ABSTRACT
Somatic embryogenesis (SE) is an important avenue for regeneration of many plants. Although documented over half a century ago, the process of SE remains poorly understood and many factors impact upon competence for SE. We recently reported that a Glycine max ortholog of a MADS-domain transcription factor that promotes SE in Arabidopsis also enhances SE in soybean. We recently assessed transcriptomes in 35Spro:GmAGL15 compared to control during an early time-course of SE and in response to 35Spro:AtAGL15. We expand here upon discussion of the types of genes regulated by overexpression of AGL15 and characterize the step of SE that may be affected by altered accumulation of AGL15.
KEYWORDS: Arabidopsis thaliana, competence, gene regulation, Glycine max, MADS-domain, microarray, NAC, quiescent center, root meristem, WRKY
Abbreviations
- AtAGL15
Arabidopsis thaliana AGAMOUS-Like15
- GmAGL15
Glycine max AGL15
- NAC
for Petunia NAM and for Arabidopsis ATAF1, ATAF2, and CUC2
- QC
quiescent center
- SAM
shoot apical meristem
- SE
somatic embryogenesis
- TF
transcription factor
- WT
wild type
Plasticity in development is an important feature of plants allowing them to cope with biotic and abiotic stresses. This includes postembryonic development from the meristems that allows the plant to control development in response to environmental cues, as well as regenerative processes such as tissue regeneration upon wounding. Somatic embryogenesis (SE) has been proposed to be an extreme response to stress where single cells are able to regenerate the entire plant (for a review see ref. 1). Somatic embryogenesis, as well as organogenesis, are important modes of plant regeneration for biotechnology, but are not well understood, and this is especially true for SE that was first described over half a century ago but was still a featured question in Science magazine's “What don't we know?” in 2005.2 Both regeneration processes are believed to occur by an inducing signal leading to changes in differentiation, followed by reprogramming to new fates. Somatic embryos differ from organogenesis in that bipolar structures develop from individual cells and the resulting embryo has no vascular connections with the explant tissue.3 Commonly, the synthetic auxin 2,4-D is present in the medium to induce SE and this compound may act as an auxin, induce production of endogenous auxin, but is also commonly considered to be a stressing agent. The process of explant preparation (wounding) is also a stressing agent. For SE, during initiation proembryos start developing, followed by maturation during which storage products accumulate (for a review see ref. 4). A variety of factors including but not limited to species, explant source, and developmental stage impact upon competence for SE. Competence can vary even between particular cultivars of a species and gene expression changes have been documented between embryogenic, and less embryogenic cultivars (for an example, see ref. 5). SE may be direct, where cells give rise to SEs, or indirect where an intervening callus phase exists.
The MADS-domain transcription factor AGAMOUS-Like15 (AGL15) has been shown to promote SE in 2 systems in Arabidopsis thaliana (At) as well as in Glycine max (Gm). One system involves culturing immature wounded zygotic embryos of 35Spro:AtAGL15 or controls on hormone free medium. In some less embryogenic ecotypes (Wassilewskija, Ws) the 35S promoter increases the number of explants with embryos compared to wild type (WT) but for the more embryogenic Columbia (Col) ecotype there was no difference at this stage. However, for both ecotypes, the 35Spro:AtAGL15 are able to proliferate as embryo tissue for extensive time periods (the oldest cultures will be 20 y old as of November 2016). Nontransgenic controls or loss-of-function in agl15 and the related agl18 cannot maintain this development.6,7 Because 2,4-D causes expression of AtAGL15,8 expression via the 35S promoter may eliminate the need for this commonly used inducer of SE from Arabidopsis zygotic explants. However, in the other SE system we have used, 2,4-D is needed to induce SE. In this system pioneered by Mordhorst et al.,9 mature seeds are allowed to complete germination in a liquid media with 4.5 μM 2,4-D, there is no deliberate wounding, and within 21 days, a fraction will have SEs at the apical meristem. 35Spro:AtAGL15 approximately doubles the number of seedlings with this development compared to WT, whereas agl15 agl18 shows about half the development as in WT. Proliferation of 35Spro:AtAGL15 SE tissue is also more extensive.6, 7 In soybean, immature zygotic cotyledon explants are placed onto medium with 2,4-D and 35Spro:GmAGL15 was found to significantly increase SE, both initiation and longer term proliferation.10
To understand how AGL15 promotes SE, we undertook expression microarray experiments in both species. For Arabidopsis the samples were 35Spro:AtAGL15, Col, WT, and agl15 agl18 cultured for 10 d in the SAM SE system.11 This is prior to any obvious embryo development but long after exposure to 2,4-D. For the soybean experiment explants (0 d after culture, dac) were compared to 3 and 7 dac for 35Spro:GmAGL15 compared to Jack WT. This allowed not only comparison between 35Spro:GmAGL15 and Jack, but also a short timecourse before and after exposure to 2,4-D. Again this is long before any obvious embryo or callus development.
Interestingly, the transcriptome in the explants of 35Spro:GmAGL15 resembled the transcriptome of WT after 3 dac on 2,4-D 10 however the endogenous indole-3-acetic acid content was actually lower in the overexpressors (submitted). The list of genes with increased transcript at 0 dac in 35Spro:GmAGL15 compared to Jack and also increased transcript in Jack at 3 dac on 2,4-D compared to the explants was overrepresented for genes involved in stress response.10 Because stress is an inducing factor for SE, and is believed to function at a change in developmental status, we examined the overlap between our gene lists and those collected by others studying callus formation which has been considered a dedifferentiation event with some recent results indicating that differentiation to a root meristem identity may be a more accurate description.12
Gm/AtAGL15 may promote SE by influencing dedifferentiation, an early step in regeneration
Grafi et al.13 surveyed datasets to generate a list of transcription factors associated with cells undergoing de-differentiation that are perhaps entering a stem cell like state. These transcription factors fell into 3 families: ANAC, WRKY and b-ZIPs. Intriguingly, 42% (8 of 19) of the ANACs associated with dedifferentiation were expressed in response to 35Spro:AtAGL15 and 3 of these may be directly regulated. Two additional genes showed significant decrease in transcript in response to 35Spro:AtAGL15, but these also were reduced in the agl15 agl18 double mutant relative to Col WT. Four of the 19 ANACs associated with dedifferentiation (21%) had putative Glycine max orthologs that were expressed in response to 35Spro:GmAGL15 compared to Jack WT in the explant tissue prior to placement on the SE 2,4-D containing induction medium. Because only 9 of the ANACs have predicted orthologs on the Affymetrix soybean genome array, this fraction of responsive genes is likely an underestimate and considering this up to 44% of the dedifferentiation-associated soybean ANAC genes present on the chip are expressed in response to 35Spro:GmAGL15 in explant tissue (presence of putative orthologs was determined by the Affychip annotation file comparing the Soybean Genome v 1.1 to Arabidopsis TAIR10 from Soybase, http://soybase.org/AffyChip/AffychipAnnotation_Glyma1.1.txt?Submit=Submit). Orthologs for all 4 of these genes also showed increased transcript accumulation when comparing Jack WT after 3 dac in SE inductive medium compared to the explants. Additional orthologs in soybean as well as 2 other ANAC “dedifferentiation-associated” genes were also expressed in Jack after 3 dac. These data are summarized in Table 1. A sixth gene (At1g69490) had a soybean ortholog that was significantly repressed in 35Spro:GmAGL15 explants compared to Jack WT. It was significantly expressed at 3 dac and had another ortholog that with significantly more transcript in 35Spro:GmAGL15 than Jack at 3 dac. Notably, ANAC2 (At1g01720) and 5 potential soybean orthologs of ANAC2 are expressed in response to increased AGL15 (35Spro:At/GmAGL15), as well as the majority of soybean orthologs being expressed in response to 3 dac in control tissue. ANAC2, normally expressed in meristems, is upregulated in response to a number of different stresses in leaves and may be involved in cells acquiring stem cell like features.14 Stress caused by dark-induced premature senescence that up-regulates ANAC2, also leads to increased callus formation, possibly by dedifferentiation mediated by ANAC2.14
Table 1.
Responses of genes encoding ANAC family transcription factors associated with dedifferentiation to AGL15 accumulation in Arabidopsis and soybean. Ranges indicate low and high values from multiple probe sets assigned to soybean loci (Glyma). All results are significant at least at P < 0.05 and for which there is at least a 1.5-fold change.
|
35S:GmAGL15 compared to Jack WT at days after culture (dac): |
Jack, WT |
35Spro:GmAGL15 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| AGI | 35S:AtAGL15 compared to Col, WT | Glyma (v1.1) | 0 | 3 | 7 | 3/0 dac | 7/3 dac | 3/0 dac | 7/3 dac |
| At1g01720 | 5.97 | 04g38560 | 3.0–3.5 | 4.5–5.4 | 1.8–2.0 | ||||
| 05g32850 | 3.9–4.2 | 8.8–9.4 | 2.9–3.2 | ||||||
| 06g11970 | 1.7 | ||||||||
| 06g16440 | 2.6–2.9 | 1.6 | |||||||
| 14g24220 | 1.7 | 2.1 | 1.6 | ||||||
| At1g52890 | 4.86 | ||||||||
| At1g69490 | 2.18 | 01g06150 | 2.0 | 13.5 | 10.9–12.7 | ||||
| 02g12220 | 4.4 | 9.9–10.4 | |||||||
| 07g35630 | 0.6 | 3.2–3.7 | 2.2–2.3 | ||||||
| 20g04400 | 2.6 | 2.0 | |||||||
| At2g43000 | 18g13574 | 2.8 | 11.2 | ||||||
| At3g04070 | 16g04740 | 4.6 | 2.8 | 4.8 | |||||
| At3g10500 | 20g33390 | 1.8 | |||||||
| At3g15500 | 2.38 | 13g35550 | 3.0 | 2.1–57.7 | 1.7–2.1 | 1.8–18.5 | 2.0 | ||
| At3g49530 | 1.54 | 07g05351 | |||||||
| At4g27410 | 06g38410 | 3.2–3.7 | 3.1 | ||||||
| 12g22880 | 1.6 | 2.2 | 1.6 | 2.6–6.5 | 2.3–4.5 | 1.7–16.0 | 2.4–3.4 | ||
| At5g09330 | 05g32470 | 1.6 | |||||||
| At5g24590 | 1.57 | ||||||||
| At5g39610 | 1.86 | ||||||||
| At5g63790 | 2.81 | ||||||||
The second class of transcription factors Grafi et al.13 highlight are members of the WRKY family. WRKY TFs are often involved in stress responses.15 As summarized in Table 2, of the 13 WRKY genes identified as associated with dedifferentiation, 3 are expressed in response to 35Spro:AtAGL15 in Arabidopsis and may be directly regulated based on ChIP-chip data. Six (46% or if counting only the WRKY's represented on the soybean array the number is 6 of 12 or 50%) have potential orthologs that are expressed in response to AGL15 accumulation in soybean, all of which have family members that are also up-regulated in non-transgenic tissue in response to 3 dac on the SE induction medium.
Table 2.
Response of genes encoding WRKY family transcription factors associated with dedifferentiation to AGL15 accumulation in Arabidopsis and soybean. See Table 1 for remainder of the explanation.
|
35S:GmAGL15 compared to Jack WT at days after culture (dac): |
Jack, WT |
35Spro:GmAGL15 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| AGI | 35S:AtAGL15 compared to Col, WT | Glyma (v1.1) | 0 | 3 | 7 | 3/0 dac | 7/3 dac | 3/0 dac | 7/3 dac |
| At1g13960 | 01g06550 | 8.0 | 1.8 | 6.9 | 2.1 | ||||
| 02g12490 | 1.9 | ||||||||
| At1g62300 | 07g39250 | 1.7–3.2 | 5.2–28.6 | 2.0–2.3 | 2.5–6.8 | 2.1 | |||
| 09g00820 | 3.6–3.8 | 7.8–11.1 | 3.0–6.4 | ||||||
| 13g38630 | 4.2–33.3 | 1.6 | 3.7–29.6 | 1.6 | |||||
| 15g11680 | 4.2–5.5 | 6.4–12.1 | |||||||
| At1g80840 | 2.07 | 06g06530 | 6.6 | ||||||
| 07g02630 | 5.0 | 10.8 | 2.5 | 2.3 | |||||
| 08g23380 | 4.6–5.6 | 5.0–9.2 | 3.3 | ||||||
| 13g44730 | 4.5 | ||||||||
| 14g11920 | 2.2 | 2.5 | |||||||
| 15g00570 | 10.5 | 3.4 | |||||||
| 17g33891 | 4.3 | 4.2 | 3.1 | ||||||
| At2g23320 | 0.47 | 01g36900 | 2.6 | 5.0 | |||||
| 11g05650 | 2.7 | 2.8 | 1.3 | ||||||
| 17g18480 | 1.8–2.0 | ||||||||
| At2g38470 | 2.58 | 01g31921 | 10.6 | 0.1 | |||||
| 02g39870 | 2.2–4.2 | 2.6–4.5 | |||||||
| 11g29720 | 4.6 | 6.4–62.2 | 1.9–2.2 | 4.8 | |||||
| 18g06360 | 86.3 | ||||||||
| At3g56400 | 04g40130 | 3.8 | 1.8 | 42 | 18.5 | ||||
| 09g41050 | 2.2–3.7 | 3.0–4.3 | 3.2–5.4 | ||||||
| 18g44560 | 3.0 | 5.2 | 5.8 | 17.9 | |||||
| At4g01250 | 3.13 | 18g47300 | 0.4 | 0.4 | |||||
| At4g18170 | 02g45530 | 0.4 | 9.7 | 5.9 | 3.5 | 16.5 | |||
| At4g31550 | 04g08060 | 3.4 | |||||||
| 06g08120 | 1.9–4.2 | 2.1 | 1.8–2.3 | ||||||
| 13g00380 | 1.5 | 0.6 | 1.6 | 1.7–2.1 | 0.5–0.6 | 2.0–2.9 | |||
| 17g06450 | 1.9 | ||||||||
| 17g29190 | 1.8 | ||||||||
| At5g13080 | 08g01430 | 1.6 | |||||||
| 19g26400 | 55.0 | 2.0 | 28.7 | ||||||
The last class of transcription factors discussed in Grafi et al.13 are b-ZIP family members. Two of the 12 show increased transcript accumulation in response to 35Spro:AtAGL15 in Arabidopsis, but only one of 5 that have a potential ortholog on the soybean chip was expressed at 0 dac in response to 35Spro:GmAGL15. An ortholog to a second b-ZIP (At5g28770) showed significant increased transcript in 35Spro:GmAGL15 at 7 dac compared to 3 dac. None were repressed in response to 35Spro:AGL15 in Arabidopsis or soybean.
As a summary, an extraordinary number of the ANAC, WRKY and b-ZIP transcription factors identified as expressed in dedifferentiating tissue are also responding to 35Spro:GmAGL15 that promotes SE (11 out of the 26 genes with potential orthologs on the soybean genome array). The majority of these also show increased transcript accumulation in Jack WT tissue cultured for 3 d on SE induction medium compared to explants. While only about one-third (13/44) of the TFs associated with dedifferentiation show significant increased transcript in 35Spro:AtAGL15 compared to Col WT in the SAM SE system, it should be recalled that tissue sampling was after 10 d in induction culture and it is likely that the earliest stages of SE development were missed. More than one half of the ANAC and WRKY genes have up to 6 orthologous genes in soybean that show increased transcript abundance in response to 35Spro:GmAGL15 at 0 dac or induction of Jack WT tissue for 3 d. There are very few instances of significantly reduced transcript with only one WRKY showing repression in response to 35Spro:AtAGL15 in Arabidopsis (At2g30250), and one ANAC showing repression in response to 35Spro:GmAGL15 compared to control explants (0 dac). In addition, only one b-ZIP and one WRKY showed repression in response to 3 d on induction medium compared to explants in Jack WT tissue. These observations indicate that the 35Spro:GmAGL15 may be promoting SE by upregulating genes involved in dedifferentiation, a step in the SE process, and may therefore be functioning very early. Placement of Jack WT on 2,4-D SE induction medium also upregulates genes involved in dedifferentiation.
To look more broadly than just at the TFs associated with dedifferentiation, we looked further at data presented in Damri et al.,16 that was part of the analysis by Grafi et al. 13 These genes were responsive to protoplasting and include additional genes with putative soybean orthologs that respond to 35Spro:GmAGL15 in explants or in response to 3 dac induction of SE in Jack WT (ref. 16, Supplemental Table 2). These include 13 genes encoding products involved in photosynthesis that show reduced transcript abundance in 3 dac Jack compared to explants with only one gene showing an opposite response where it is upregulated during protoplasting but shows reduced transcript in response to incubation of Jack WT for 3 dac. Seven also have reduced transcript in 35Spro:GmAGL15 compared to Jack WT explants prior to culture. Of the transcription factors and chromatin remodeling factors that were not flagged as associated with dedifferentiation in Grafi et al.,13 but that do respond to protoplasting (Supplemental Tables 1 and 3 in Damri et al.,16 without the ANACs, WRKYs and b-ZIP previously discussed), 9 are regulated in a similar manner in response to 35Spro:GmAGL15 in explants compared to Jack WT (7 expressed and 2 repressed). Only one TF shows an opposite pattern. The majority with predicted Glycine max orthologs also respond congruently in Jack comparing 3 dac to 0 dac compared to protoplasting with 4 repressed and 8 expressed. Six show opposite modes of regulation where they are expressed in response to protoplasting but repressed in response to 3 dac compared to Jack WT explants or vice versa. Thus, there is similarity between gene regulation during protoplasting and very early stages of SE induction in soybean.
Does Gm/AtAGL15 promote a root meristem identity during early SE?
Recent work has indicated that dedifferentiated tissue may not be truly completely dedifferentiated because the transcriptome of callus resembles that of root meristems regardless of the origin of the explant tissue.17 We focus our discussion on transcriptome of 0 dac 35Spro:GmAGL15 compared to Jack WT to draw conclusions as to how increased AGL15 potentiates SE at the earliest stages. To compare to root expressed genes, we initially looked at the list of genes associated with the quiescent center (QC) of the root (the AGL42 list in Brady et al.18), because these QC cell transcripts were most prevalent in callus derived from different origins (31.5% of the 90 genes on this list; ref. 17).
As summarized in Table 3 and Fig. 1, 13 of the 90 QC genes (14%) had potential soybean orthologs that were significantly down-regulated at 0 dac in 35Spro:GmAGL15 compared to Jack WT and only one showed a significant upregulation (1%). Because only 48 of the 90 QC associated genes had potential soybean orthologs of the Arabidopsis genes, 14% is likely an underestimate of genes in this data set regulated by GmAGL15. Of those with putative orthologs present, 27% (13/48) were downregulated by 35Spro:GmAGL15 at 0 dac. Eight of these genes showed a decrease after 3 dac on induction medium for Jack compared to 0 dac and one was up regulated. At 3 dac, comparing 35Spro:GmAGL15 to Jack, 6 QC transcripts were reduced in the overexpression tissue, but orthologs of 7 QC genes were now upregulated. At 7 dac, 2 soybean orthologs to one gene were down-regulated while one ortholog to another gene was expressed in response to 35Spro:GmAGL15. More than one-third (35/90) of the total list of QC genes showed differential expression in Jack cultured 3 dac on induction medium compared to explants but of these 6 (17%) were upregulated and 28 (80%) downregulated. One gene had a soybean ortholog that was significantly upregulated and another ortholog that was downregulated.
Table 3.
Genes from the root QC list (Brady et al.18 Supplemental Table 2, AGL42 tab) with putative orthologs in Glycine max that respond to increased GmAGL15 or to time in culture. Data are presented as the number expressed or repressed out of the total responsive. Some Arabidopsis genes had multiple orthologs in soybean, some of which were expressed and others repressed in the comparisons and these are listed in “both/total.” Percentage of the total genes in each category is in parentheses.
|
35Spro:GmAGL15 compared to Jack WT |
Jack, WT timecourse |
35Spro:GmAGL15 timecourse |
|||||
|---|---|---|---|---|---|---|---|
| QC Genes | 0 dac | 3 dac | 7 dac | 3dac/0dac | 7dac/3dac | 3dac/0dac | 7dac/3dac |
| Expressed/total (%) | 1/14 (7%) | 7/13 (54%) | 1/2 (50%) | 6/35 (17%) | 8/23 (35%) | 6/29 (21%) | 12/27 (44%) |
| Repressed/total (%) | 13/14 (93%) | 6/13 (46%) | 1/2 (50%) | 28/35 (80%) | 15/23 (65%) | 22/29 (76%) | 15/27 (56%) |
| Both/total (5) | 0/14 (0%) | 0/13 (0%) | 0/2 (0%) | 1/35 (3%) | 0/23 (0%) | 1/29 (3%) | 0/27 (0%) |
Figure 1.
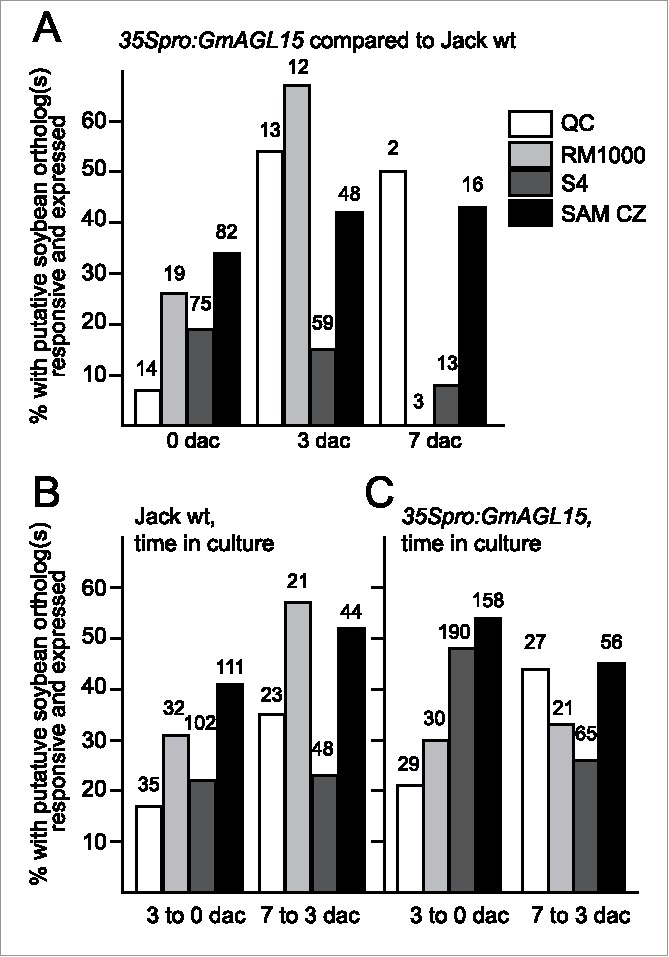
Transcript accumulation patterns for putative soybean orthologs of genes in Arabidopsis data sets corresponding to root and SAM transcriptomes. Data shown are percentage of the total responsive soybean orthologs that are expressed in response to 35Spro:GmAGL15 at 0, 3, and 7 dac (A) or to time in SE induction culture (B) for Jack wt and (C) for 35Spro:GmAGL15). The numbers above the bars indicates the total number of Arabidopsis genes with at least one putative soybean ortholog responsive to 35Spro:GmAGL15 or time in culture.
Comparing 7 dac Jack to 3 dac Jack, an increased fraction of the QC genes with orthologs present and showing significant change (P < 0.05 and <0.67 or >1.5 fold) were up regulated (8/23 = 35%) with 15 (65%) of the 23 down-regulated. Thus with continuing time on the induction medium, an increase number of orthologs of the QC genes are expressed, but at least initially, if anything, the QC genes are repressed in 35Spro:GmAGL15 compared to Jack explants and in the early timecourse of Jack (3 dac compared to explants). Likewise, in the timecourse of the 35Spro:GmAGL15, a greater fraction of QC genes with soybean orthologs present showed increased expression (21% comparing 3 dac to explants and 44% comparing 7 dac to 3 dac). However, the majority were repressed.
To extend these observations, we also looked at the list of genes preferentially expressed in protoxylem and 2/3 metaxylem (list S4 in Brady et al. 18) and lateral root primordia initials (list RM1000 in Brady et al. 18). Genes in these lists were also overrepresented as expressed in callus derived from various explants with 22.1% of the RM1000 genes and 18.8% of the S4 genes expressed in callus.17 Soybean transcript accumulation from the RM1000 genes shows roughly the same pattern as the QC list with initially (e.g. 35Spro:GmAGL15 compared to Jack WT at 0 dac, or Jack WT at 3 dac compared to explants) the majority of the genes repressed (63% for both comparisons). Some of the Arabidopsis RM1000 genes had multiple soybean orthologs some of which were expressed and others repressed in response to 35Spro:GmAGL15 or for Jack WT with 3 dac, and these accounted for 11 and 6% respectively, leaving 26 and 31% respectively where orthologs were only expressed (Fig. 1). The fraction of responsive orthologous genes expressed increased at 3 dac (35Spro:GmAGL15 compared to Jack WT) or comparing Jack WT at 7 dac to 3 dac, with more than half of the responsive genes expressed in these comparisons. For the S4 gene list, the majority of the responsive genes with soybean orthologs remained repressed in all comparisons. This data is summarized in Fig. 1.
The microarray experiment in Arabidopsis was performed on SAM SE 10 dac tissue.11 While this was far before any obvious SE development, it was also long after exposure to the induction medium containing the 2,4-D. Considering only genes that show significant (P < 0.05 and at least 1.5-fold) changes in transcript between 35Spro:AtAGL15 and Col WT, 5 QC genes are expressed while 4 genes are repressed. While all 4 repressed genes show no significant change in the agl15 agl18 double mutant compared to Col WT, 2 of the “expressed” genes also have significantly increased transcript in the loss-of-function that produces less SAM SE than Col WT and thus there is not a correlation between at transcript accumulation from these genes and SAM SE. If one considers genes as expressed as either increased in 35Spro:AtAGL15 or decreased in agl15 agl18 compared to Col WT, with either a consistent or no significant change in the other comparison, 29% (26 of 90) of the QC genes are upregulated by AGL15/18. By these criteria only 10% (9) showed significant down-regulation (e.g., decreased transcript in 35Spro:AtAGL15 or increased transcript in agl15 agl18 compared to Col WT) in response to AGL15/18 accumulation. For the genes listed in the root dataset corresponding to lateral root primordia initials (RM1000), 32 of the 114 genes are expressed while 16 are repressed (28 and 14% respectively). For the S4 gene list, 233 (23%) are expressed and 76 (7%) are repressed by AtAGL15.
In summary, although root associated genes are initially repressed in soybean in explants with increased GmAGL15 or comparing 3 dac to explants, the fraction expressed generally increases with time. In Arabidopsis where only a stage after 10 dac of induction of SE by 2,4-D was sampled, 2 to 3 times more root associated genes are expressed than repressed. These results may support the idea that SE at least eventually may take on root meristem-like features.
How do QC markers and transcription factors associated with dedifferentiation respond in other dedifferentiation systems?
A number of studies have examined transcriptomes in response to dedifferentiation including (but not limited to) those of Xu et al.19 where a timecourse comparing explant tissue to tissue placed for 12, 24, 48 and 96 hours on callus-induction medium that contains 2,4-D (2.2 μM) and kinetin (0.2 μM). The explants were aerial or root fragments from 10 day old seedlings. Like Sugimoto et al.,17 they found root marker genes were upregulated during callus induction. These included SCR, and SHR as well a number of genes involved in lateral root development. This later group includes several LATERAL ORGAN BOUNDARIES DOMAIN (LBD) members that were also found to lead to spontaneous callus formation without the need to supply exogenous hormones when ectopically expressed.20 This LBD induced callus also had features similar to root meristems including expression of PLETHORA1 and WOX5. Loss of function of other members of the PLETHORA family (PLT3, PLT5, and PLT7) are able to form callus but the callus is not competent to generate shoots. PLT5 and PLT7, besides being necessary for shoot regeneration are also sufficient to induce shoots without exogenous hormones.21 None of these genes show a significant change increase in transcript accumulation in 35Spro:At/GmAGL15 compared to control tissue with the exception of one putative soybean ortholog of a PLT that is 1.5-fold increased comparing 35Spro:GmAGL15 explants to control tissue. Two other PLT orthologs are significantly down at this stage. There also does not appear to be much overlap between root markers (according to Brady et al.18, sum of AGL42, RM1000 and S4 lists) and the list of “dedifferentiation-associated” TFs13 with only 2 genes on both lists (At1g69490 and At4g34000). For the data sets of Xu et al.19 all of the dedifferentiation TFs that show a significant change in transcript accumulation during callus induction are down regulated when roots are used as explants (23 of the list of 44 TF genes), but 8 are upregulated when the explants are derived from shoots. In general, there does not appear to be much correlation between the TFs associated with dedifferentiation and at least some types of de- or redifferentiation. Although amount of time on hormone containing medium could make a difference as found for AGL15, the dataset in ref. 19 included early timepoints (24 and 49 hours).
Another gene that when expressed via a 35S promoter that causes dedifferentiation without the need for exogenous hormones is WOUND INDUCED DEDIFFERENTIATION 1 (WIND1). This AP2/ERF TF does not cause root meristem marker expression and in fact some markers are downregulated.22 Of the QC list of Brady et al.,19 4 genes are upregulated by 35Spro:WIND1, and 14 genes are downregulated. Interestingly twelve of the TF genes associated with dedifferentiation are significantly upregulated by 35Spro:WIND1, but none are repressed. In this way, SE induced by ectopic expression of At/GmAGL15 seems more similar to the mode of regeneration competency induced by WIND1 than functioning via a root meristem pathway, at least initially. To further investigate, we compared our results to transcriptome data in response to 35Spro:WIND1.23 While both our data and the data set upregulated by 35Spro:WIND1, had many of the TFs identified by Grafi et al. 13 as associated with dedifferentiation, in general there is not much overlap between genes expressed or repressed in response to 35Spro:WIND1 and genes responsive to 35Spro:GmAGL15 compared to Jack WT at any of the timepoints analyzed. For the 35Spro:WIND1 expressed list, only 1, 0, and 4 putative soybean orthologs are expressed in response to 35Spro:GmAGL15 compared to Jack WT at 0, 3 and 7 dac (Table 4). However, there is there is marked overlap with the timecourse of Jack WT or 35Spro:GmAGL15 in culture. For the genes expressed by 35Spro:WIND1, 123 to 235, depending on genotype and timepoints compared, have orthologs in soybean that respond to time in culture and the majority are expressed. Thus there is greater overlap with the genes expressed in response to 35Spro:WIND1 with culture on SE induction medium than comparing increased GmAGL15 to Jack WT regardless of culture on SE induction medium or not with at least 4.5 times the number of genes responsive to culturing than to the 35Spro:GmAGL15 transgene. Similar results were obtained comparing out data to the gene list repressed by 35Spro:WIND1 (Table 4). This likely reflects the fact that WIND1 is expressed in response to wounding 23 that is part of the culture process for soybean, but these results also suggest that 35Spro:GmAGL15 is not promoting SE by ectopically regulating genes involved in wound induced dedifferentiation. In addition, WIND1 is also not regulated in a manner by At/GmAGL15 consistent with SE, and none of the SAM markers (WUSCHEL (WUS), SHOOT MERISTEMLESS (STM) and CLAVATA3 (CLV3)) that show increased transcript accumulation in 35Spro:WIND1 compared to control, are consistently expressed in a manner to support roles in SE in our systems. Interestingly WUS is important in a number of regeneration processes, including formation of shoots, conversion of lateral root primordia to shoot meristems, and maintenance of stem cells during SE.21, 24-26 When WUS is present with auxin, SE form in culture. In this later case AGL15 was shown to be expressed.25
Table 4.
Genes responsive to 35Spro:WIND1 (Iwase et al. 23) with putative orthologs in Glycine max that respond to increased GmAGL15 or to time in culture. See Table 3 for the remainder of the explanation.
|
35Spro:GmAGL15 compared to Jack WT |
Jack, WT timecourse |
35Spro:GmAGL15 timecourse |
|||||
|---|---|---|---|---|---|---|---|
| 0 dac | 3 dac | 7 dac | 3dac/0dac | 7dac/3dac | 3dac/0dac | 7dac/3dac | |
| 35Spro:WIND1 upregulated | |||||||
| Expressed/total (%) | 1/3 (33%) | 0/2 (0%) | 4/26 (15%) | 136/230 (59%) | 82/123 (67%) | 123/235 (52%) | 68/134 (51%) |
| Repressed/total (%) | 2/3 (67%) | 2/2 (100%) | 22/26 (85%) | 78/230 (34%) | 31/123 (25%) | 88/235 (37%) | 57/134 (42%) |
| Both/total (%) | 0/3 (0%) | 0/2 (0%) | 0/26 (0%) | 16/230 (7%) | 10/123 (8%) | 24/235 (10%) | 9/134 (7%) |
| 35Spro:WIND1 downregulated | |||||||
| Expressed/total (%) | 3/8 (38%) | 0/0 (0%) | 14/45 (31%) | 67/250 (27%) | 85/145 (59%) | 79/246 (32%) | 69/160 (43%) |
| Repressed/total (%) | 5/8 (62%) | 4/4 (100%) | 31/45 (69%) | 169/250 (67%) | 59/145 (41%) | 145/246 (59%) | 85/160 (53%) |
| Both/total (%) | 0/8 (0%) | 0/4 (0%) | 0/45 (0%) | 14/250 (6%) | 1/145 (0%) | 22/246 (9%) | 6/160 (4%) |
While WUS, STM and CLV3 show increased transcript accumulation in 35Spro:WIND1 compared to the control, reporter constructs derived from these genes are not expressed in callus, regardless of origin in the study performed by Sugimoto et al. where callus was found to have similarities to root meristems.17, 22 Although these particular SAM markers were not expressed, genes associated with the central zone (CZ) of the SAM were significantly represented in the callus expressed data of Sugimoto et al.17 with 108 of the 751 CZ genes up-regulated in callus (14.4%). We compared our data with the SAM CZ dataset Sugimoto et al. 17 used for comparison. For those genes with a putative ortholog in soybean responsive to 35Spro:GmAGL15 or time in culture, at least one third of the total number with responsive orthologs was expressed in response to 35Spro:GmAGL15 at all timepoints (Fig. 1). About half were expressed and half repressed comparing time in culture (Fig. 1). For the Arabidopsis experiment, 216 of the 682 genes on the SAM CZ list were expressed (31.7%) and 43 were repressed (6%).
In conclusion, much about the processes of regeneration remains poorly understood in plants and in animals, but a theme in both involves stress responses.27 There appear to be different routes of changes in developmental status in Arabidopsis. At/GmAGL15 appears to induce expression of many stress related genes including a large fraction of TFs associated with dedifferentiation. Increased accumulation of these transcripts occurs in 35Spro:GmAGL15 compared to Jack WT explants, as well as in Jack WT cultured on SE induction medium for 3 d compared to explants, suggesting GmAGL15 may affect very early stages of SE with response of root meristem related genes occurring later. Overlap with genes regulated in response to 35Spro:WIND1 is more extensive after wounding and/or exposure to 2,4-D than simply in response to the presence of the 35Spro:GmAGL15 transgene with a large number responsive by 3 dac. Genes associated with root meristems generally become increasingly expressed with time in culture, at least for QC and RM1000 gene lists.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Jeanne Hartman, Olivia Jones, and Rachel Mueller (University of Kentucky) for assistance with cultures and Donna Wall at the UK Microarray Core Facility for assistance with the transcriptome work. We would also like to thank our 2 anonymous reviewers for helpful comments.
Funding
This work was supported by the United Soybean Board (0282/1282/2282) and the National Science Foundation (IOS-0922845). This is publication No. 16-06-029 of the Kentucky Agricultural Experiment Station and is published with the approval of the Director. This work is supported by the National Institute of Food and Agriculture, US. Department of Agriculture, Hatch 0231956.
References
- 1.Feher A. Somatic embryogenesis - Stress-induced remodeling of plant cell fate. Biochim Biophys Acta 2015; 1849:385-402; PMID:25038583; http://dx.doi.org/ 10.1016/j.bbagrm.2014.07.005 [DOI] [PubMed] [Google Scholar]
- 2.Vogel G. What Don't We Know? How does a single somatic cell become a whole plant. Science 2005; 309:86; PMID:15994532; http://dx.doi.org/ 10.1126/science.309.5731.86 [DOI] [PubMed] [Google Scholar]
- 3.Kumar N, Reddy MP. In vitro plant propagation: a review. J Forest Sci 2011; 27:61-72; http://pdf.medrang.co.kr/Jfs/2011/027/Jfs027-02-01.pdf [Google Scholar]
- 4.Zavattieri MA, Federico AM, Lima M, Sabino R, Arnholdt-Schmitt B. Induction of somatic embryogenesis as an example of stress-related plant reactions. Electronic J Biotech 2010; 13; http://dx.doi.org/ 10.2225/vol13-issue1-fulltext-4 [DOI] [Google Scholar]
- 5.Imin N, Goffard N, Nizamidin M, Rolfe BG. Genome-wide transcriptional analysis of super-embryogenic Medicago truncatula explant cultures. BMC Plant Biol 2008; 8:110; PMID:18950541; http://dx.doi.org/ 10.1186/1471-2229-8-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harding EW, Tang W, Nichols KW, Fernandez DE, Perry SE. Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. Plant Physiol 2003; 133:653-63; PMID:14512519; http://dx.doi.org/ 10.1104/pp.103.023499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thakare D, Tang W, Hill K, Perry SE. The MADS-domain transcriptional regulator AGAMOUS-LIKE15 promotes somatic embryo development in Arabidopsis and soybean. Plant Physiol 2008; 146:1663-72; PMID:18305206; http://dx.doi.org/ 10.1104/pp.108.115832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu C, Perry SE. Control of expression and autoregulation of AGL15, a member of the MADS-box family. Plant J 2005; 41:583-94; PMID:15686521; http://dx.doi.org/ 10.1111/j.1365-313X.2004.02320.x [DOI] [PubMed] [Google Scholar]
- 9.Mordhorst AP, Voerman KJ, Hartog MV, Meijer EA, van-Went J, Koornneef M, de Vries SC. Somatic embryogenesis in Arabidopsis thaliana is facilitated by mutations in genes repressing meristematic cell divisions. Genetics 1998; 149:549-63; PMID:9611173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng Q, Perry SE. Alterations in the transcriptome of soybean in response to enhanced somatic embryogenesis promoted by orthologs of Agamous-like15 and Agamous-like18. Plant Physiol 2014; 164:1365-77; PMID:24481137; http://dx.doi.org/ 10.1104/pp.113.234062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng Y, Ren N, Wang H, Stromberg AJ, Perry SE. Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 2009; 21:2563-77; PMID:19767455; http://dx.doi.org/ 10.1105/tpc.109.068890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugimoto K, Gordon SP, Meyerowitz EM. Regeneration in plants and animals: dedifferentiation, transdifferentiation, or just differentiation? Trends Cell Biol 2011; 21:212-8; PMID:21236679; http://dx.doi.org/ 10.1016/j.tcb.2010.12.004 [DOI] [PubMed] [Google Scholar]
- 13.Grafi G, Chalifa-Caspi V, Nagar T, Plaschkes I, Barak S, Ransbotyn V. Plant response to stress meets dedifferentiation. Planta 2011; 233:433-8; PMID:21312042; http://dx.doi.org/ 10.1007/s00425-011-1366-3 [DOI] [PubMed] [Google Scholar]
- 14.Florentin A, Damri M, Grafi G. Stress induces plant somatic cells to acquire some features of stem cells accompanied by selective chromatin reorganization. Dev Dynam 2013; 242:1121-33; PMID:23798027; http://dx.doi.org/ 10.1002/dvdy.24003 [DOI] [PubMed] [Google Scholar]
- 15.Chen LG, Song Y, Li SJ, Zhang LP, Zou CS, Yu DQ. The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta 2012; 1819:120-8; PMID:21964328; http://dx.doi.org/ 10.1016/j.bbagrm.2011.09.002 [DOI] [PubMed] [Google Scholar]
- 16.Damri M, Granot G, Ben-Meir H, Avivi Y, Plaschkes I, Chalifa-Caspi V, Wolfson M, Fraifeld V, Grafi G. Senescing cells share common features with dedifferentiating cells. Rejuv Res 2009; 12:435-43; PMID:20041737; http://dx.doi.org/ 10.1089/rej.2009.0887 [DOI] [PubMed] [Google Scholar]
- 17.Sugimoto K, Jiao Y, Meyerowitz EM. Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev Cell 2010; 18:463-71; PMID:20230752; http://dx.doi.org/ 10.1016/j.devcel.2010.02.004 [DOI] [PubMed] [Google Scholar]
- 18.Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 2007; 318:801-6; PMID:17975066; http://dx.doi.org/ 10.1126/science.1146265 [DOI] [PubMed] [Google Scholar]
- 19.Xu K, Liu J, Fan M, Xin W, Hu Y, Xu C. A genome-wide transcriptome profiling reveals the early molecular events during callus initiation in Arabidopsis multiple organs. Genomics 2012; 100:116-24; PMID:22664253; http://dx.doi.org/ 10.1016/j.ygeno.2012.05.013 [DOI] [PubMed] [Google Scholar]
- 20.Fan M, Xu C, Xu K, Hu Y. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res 2012; 22:1169-80; PMID:22508267; http://dx.doi.org/ 10.1038/cr.2012.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kareem A, Durgaprasad K, Sugimoto K, Du Y, Pulianmackal AJ, Trivedi ZB, Abhayadev PV, Pinon V, Meyerowitz EM, Scheres B, et al.. PLETHORA genes control regeneration by a two-step mechanism. Current Biol 2015; 25:1017-30; PMID:25819565; http://dx.doi.org/ 10.1016/j.cub.2015.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwase A, Mita K, Nonaka S, Ikeuchi M, Koizuka C, Ohnuma M, Ezura H, Imamura J, Sugimoto K. WIND1-based acquisition of regeneration competency in Arabidopsis and rapeseed. J Plant Res 2015; 128:389-97; PMID:25810222; http://dx.doi.org/ 10.1007/s10265-015-0714-y [DOI] [PubMed] [Google Scholar]
- 23.Iwase A, Mitsuda N, Koyama T, Hiratsu K, Kojima M, Arai T, Inoue Y, Seki M, Sakakibara H, Sugimoto K, et al.. The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Current Biol 2011; 21:508-14; PMID:21396822; http://dx.doi.org/ 10.1016/j.cub.2011.02.020 [DOI] [PubMed] [Google Scholar]
- 24.Chatfield SP, Capron R, Severino A, Penttila PA, Alfred S, Nahal H, Provart NJ. Incipient stem cell niche conversion in tissue culture: using a systems approach to probe early events in WUSCHEL-dependent conversion of lateral root primordia into shoot meristems. Plant J 2013; 73:798-813; PMID:23181633; http://dx.doi.org/ 10.1111/tpj.12085 [DOI] [PubMed] [Google Scholar]
- 25.Gallois J-L, Nora FR, Mizukami Y, Sablowski R. WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev 2004; 18:375-80; PMID:15004006; http://dx.doi.org/ 10.1101/gad.291204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su YH, Zhao XY, Liu YB, Zhang CL, O'Neill SD, Zhang XS. Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. Plant J 2009; 59:448-60; PMID:19453451; http://dx.doi.org/ 10.1111/j.1365-313X.2009.03880.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grafi G, Barak S. Stress as a fundamental theme in cell plasticity. Biochim Biophys Acta 2015; 1849:369-70; PMID:25464033; http://dx.doi.org/ 10.1016/j.bbagrm.2014.11.001 [DOI] [PubMed] [Google Scholar]


