67 In addition, ergosterol enriched in lipid rafts has been implicated in many cellular processes, such as sporulation, pheromone signaling, polarization and membrane fusion during mating and endocytosis in S. cerevisiae.68,69 Meanwhile, a highly polarized ergosterol-rich domain which splits the hyphae into several cavities is formed specifically during hyphal growth. The inhibition of either sterol biosynthesis with azoles or sphingolipid biosynthesis with myriocin leads to a loss of ergosterol polarization and disruptions in hyphae morphogenesis, suggesting that lipid rafts are necessary in filamentation.63 Recently, O’Meara et al. illuminated the correlation between cell morphology and sterol synthesis using the C. albicans gene replacement and conditional expression (GRACE) collection. They identified that transcriptional disruption or repression of genes involved in the early phases of ergosterol synthesis, up to the production of episterol which was generated by ERG2, resulted in a hyphal growth deficiency, yet repression of genes functioning in the process of episterol to ergosterol modification did not.70 This result from ERG gene deficient strains was confirmed using the ergosterol biosynthesis inhibitors terbinafine and fluconazole, as well as with amphotericin B which binds to ergosterol directly.71 But the exact mechanisms of the antifungal-modulated morphogenesis phenotype have several explanations. One is that C. albicans cells treated with various azoles produce increased levels of the quorum sensing molecule farnesol, which suppresses the transition from yeast to hyphae.72 The other is that blocking of sterol biosynthesis with azoles results in a loss of the ergosterol polarization needed to form the highly ergosterol-riched domains found in the cell tips.61 Morphogenesis is a critical part of biofilm formation.73 Biofilms generally possess distinct sterol patterns in diverse phases compared with planktonic cells. Sterol analyses show that ergosterol levels are significantly decreased in both intermediate (12-30h) and mature phases (31-72h), compared to those in early-phase (0-11h) biofilms, and this is connected to the decreased expression of ergosterol biosynthetic genes (ERG25, ERG11, and ERG6).74-76
Ergosterol exists in many membranes of organelles. But its role is not confined to that of a constituent part of membranes as it forms the basis of maintaining the localization and activities of some enzymes within the membrane. Vacuole as the largest organelle in yeast cells plays an important role in several cellular functions such as environment stresses response and the yeast-hyphae switch. To function normally, the vacuole need to maintain the intravacuolar homeostasis of pH and ion, which is regulated by the vacuolar proton-translocating ATPase.77 Cells with a depletion of ERG2 or ERG24, or treated with azoles or morpholines, reduce the degree of vacuolar acidification and exhibit an abnormal morphology of the vacuole. Meanwhile, erg2Δ/erg2Δ and erg24Δ/erg24Δ mutant strains have similar phenotypes to vacuolar deficient mutants in being hypersensitive to elevated temperature and to high ionic or osmotic stress.78 Moreover, in the ERG24-defiecient strains, the V-ATPase that is required for the generation of a pH gradient in the vacuole and cytoplasm was mislocalized and reduced in abundance in the vacuolar membrane.79 Surprisingly, ergosterol depletion diminishes the function of V-ATPase in the vacuolar membrane but has no influence on Pma1, which encodes a plasma membrane H+-ATPase. This research demonstrates that a specific lipid composition is required for normal membrane protein function. For other organelles, the relationship between the mitochondrial dysfunction and the decreased fluconazole susceptibility in C. albicans has been reported by several literatures. For instance, the inhibitors of mitochondrial complex I (CI) or complex V (CV) can always improve the outcome of fluconazole treatment in patients or the lab isolates. And the CI null mutants showed hypersusceptibility to fluconazole. In C. albicans, mitochondria are power houses which are needed for many cellular processes, such as drug susceptibility, cell wall integrity, phospholipid homeostasis, and virulence.80 On one hand, mitochondrial dysfunction always cause downregulation of transporter genes (CDR1 and CDR2) and the ergosterol synthesis gene family, which in turn decrease the cellular ergosterol levels. On the other hand, dysfunctional mitochondria deregulates iron metabolism caused by perturbed Fe-S cluster biogenesis.81 Iron is a necessary cofactor for essential cellular processes as well as its toxicity (via hydroxyl radicals produced by the Fenton reaction) to proteins, lipids, and nucleic acids. C. albicans have a tight iron regulation system including 3 major transcriptional regulators, Sef1p, Sfu1p, and the Hap43p-associated CCAAT-binding complex. In addition, iron concentration is a significant signal to induce morphological transformation in C. albicans.82 Iron deprivation could increase the membrane fluidity and permeability, and reduce drug sensitivity to azoles by decreasing ERG1, ERG2, ERG11 and ERG25 transcription and ergosterol levels.83,84 There are few studies about the influence of ergosterol on other organelles in organisms other than in S. cerevisiae. In S. cerevisiae, ERG-deficient mutants exhibit defects in endocytosis, vacuolar fusion, mitochondrial morphogenesis and maintenance.85,86 Some ERG-deficient strains even show perturbations in ion uptake and utilization that are similar to those in C. albicans. Ca2+ and Mg2+ can change the growth rate of the erg24Δ/erg24Δ mutant. Li+ and Na+ are absorbed more easily in the erg6Δ/erg6Δ mutant.87,88 These functions of ergosterol demonstrated in S. cerevisiae are still obscure in C. albicans. We need to be cautious about how ergosterol affects the activity of enzymes on the membranes and the other membrane functions besides vacuolar acidification.
Perspectives
Since the first azoles targeting ERG11 became available in 1960s, extensive research has been carried out to find new chemicals to inhibit the synthesis of ergosterol. Fluconazole does not have an antifungal effect as strong as amphotericin B, the latter which destroys the cell membrane through extracting ergosterol from lipid bilayers and is fungicidal.89 Its antifungal mechanisms include ergosterol depletion, aberrant sterol accumulation and the hypothesis of unbalanced proportion between ergosterol and other sterols.79 The fungistatic effects may result from the comprehensive factors, but there is no doubt that lipid mobilization plays an important role in C. albicans fighting against pressures from the outside environment. Due to the fact that C. albicans cannot take up sterols from external sources under anaerobic conditions, it is likely that studying the storage and metabolism of sterols in pathogenic fungi will be informative. Lipid droplets are a particularly interesting area for future investigation. By inhibiting the storage or release of sterols and synergizing with the sterol synthesis inhibition, we may find new strategies to kill fungi and solve the problems of azole resistance. Apart from the critical role of ergosterol, the regulation of the ERG genes exhibits cross talk with many other pathways, such as those controlling morphological transformation, biofilm formation, and GPI-anchor synthesis. Identifying the key signaling molecular process that adjusts the proportions of various kinds of sterols and what is the co-regulator that coordinates lipid metabolism and other biological processes will be critical. This information should bring us new ideas to further exploit the sterol synthesis pathway as an antifungal target.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Malcolm Whiteway (Concordia University) for critical reading of our manuscript. The authors would like to apologize to all researchers whose important works were unable to be cited because of space limitations.
Funding
L.Y. is supported by Natural Science Foundation of China (81470158). Y.J. is supported by China National 973 Program (2013CB531602) and Natural Science Foundation of China (81330083).
References
- [1].Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med 2012; 4:165rv13; PMID:23253612 [DOI] [PubMed] [Google Scholar]
- [2].Martins N, Ferreira IC, Barros L, Silva S, Henriques M. Candidiasis: predisposing factors, prevention, diagnosis and alternative treatment. Mycopathologia 2014; 177:223-40; PMID:24789109; http://dx.doi.org/ 10.1007/s11046-014-9749-1 [DOI] [PubMed] [Google Scholar]
- [3].Parker JE, Warrilow AG, Price CL, Mullins JG, Kelly DE, Kelly SL. Resistance to antifungals that target CYP51. J Chem Biol 2014; 7:143-61; PMID:25320648; http://dx.doi.org/ 10.1007/s12154-014-0121-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ostrosky-Zeichner L, Casadevall A, Galgiani JN, Odds FC, Rex JH. An insight into the antifungal pipeline: selected new molecules and beyond. Nat Rev Drug Discov 2010; 9:719-27; PMID:20725094; http://dx.doi.org/ 10.1038/nrd3074 [DOI] [PubMed] [Google Scholar]
- [5].Solomon SL, Oliver KB. Antibiotic resistance threats in the United States: stepping back from the brink. Am Fam Physician 2014; 89:938-41; PMID:25162160 [PubMed] [Google Scholar]
- [6].Xie JL, Polvi EJ, Shekhar-Guturja T, Cowen LE. Elucidating drug resistance in human fungal pathogens. Future Microbiol 2014; 9:523-42; PMID:24810351; http://dx.doi.org/ 10.2217/fmb.14.18 [DOI] [PubMed] [Google Scholar]
- [7].Noel T. The cellular and molecular defense mechanisms of the Candida yeasts against azole antifungal drugs. J Mycol Med 2012; 22:173-8; PMID:23518020; http://dx.doi.org/ 10.1016/j.mycmed.2012.04.004 [DOI] [PubMed] [Google Scholar]
- [8].Kontoyiannis DP, Sagar N, Hirschi KD. Overexpression of Erg11p by the regulatable GAL1 promoter confers fluconazole resistance in Saccharomyces cerevisiae. Antimicrob Agents Chemother 1999; 43:2798-800; PMID:10543768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Coste A, Selmecki A, Forche A, Diogo D, Bougnoux ME, d'Enfert C, Berman J, Sanglard D. Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryot Cell 2007; 6:1889-904; PMID:17693596; http://dx.doi.org/ 10.1128/EC.00151-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Schubert S, Rogers PD, Morschhauser J. Gain-of-function mutations in the transcription factor MRR1 are responsible for overexpression of the MDR1 efflux pump in fluconazole-resistant Candida dubliniensis strains. Antimicrob Agents Chemother 2008; 52:4274-80; PMID:18809934; http://dx.doi.org/ 10.1128/AAC.00740-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Prasad R, Kapoor K. Multidrug resistance in yeast Candida. Int Rev of Cytol 2005; 242:215-48; PMID:15598470; http://dx.doi.org/ 10.1016/S0074-7696(04)42005-1 [DOI] [PubMed] [Google Scholar]
- [12].Liu TT, Znaidi S, Barker KS, Xu L, Homayouni R, Saidane S, Morschhäuser J, Nantel A, Raymond M, Rogers PD. Genome-wide expression and location analyses of the Candida albicans Tac1p regulon. Eukaryot Cell 2007; 6:2122-38; PMID:17905926; http://dx.doi.org/ 10.1128/EC.00327-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dunkel N, Blass J, Rogers PD, Morschhauser J. Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol Microbiol 2008; 69:827-40; PMID:18577180; http://dx.doi.org/ 10.1111/j.1365-2958.2008.06309.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Karababa M, Valentino E, Pardini G, Coste AT, Bille J, Sanglard D. CRZ1, a target of the calcineurin pathway in Candida albicans. Mol Microbiol 2006; 59:1429-51; PMID:16468987; http://dx.doi.org/ 10.1111/j.1365-2958.2005.05037.x [DOI] [PubMed] [Google Scholar]
- [15].Nett JE, Sanchez H, Cain MT, Andes DR. Genetic basis of Candida biofilm resistance due to drug-sequestering matrix glucan. J Infec Dis 2010; 202:171-5; PMID:20497051; http://dx.doi.org/ 10.1086/651200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Daum G, Wagner A, Czabany T, Athenstaedt K. Dynamics of neutral lipid storage and mobilization in yeast. Biochimie 2007; 89:243-8; PMID:16919863; http://dx.doi.org/ 10.1016/j.biochi.2006.06.018 [DOI] [PubMed] [Google Scholar]
- [17].Abe F, Usui K, Hiraki T. Fluconazole modulates membrane rigidity, heterogeneity, and water penetration into the plasma membrane in Saccharomyces cerevisiae. Biochemistry 2009; 48:8494-504; PMID:19670905; http://dx.doi.org/ 10.1021/bi900578y [DOI] [PubMed] [Google Scholar]
- [18].Sgherri C, Porta A, Castellano S, Pinzino C, Quartacci MF, Calucci L. Effects of azole treatments on the physical properties of Candida albicans plasma membrane: a spin probe EPR study. Biochimica Et Biophysica Acta 2014; 1838:465-73; PMID:24184423; http://dx.doi.org/ 10.1016/j.bbamem.2013.10.015 [DOI] [PubMed] [Google Scholar]
- [19].Espenshade PJ, Hughes AL. Regulation of sterol synthesis in eukaryotes. Annu Rev of Genet 2007; 41:401-27; PMID:17666007; http://dx.doi.org/ 10.1146/annurev.genet.41.110306.130315 [DOI] [PubMed] [Google Scholar]
- [20].Bard M, Sturm AM, Pierson CA, Brown S, Rogers KM, Nabinger S, Eckstein J, Barbuch R, Lees ND, Howell SA, et al.. Sterol uptake in Candida glabrata: rescue of sterol auxotrophic strains. Diagn Microbiol Infect Dis 2005; 52:285-93; PMID:15893902; http://dx.doi.org/ 10.1016/j.diagmicrobio.2005.03.001 [DOI] [PubMed] [Google Scholar]
- [21].Zavrel M, Hoot SJ, White TC. Comparison of sterol import under aerobic and anaerobic conditions in three fungal species, Candida albicans, Candida glabrata, and Saccharomyces cerevisiae. Eukaryot Cell 2013; 12:725-38; PMID:23475705; http://dx.doi.org/ 10.1128/EC.00345-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Martel CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AG, Rolley N, Kelly DE, Kelly SL. Identification and characterization of four azole-resistant erg3 mutants of Candida albicans. Antimicrob Agents Chemother 2010; 54:4527-33; PMID:20733039; http://dx.doi.org/ 10.1128/AAC.00348-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Theesfeld CL, Hampton RY. Insulin-induced gene protein (INSIG)-dependent sterol regulation of Hmg2 endoplasmic reticulum-associated degradation (ERAD) in yeast. J Biol Chem 2013; 288:8519-30; PMID:23306196; http://dx.doi.org/ 10.1074/jbc.M112.404517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pasrija R, Krishnamurthy S, Prasad T, Ernst JF, Prasad R. Squalene epoxidase encoded by ERG1 affects morphogenesis and drug susceptibilities of Candida albicans. J Antimicrob Chemother 2005; 55:905-13; PMID:15845783; http://dx.doi.org/ 10.1093/jac/dki112 [DOI] [PubMed] [Google Scholar]
- [25].Kennedy MA, Johnson TA, Lees ND, Barbuch R, Eckstein JA, Bard M. Cloning and sequencing of the Candida albicans C-4 sterol methyl oxidase gene (ERG25) and expression of an ERG25 conditional lethal mutation in Saccharomyces cerevisiae. Lipids 2000; 35:257-62; PMID:10783002; http://dx.doi.org/ 10.1007/s11745-000-0521-2 [DOI] [PubMed] [Google Scholar]
- [26].Aaron KE, Pierson CA, Lees ND, Bard M. The Candida albicans ERG26 gene encoding the C-3 sterol dehydrogenase (C-4 decarboxylase) is essential for growth. FEMS Yeast Res 2001; 1:93-101; PMID:12702354; http://dx.doi.org/ 10.1111/j.1567-1364.2001.tb00020.x [DOI] [PubMed] [Google Scholar]
- [27].Pierson CA, Jia N, Mo C, Lees ND, Sturm AM, Eckstein J, Barbuct R, Bard M. Isolation, characterization, and regulation of theCandidaalbicansERG27gene encoding the sterol 3-keto reductase. Med Mycol 2004; 42:461-73; PMID:15552648; http://dx.doi.org/ 10.1080/1369378032000141471 [DOI] [PubMed] [Google Scholar]
- [28].Mukhopadhyay K, Prasad T, Saini P, Pucadyil TJ, Chattopadhyay A, Prasad R. Membrane sphingolipid-ergosterol interactions are important determinants of multidrug resistance in Candida albicans. Antimicrob Agents Chemother 2004; 48:1778-87; PMID:15105135; http://dx.doi.org/ 10.1128/AAC.48.5.1778-1787.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Martel CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AG, Kelly DE, Kelly SL. A clinical isolate of Candida albicans with mutations in ERG11 (encoding sterol 14alpha-demethylase) and ERG5 (encoding C22 desaturase) is cross resistant to azoles and amphotericin B. Antimicrob Agents Chemother 2010; 54:3578-83; PMID:20547793; http://dx.doi.org/ 10.1128/AAC.00303-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jensen-Pergakes KL, Kennedy MA, Lees ND, Barbuch R, Koegel C, Bard M. Sequencing, disruption, and characterization of the Candida albicans sterol methyltransferase (ERG6) gene: drug susceptibility studies in erg6 mutants. Antimicrob Agents Chemother 1998; 42:1160-7; PMID:9593144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jia N, Arthington-Skaggs B, Lee W, Pierson CA, Lees ND, Eckstein J, Barbuch R, Bard M. Candida albicans Sterol C-14 reductase, encoded by the ERG24 gene, as a potential antifungal target site. Antimicrob Agents Chemother 2002; 46:947-57; PMID:11897574; http://dx.doi.org/ 10.1128/AAC.46.4.947-957.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Martel CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AG, Kelly DE, Kelly SL. A clinical isolate of Candida albicans with mutations in ERG11 (encoding sterol 14alpha-demethylase) and ERG5 (encoding C22 desaturase) is cross resistant to azoles and amphotericin B. Antimicrob Agents Chemother 2010; 54:3578-83; PMID:20547793; http://dx.doi.org/ 10.1128/AAC.00303-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sanglard D, Ischer F, Parkinson T, Falconer D, Bille J. Candida albicans mutations in the ergosterol biosynthetic pathway and resistance to several antifungal agents. Antimicrob Agents Chemother 2003; 47:2404-12; PMID:12878497; http://dx.doi.org/ 10.1128/AAC.47.8.2404-2412.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Keller P, Muller C, Engelhardt I, Hiller E, Lemuth K, Eickhoff H, Wiesmüller KH, Burger-Kentischer A, Bracher F, Rupp S. An antifungal benzimidazole derivative inhibits ergosterol biosynthesis and reveals novel sterols. Antimicrob Agents Chemother 2015; 59:6296-307; PMID:26248360; http://dx.doi.org/ 10.1128/AAC.00640-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pierson CA, Eckstein J, Barbuch R, Bard M. Ergosterol gene expression in wild-type and ergosterol-deficient mutants of Candida albicans. Med Mycol 2004; 42:385-9; PMID:15473366; http://dx.doi.org/ 10.1080/13693780410001712016 [DOI] [PubMed] [Google Scholar]
- [36].Liu TT, Lee RE, Barker KS, Lee RE, Wei L, Homayouni R, Rogers PD. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob Agents Chemother 2005; 49:2226-36; PMID:15917516; http://dx.doi.org/ 10.1128/AAC.49.6.2226-2236.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Arthington-Skaggs BA1, Jradi H, Desai T, Morrison CJ. Quantitation of ergosterol content: novel method for determination of fluconazole susceptibility of Candida albicans. J Clin Microbiol 1999; 10:3332-7; PMID:10488201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Morio F, Pagniez F, Lacroix C, Miegeville M, Le Pape P. Amino acid substitutions in the Candida albicans sterol Delta5,6-desaturase (Erg3p) confer azole resistance: characterization of two novel mutants with impaired virulence. J Antimicrob Chemother 2012; 67:2131-8; PMID:22678731; http://dx.doi.org/ 10.1093/jac/dks186 [DOI] [PubMed] [Google Scholar]
- [39].Bard M, Lees ND, Barbuch RJ, Sanglard D. Characterization of a cytochrome P450 deficient mutant of Candida albicans. Biochem Biophys Res Commun 1987; 147:794-800; PMID:3307785; http://dx.doi.org/ 10.1016/0006-291X(87)91000-X [DOI] [PubMed] [Google Scholar]
- [40].Shapiro RS, Robbins N, Cowen LE. Regulatory circuitry governing fungal development, drug resistance, and disease. MMBR 2011; 75:213-67; PMID:21646428; http://dx.doi.org/ 10.1128/MMBR.00045-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cruz MC, Goldstein AL, Blankenship JR, Del Poeta M, Davis D, Cardenas ME, Perfect JR, McCusker JH, Heitman J. Heitman J. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J 2002; 21:546-59; PMID:11847103; http://dx.doi.org/ 10.1093/emboj/21.4.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Golabek K, Strzelczyk JK, Owczarek A, Cuber P, Slemp-Migiel A, Wiczkowski A. Selected mechanisms of molecular resistance of Candida albicans to azole drugs. Acta Biochimica Polonica 2015; 62:247-51; PMID:25901298; http://dx.doi.org/ 10.18388/abp.2014_940 [DOI] [PubMed] [Google Scholar]
- [43].Liu JY, Shi C, Wang Y, Li WJ, Zhao Y, Xiang MJ. Mechanisms of azole resistance in Candida albicans clinical isolates from Shanghai, China. Res in Microbiol 2015; 166:153-61; PMID:25748216; http://dx.doi.org/ 10.1016/j.resmic.2015.02.009 [DOI] [PubMed] [Google Scholar]
- [44].Wang H, Kong F, Sorrell TC, Wang B, McNicholas P, Pantarat N, Ellis D, Xiao M, Widmer F, Chen SC. Rapid detection of ERG11 gene mutations in clinical Candida albicans isolates with reduced susceptibility to fluconazole by rolling circle amplification and DNA sequencing. BMC Microbiology 2009; 9:167; PMID:19682357; http://dx.doi.org/ 10.1186/1471-2180-9-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Eddouzi J, Parker JE, Vale-Silva LA, Coste A, Ischer F, Kelly S, Manai M, Sanglard D. Molecular mechanisms of drug resistance in clinical Candida species isolated from Tunisian hospitals. Antimicrob Agents Chemother 2013; 57:3182-93; PMID:23629718; http://dx.doi.org/ 10.1128/AAC.00555-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Flowers SA, Barker KS, Berkow EL, Toner G, Chadwick SG, Gygax SE, Morschhäuser J, Rogers PD. Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot Cell 2012; 11:1289-99; PMID:22923048; http://dx.doi.org/ 10.1128/EC.00215-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kim KY, Shin YK, Park JC, Kim JH, Yang H, Han DM, Paik YK. Molecular cloning and biochemical characterization of Candida albicans acyl-CoA:sterol acyltransferase, a potential target of antifungal agents. Biochem Biophys Res Commun 2004; 319:911-9; PMID:15184069; http://dx.doi.org/ 10.1016/j.bbrc.2004.05.076 [DOI] [PubMed] [Google Scholar]
- [48].Beller M, Thiel K, Thul PJ, Jackle H. Lipid droplets: a dynamic organelle moves into focus. FEBS Lett 2010; 584:2176-82; PMID:20303960; http://dx.doi.org/ 10.1016/j.febslet.2010.03.022 [DOI] [PubMed] [Google Scholar]
- [49].Ishida K, Fernandes Rodrigues JC, Cammerer S, Urbina JA, Gilbert I, de Souza W, Rozental S. Synthetic arylquinuclidine derivatives exhibit antifungal activity against Candida albicans, Candida tropicalis and Candida parapsilopsis. Ann Clin Microbiol Antimicrob 2011; 10:3; PMID:21255433; http://dx.doi.org/ 10.1186/1476-0711-10-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Debelyy MO, Thoms S, Connerth M, Daum G, Erdmann R. Involvement of the Saccharomyces cerevisiae hydrolase Ldh1p in lipid homeostasis. Eukaryot Cell 2011; 10:776-81; PMID:21478434; http://dx.doi.org/ 10.1128/EC.05040-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Klug L, Daum G. Yeast lipid metabolism at a glance. FEMS Yeast Res 2014; 14:369-88; PMID:24520995; http://dx.doi.org/ 10.1111/1567-1364.12141 [DOI] [PubMed] [Google Scholar]
- [52].Culakova H, Dzugasova V, Valencikova R, Gbelska Y, Subik J. Stress response and expression of fluconazole resistance associated genes in the pathogenic yeast Candida glabrata deleted in the CgPDR16 gene. Microbiol Res 2015; 174:17-23; PMID:25946325; http://dx.doi.org/ 10.1016/j.micres.2015.03.004 [DOI] [PubMed] [Google Scholar]
- [53].Bien CM, Espenshade PJ. Sterol regulatory element binding proteins in fungi: hypoxic transcription factors linked to pathogenesis. Eukaryot Cell 2010; 9:352-9; PMID:20118213; http://dx.doi.org/ 10.1128/EC.00358-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Maguire SL, Wang C, Holland LM, Brunel F, Neuveglise C, Nicaud JM, Zavrel M, White TC, Wolfe KH, Butler G. Zinc finger transcription factors displaced SREBP proteins as the major Sterol regulators during Saccharomycotina evolution. PLoS Genet 2014; 10:e1004076; PMID:24453983; http://dx.doi.org/ 10.1371/journal.pgen.1004076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Silver PM, Oliver BG, White TC. Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot Cell 2004; 3:1391-7; PMID:15590814; http://dx.doi.org/ 10.1128/EC.3.6.1391-1397.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].MacPherson S, Akache B, Weber S, De Deken X, Raymond M, Turcotte B. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob Agents Chemother 2005; 49:1745-52; PMID:15855491; http://dx.doi.org/ 10.1128/AAC.49.5.1745-1752.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hoot SJ1, Brown RP, Oliver BG, White TC. The UPC2 promoter in Candida albicans contains two cis-acting elements that bind directly to Upc2p, resulting in transcriptional autoregulation. Eukaryot Cell 2010; 9:1354-1362; PMID:20656915; http://dx.doi.org/ 10.1128/EC.00130-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Gallo-Ebert C, Donigan M, Stroke IL, Swanson RN, Manners MT, Francisco J, Toner G, Gallagher D, Huang CY, Gygax SE, et al.. Novel antifungal drug discovery based on targeting pathways regulating the fungus-conserved Upc2 transcription factor. Antimicrob Agents Chemother 2014; 58:258-66; PMID:24145546; http://dx.doi.org/ 10.1128/AAC.01677-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Sellam A, Tebbji F, Nantel A. Role of Ndt80p in sterol metabolism regulation and azole resistance in Candida albicans. Eukaryot Cell 2009; 8:1174-83; PMID:19542309; http://dx.doi.org/ 10.1128/EC.00074-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Sasse C, Schillig R, Dierolf F, Weyler M, Schneider S, Mogavero S, Rogers PD, Morschhäuser J. The transcription factor Ndt80 does not contribute to Mrr1-, Tac1-, and Upc2-mediated fluconazole resistance in Candida albicans. PLoS One 2011; 6:1-9; PMID:21980509; http://dx.doi.org/ 10.1371/journal.pone.0025623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Prasad T, Hameed S, Manoharlal R, Biswas S, Mukhopadhyay CK, Goswami SK, Prasad R. Morphogenic regulator EFG1 affects the drug susceptibilities of pathogenic Candida albicans. FEMS Yeast Res 2010; 10:587-96; PMID:20491944 [DOI] [PubMed] [Google Scholar]
- [62].Schneiter R, Brugger B, Sandhoff R, Zellnig G, Leber A, Lampl M, Athenstaedt K, Hrastnik C, Eder S, Daum G, et al.. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J Cell Biol 1999; 146:741-54; PMID:10459010; http://dx.doi.org/ 10.1083/jcb.146.4.741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kartsonis NA, Nielsen J, Douglas CM. Caspofungin: the first in a new class of antifungal agents. Drug resistance updates: reviews and commentaries in antimicrobial and anticancer chemotherapy 2003; 6:197–218. [DOI] [PubMed] [Google Scholar]
- [64].Martin SW, Konopka JB. Lipid raft polarization contributes to hyphal growth in Candida albicans. Eukaryot Cell 2004; 3:675-84; PMID:15189988; http://dx.doi.org/ 10.1128/EC.3.3.675-684.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Mollinedo F. Lipid raft involvement in yeast cell growth and death. Front in Oncol 2012; 2:140; PMID:23087902; http://dx.doi.org/ 10.3389/fonc.2012.00140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Insenser M, Nombela C, Molero G, Gil C. Proteomic analysis of detergent-resistant membranes from Candida albicans. Proteomics 2006; 6(Suppl 1):S74-81; PMID:16534748; http://dx.doi.org/ 10.1002/pmic.200500465 [DOI] [PubMed] [Google Scholar]
- [67].Pasrija R1, Panwar SL, Prasad R. Multidrug transporters CaCdr1p and CaMdr1p of Candida albicans display different lipid specificities: both ergosterol and sphingolipids are essential for targeting of CaCdr1p to membrane rafts. Antimicrob Agents Chemother 2008; 52:694-704; PMID:18056285; http://dx.doi.org/ 10.1128/AAC.00861-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Thorpe GW, Fong CS, Alic N, Higgins VJ, Dawes IW. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc Natl Acad Sci USA 2004; 101:6564-9; PMID:15087496; http://dx.doi.org/ 10.1073/pnas.0305888101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Umebayashi K, Nakano A. Ergosterol is required for targeting of tryptophan permease to the yeast plasma membrane. J Cell Biol 2003; 161:1117-31; PMID:12810702; http://dx.doi.org/ 10.1083/jcb.200303088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].O'Meara TR, Veri AO, Ketela T, Jiang B, Roemer T, Cowen LE. Global analysis of fungal morphology exposes mechanisms of host cell escape. Nat Commun 2015; 6:6741; PMID:25824284; http://dx.doi.org/ 10.1038/ncomms7741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Odds FC, Cockayne A, Hayward J, Abbott AB. Effects of imidazole- and triazole-derivative antifungal compounds on the growth and morphological development of Candida albicans hyphae. J Gen Microbiol 1985; 131:2581-9; PMID:2999296 [DOI] [PubMed] [Google Scholar]
- [72].Hornby JM, Kebaara BW, Nickerson KW. Farnesol biosynthesis in Candida albicans: cellular response to sterol inhibition by zaragozic acid B. Antimicrob Agents Chemother 2003; 47:2366-9; PMID:12821501; http://dx.doi.org/ 10.1128/AAC.47.7.2366-2369.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Blankenship JR, Mitchell AP. How to build a biofilm: a fungal perspective. Curr Opin Microbiol 2006; 9:588-94; PMID:17055772; http://dx.doi.org/ 10.1016/j.mib.2006.10.003 [DOI] [PubMed] [Google Scholar]
- [74].Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol 2001; 183:5385-94; PMID:11514524; http://dx.doi.org/ 10.1128/JB.183.18.5385-5394.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Mukherjee PK, Chandra J, Kuhn DM, Ghannoum MA. Mechanism of fluconazole resistance in Candida albicans biofilms: phase-specific role of efflux pumps and membrane sterols. Infect Immun 2003; 71:4333-40; PMID:12874310; http://dx.doi.org/ 10.1128/IAI.71.8.4333-4340.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Garcia-Sanchez S, Aubert S, Iraqui I, Janbon G, Ghigo JM, d'Enfert C. Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryot Cell 2004; 3:536-45; PMID:15075282; http://dx.doi.org/ 10.1128/EC.3.2.536-545.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Olsen I. Attenuation of Candida albicans virulence with focus on disruption of its vacuole functions. J Oral Microbiol 2014; 6:23898; PMID:24765242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Luna-Tapia A, Peters BM, Eberle KE, Kerns ME, Foster TP, Marrero L, Noverr MC, Fidel PL Jr, Palmer GE. ERG2 and ERG24 are required for normal vacuolar physiology as well as Candida albicans pathogenicity in a murine model of disseminated but not vaginal candidiasis. Eukaryot Cell 2015; 14(10):1006-16; PMID:26231054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Zhang YQ, Gamarra S, Garcia-Effron G, Park S, Perlin DS, Rao R. Requirement for ergosterol in V-ATPase function underlies antifungal activity of azole drugs. PLoS Pathog 2010; 6:e1000939; PMID:20532216; http://dx.doi.org/ 10.1371/journal.ppat.1000939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Sun N, Fonzi W, Chen H, She X, Zhang L, Zhang L, Calderone R. Azole susceptibility and transcriptome profiling in Candida albicans mitochondrial electron transport chain complex I mutants. Antimicrob Agents Chemother 2013; 57:532-42; PMID:23147730; http://dx.doi.org/ 10.1128/AAC.01520-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Thomas E, Roman E, Claypool S, Manzoor N, Pla J, Panwar SL. Mitochondria influence CDR1 efflux pump activity, Hog1-mediated oxidative stress pathway, iron homeostasis, and ergosterol levels in Candida albicans. Antimicrob Agents Chemother 2013; 57:5580-99; PMID:23979757; http://dx.doi.org/ 10.1128/AAC.00889-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Chen C, Pande K, French SD, Tuch BB, Noble SM. An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe 2011; 10:118-35; PMID:21843869; http://dx.doi.org/ 10.1016/j.chom.2011.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Prasad T, Chandra A, Mukhopadhyay CK, Prasad R. Unexpected link between iron and drug resistance of Candida spp.: iron depletion enhances membrane fluidity and drug diffusion, leading to drug-susceptible cells. Antimicrob Agents Ch 2006; 50:3597-606; PMID:16954314; http://dx.doi.org/ 10.1128/AAC.00653-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Hameed S, Dhamgaye S, Singh A, Goswami SK, Prasad R. Calcineurin signaling and membrane lipid homeostasis regulates iron mediated multidrug resistance mechanisms in Candida albicans. PloS one 2011; 6:e18684; PMID:21533276; http://dx.doi.org/ 10.1371/journal.pone.0018684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Zhang YQ, Rao R. Beyond ergosterol: linking pH to antifungal mechanisms. Virulence 2010; 1:551-4; PMID:21178501; http://dx.doi.org/ 10.4161/viru.1.6.13802 [DOI] [PubMed] [Google Scholar]
- [86].Heese-Peck A. Multiple functions of sterols in yeast endocytosis. Mole Biolo Cell 2002; 13:2664-80; PMID:12181337; http://dx.doi.org/ 10.1091/mbc.E02-04-0186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Crowley JH, Tove S, Parks LW. A calcium-dependent ergosterol mutant of Saccharomyces cerevisiae. Curr Genet 1998; 34:93-9; PMID:9724410; http://dx.doi.org/ 10.1007/s002940050371 [DOI] [PubMed] [Google Scholar]
- [88].Welihinda AA, Beavis AD, Trumbly RJ. Mutations in LIS1 (ERG6) gene confer increased sodium and lithium uptake in Saccharomyces cerevisiae. Biochimica Et Biophysica Acta 1994; 1193:107-17; PMID:8038180; http://dx.doi.org/ 10.1016/0005-2736(94)90339-5 [DOI] [PubMed] [Google Scholar]
- [89].Anderson TM, Clay MC, Cioffi AG, Diaz KA, Hisao GS, Tuttle MD, Nieuwkoop AJ, Comellas G, Maryum N, Wang S, et al.. Amphotericin forms an extramembranous and fungicidal sterol sponge. Nat Chem Biol 2014; 10:400-6; PMID:24681535; http://dx.doi.org/ 10.1038/nchembio.1496 [DOI] [PMC free article] [PubMed] [Google Scholar]


