ABSTRACT
The large dynamic range of gene expression changes accompanying floral organ abscission can be explained by a molecular positive feedback loop that regulates the process. In short, a mitogen-activated protein kinase (MAPK) cascade, positioned genetically downstream from the abscission receptor HAESA (HAE), phosphorylates the transcription factor, AGAMOUS-like 15 (AGL15), allowing HAE to be expressed. However, it is unknown which residues of AGL15 are phosphorylated and precisely how phosphorylation alters AGL15 function. Here we report that serine 231 and 257 of AGL15 are phosphorylated in floral receptacles. Effects of phosphorylation on AGL15 are discussed.
KEYWORDS: Abscission, MAPK cascade, phosphorylation, signal transduction, transcription factor
Abscission is the process in which a plant sheds unwanted organs. Arabidopsis floral organ abscission has served as an excellent model system for studying abscission. A number of genetic components have been elucidated in Arabidopsis floral abscission. INFLORESCENCE DEFICIENT IN ABSCISSION (IDA), HAE/HAE-like 2 (HSL2), NEVERSHED (NEV), MAPKK4/5, MAPK3/6 (MPK3/6), ETHYLENE INSENSITIVE 2, ETHYLENE RESPONSE 1, and CORONATINE INSENSITIVE 1 are all necessary for normal abscission.1–5 Secondary mutations in EVERSHED, CAST AWAY, and SOMATIC EMBRYOGENESIS RECEPTOR KINASE1 rescue abscission in nev mutants and secondary mutations in BREVIPEDICELLUS rescue abscission in ida and hae hsl2 mutants.6–9 Over-expression of AGL15, AGAMOUS-like 18 (AGL18), FOREVER YOUNG FLOWER, or ZINC FINGER PROTEIN 2 results in delayed abscission.10–13 A number of recent reviews thoroughly detail what is currently known about floral abscission in Arabidopsis.14,15 Recently, we described a positive feedback loop that regulates HAE, a gene that encodes a receptor-like protein kinase, expression. The positive feedback loop explains the large dynamic range of gene expression changes accompanying abscission.16 Briefly, the MADS-domain transcription factor AGL15, residing on the HAE promoter, is phosphorylated by MPK3/6, allowing more HAE to be expressed.16 Here we characterize which AGL15 amino acids are phosphorylated by the MAPK cascade and what effect phosphorylation has on AGL15 function.
MAP kinases preferentially phosphorylate serines or threonines followed by proline.17 Therefore, of the 32 serine residues and 16 theonine residues in AGL15, only S231 and S257 are putative MAPK phosphorylation sites since they are followed by proline. We tested whether these sites are phosphorylated in floral receptacles by mutating them to alanine. With protein extracts from stage 15 floral receptacles from tagged wild-type AGL15 (native promoter and genomic coding sequence of AGL15 with a C-terminal double hemaglutanin tag in the agl15-4 background) we visualize 3 isoforms when run on a Phos-tag gels (Fig. 1).16 Changing S231 and S257 to alanine reduced phosphorylation of the AGL15 B and C phosphorylated isoforms by 67% (Fig. 1B). However, the AGL15 B and C isoforms are not completely eliminated. This suggests that additional residues in AGL15 are also phosphorylated. Interestingly S231 is followed by amino acids PSS. In some cases MAPKs can phosphorylate serines that follow proline, especially when proline is 2 positions before serine.17 Since only S231 was changed to alanine and not S234 or S235, these residues could potentially account for the observed residual phosphorylation.
Figure 1.
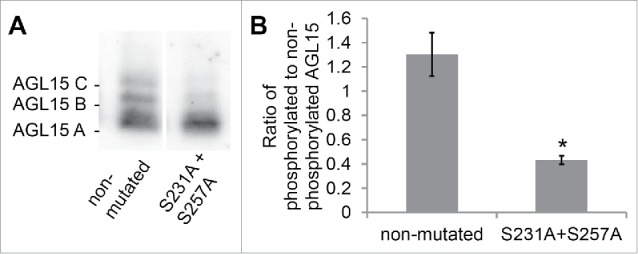
Serines S231 and S257 of AGL15 are phosphorylated in stage 15 floral receptacles. (A) AGL15 expressed as a hemagglutinin (HA) tag in stage 15 floral receptacles, driven by its native promoter, runs a multiple bands on 25 µM Phos-tag gels.16 Mutating serines 231 and 257 to alanine reduces phosphorylated isoforms of AGL15. Exposures were chosen for each lane that scaled the 2 lanes to similar intensity for the lower, non-phosphorylated band. Expression was performed in the AGL15 null mutant, agl15-4.18 (B) Quantified ratio of phosphorylated to non-phosphorylated AGL15. Three independent lines were assayed. *P < 0.05 (t-test). n = 3; SEM error bars shown. Floral receptacles (1 mm hand cut cross section of the base of the flower) were ground in liquid nitrogen and then crude protein extracts were prepared by mixing the ground tissue with 1x SDS sample buffer (1 receptacle per 12 ul SDS sample buffer). Phos-tag gel electrophoresis and Western blotting were performed as described.16 Quantification of Western blots was performed with Image Lab software (Bio-Rad). Mutations to the previously described AGL15 native promoter construct were made via the QuikChange method (Agilent).16
35S-AGL15 from floral receptacles runs as 2 bands on 12% SDS-PAGE gels that are a day old and the slower mobility band can be converted into the faster mobility isoform with phosphatase treatment.16 In contrast, 35S-AGL15 produced in mesophyll protoplasts runs as a single band indicating it is not modified in the same way as AGL15 produced in floral receptacles (Fig. 2). Changing either S231, S257 or both to aspartate causes mesophyll protoplast produced AGL15 to mimic the slower mobility phosphorylated isoform derived from floral receptacles, with the double aspartate substitution causing the larger gel shift (Fig. 2). These gel shifts further support that it is S231 and S257 that are being phosphorylated in floral receptacles. Interestingly, more AGL15 accumulates in protoplasts when either S231 or S257 is changed to either alanine or aspartate. This suggested that S231 and S257 may play some role in AGL15 protein stability or translatability.
Figure 2.
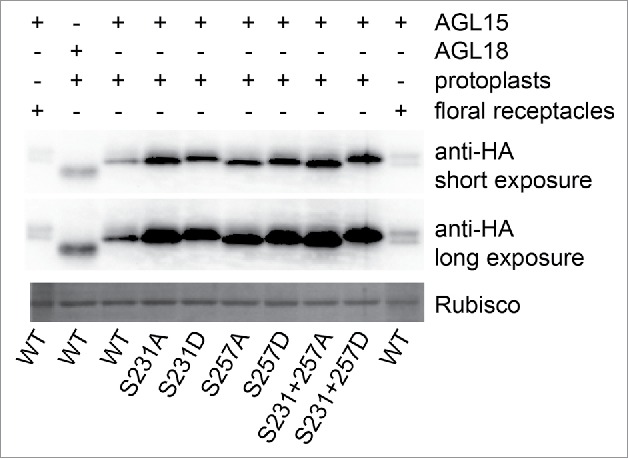
AGL15 phosphomimetics have similar mobility shifts as 35S-AGL15 from floral receptacles. AGL15 produced by the 35S promoter in floral receptacles runs as 2 bands on one day old 12% SDS-PAGE gels. The slower mobility band can be shifted to the faster mobility band by phosphatase treatment.16 However, 35S-AGL15 expressed in mesophyll protoplasts runs as a single band. Mutating either S231 or S257 to aspartate reduces AGL15 mobility with the double substitution causing the biggest mobility shift similar to phosphorylated AGL15. The experiment was repeated twice with the same result. Protoplast preparation and transfection was performed as described.19 Six hours after transfection, protoplasts were solubilized in SDS sample buffer. SDS-PAGE and Western blotting were performed as described.16 35S-AGL15 floral stage 15 receptacle extract was prepare as described in the Fig. 1 legend. cDNA from the previously described construct used to generate 35S-AGL15-double HA tagged plants was inserted into pRLT2 for protoplast studies and mutations were made via the QuikChange method (Agilent).16
In floral receptacles AGL15 overexpression reduces HAE expression 5-fold.16 We attempted to determine if changing AGL15's MAPK phosphorylation sites to alanine or aspartate affected its ability to regulate HAE expression in protoplasts. To our surprise AGL15 overexpression did not reduce HAE expression in protoplasts (Fig. 3A). This could be because HAE expression is already extremely low in mesophyll protoplasts compared to its level in floral receptacles. Even more surprisingly, changing either or both S231 and S257 to alanine or aspartate resulting in about a 2-fold increase of HAE compared to the empty vector control. Interpretation of this result is difficult since alanine and aspartate mutations have the same effect. Perhaps the simplest interpretation is that mesophyll cells and abscission zone cells are very different both morphologically and biochemically. HAE has little to no expression in mesophyll protoplasts so perhaps it is unreasonable to use mesophyll protoplasts to study abscission related events that require the presence of functional abscission machinery. AGL15S231+257A causes the greatest increase in HAE expression in protoplasts but had only one fifth the HAE expression found in stage 12 floral receptacles (before anthesis) and 145-fold less HAE expression in than in stage 15 floral receptacles (after pollination) (Fig. 3B). Our conclusion for future studies is that it is best to perform experiments in abscission zones whenever possible and avoid performing experiments designed to unravel abscission in unrelated tissues.
Figure 3.
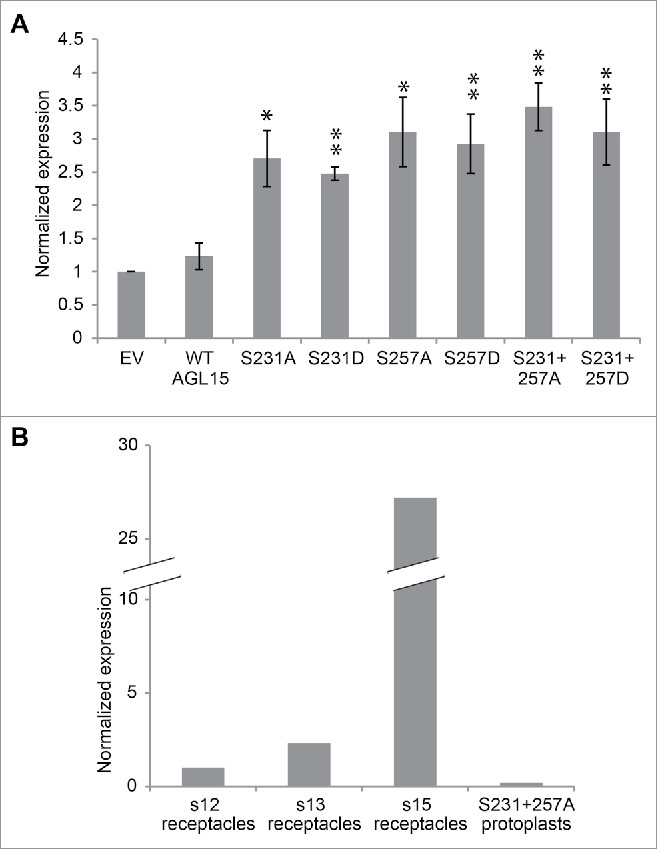
Mutating S231 and S257 of 35S-AGL15 alters HAESA expression in mesophyll protoplasts. (A) HAESA transcript accumulation in mesophyll protoplasts 6 hours after transfection with empty vector or 35S-AGL15 with indicated mutations. Protoplast were prepared as describes.19 RNA was extracted from protoplasts with Trizol (Ambion). Quantitative-PCR was performed as described.16 *P < 0.1, **P < 0.05 (t-test). n = 3; SEM error bars shown. (B) Relative expression of HAESA in floral receptacles and in transfected mesophyll protoplasts.
In conclusion, AGL15 is phosphorylated on serine 231 and 257 in floral receptacles but also has minor phosphorylation elsewhere. We speculate that AGL15 could act as a positive regulator of HAE expression once it is phosphorylated in addition to its defined role as a negative regulator of HAE expression before being phosphorylated.16 Circumstantial evidence supporting this speculation include: AGL15 still binds the HAE promoter once HAE expression has been turned on, AGL15 accumulates to a higher level after pollination than before anthesis,16 and in mesophyll protoplast mutated AGL15 can increase HAE expression slightly. Further analysis of effects of AGL15 phosphomimetic expression in floral receptacles will be necessary to conclusively answer if MAPK phosphorylation of AGL15 simply de-represses HAE expression or if it changes AGL15 from a repressor to an activator of HAE expression.
Disclosure of potential confllicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Butenko MA, Patterson SE, Grini PE, Stenvik G-E, Amundsen SS, Mandal A, Aalen RB. INFLORESCENCE DEFICIENT IN ABSCISSION controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 2003; 15:2296-307; PMID:12972671; http://dx.doi.org/ 10.1105/tpc.014365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liljegren SJ, Leslie ME, Darnielle L, Lewis MW, Taylor SM, Luo R, Geldner N, Chory J, Randazzo PA, Yanofsky MF, et al.. Regulation of membrane trafficking and organ separation by the NEVERSHED ARF-GAP protein. Development 2009; 136:1909-18; PMID:19429787; http://dx.doi.org/ 10.1242/dev.033605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho SK, Larue CT, Chevalier D, Wang H, Jinn T-L, Zhang S, Walker JC. Regulation of floral organ abscission in Arabidopsis thaliana. Proc Natl Acad Sci U S A 2008; 105:15629-34; PMID:18809915; http://dx.doi.org/ 10.1073/pnas.0805539105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patterson SE, Bleecker AB. Ethylene-Dependent and -Independent Processes Associated with Floral Organ Abscission in Arabidopsis. Plant Physiol 2004; 134:194-203; PMID:14701913; http://dx.doi.org/ 10.1104/pp.103.028027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J, Dotson B, Rey C, Lindsey J, Bleecker AB, Binder BM, Patterson SE. New Clothes for the Jasmonic Acid Receptor COI1 : Delayed Abscission, Meristem Arrest and Apical Dominance. PLOS ONE 2013; 8:e60505; PMID:23573263; http://dx.doi.org/ 10.1371/journal.pone.0060505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leslie ME, Lewis MW, Youn J-Y, Daniels MJ, Liljegren SJ. The EVERSHED receptor-like kinase modulates floral organ shedding in Arabidopsis. Development 2010; 137:467-76; PMID:20081191; http://dx.doi.org/ 10.1242/dev.041335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burr CA, Leslie ME, Orlowski SK, Chen I, Wright CE, Daniels MJ, Liljegren SJ. CAST AWAY, a membrane-associated receptor-like kinase, inhibits organ abscission in Arabidopsis. Plant Physiol 2011; 156:1837-50; PMID:21628627; http://dx.doi.org/ 10.1104/pp.111.175224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis MW, Leslie ME, Fulcher EH, Darnielle L, Healy PN, Youn J-Y, Liljegren SJ. The SERK1 receptor-like kinase regulates organ separation in Arabidopsis flowers. Plant J 2010; 62:817-28; PMID:20230490; http://dx.doi.org/ 10.1111/j.1365-313X.2010.04194.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi C-L, Stenvik G-E, Vie AK, Bones AM, Pautot V, Proveniers M, Aalen RB, Butenko MA. Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway. Plant Cell 2011; 23:2553-67; PMID:21742991; http://dx.doi.org/ 10.1105/tpc.111.084608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez DE, Heck GR, Perry SE, Patterson SE, Bleecker AB, Fang SC. The embryo MADS domain factor AGL15 acts postembryonically. Inhibition of perianth senescence and abscission via constitutive expression. Plant Cell 2000; 12:183-98; PMID:10662856; http://dx.doi.org/ 10.1105/tpc.12.2.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adamczyk BJ, Lehti-Shiu MD, Fernandez DE. The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J 2007; 50:1007-19; PMID:17521410; http://dx.doi.org/ 10.1111/j.1365-313X.2007.03105.x [DOI] [PubMed] [Google Scholar]
- 12.Chen M-K, Hsu W-H, Lee P-F, Thiruvengadam M, Chen H-I, Yang C-H. The MADS box gene, FOREVER YOUNG FLOWER, acts as a repressor controlling floral organ senescence and abscission in Arabidopsis. Plant J 2011; 68:168-85; PMID:21689171; http://dx.doi.org/ 10.1111/j.1365-313X.2011.04677.x [DOI] [PubMed] [Google Scholar]
- 13.Cai S, Lashbrook CC. Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: enhanced retention of floral organs in transgenic plants overexpressing Arabidopsis ZINC FINGER PROTEIN2. Plant Physiol 2008; 146:1305-21; PMID:18192438; http://dx.doi.org/ 10.1104/pp.107.110908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niederhuth CE, Cho SK, Seitz K, Walker JC. Letting go is never easy: abscission and receptor-like protein kinases. J Integr Plant Biol 2013; 55:1251-63; PMID:24138310; http://dx.doi.org/ 10.1111/jipb.12116 [DOI] [PubMed] [Google Scholar]
- 15.Liljegren SJ. Organ abscission: exit strategies require signals and moving traffic. Curr Opin Plant Biol 2012; 15:670-6; PMID:23047135; http://dx.doi.org/ 10.1016/j.pbi.2012.09.012 [DOI] [PubMed] [Google Scholar]
- 16.Patharkar OR, Walker JC. Floral organ abscission is regulated by a positive feedback loop. Proc Natl Acad Sci 2015; 112:2906-11; PMID:25730871; http://dx.doi.org/ 10.1073/pnas.1423595112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheridan DL, Kong Y, Parker SA, Dalby KN, Turk BE. Substrate discrimination among mitogen-activated protein kinases through distinct docking sequence motifs. J Biol Chem 2008; 283:19511-20; PMID:18482985; http://dx.doi.org/ 10.1074/jbc.M801074200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehti-Shiu MD, Adamczyk BJ, Fernandez DE. Expression of MADS-box genes during the embryonic phase in Arabidopsis. Plant Mol Biol 2005; 58:89-107; PMID:16028119; http://dx.doi.org/ 10.1007/s11103-005-4546-3 [DOI] [PubMed] [Google Scholar]
- 19.Yoo S-D, Cho Y-H, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2007; 2:1565-72; PMID:17585298; http://dx.doi.org/ 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]


