Article first published online 10 August 2016.
Key Words: smoking, microbiota, Crohn's disease, inflammatory bowel disease, sequencing
Abstract
Background:
Smoking has a negative impact on Crohn's disease (CD), but the mechanisms underlying this association are unclear. We compared the gut microbiota composition of smoking with nonsmoking patients with CD using a metagenomic approach.
Methods:
Stool samples and clinical data were collected from current smokers and nonsmokers with CD from France and the Netherlands, matched for country, gender, age, disease activity, and body mass index. Fecal DNA was sequenced on an Illumina HiSeq 2500. On average, 40 million paired-end reads were generated per sample. Gene richness and the Shannon index were computed to assess microbial diversity. Wilcoxon's signed-rank tests for paired samples were performed to detect differences between the 2 groups.
Results:
In total, 21 smoking and 21 nonsmoking patients with CD were included. Compared with nonsmoking patients, gut microbial gene richness (P = 0.01), genus diversity (P < 0.01), and species diversity (P = 0.01) were decreased in smoking patients. This was accompanied by a reduced relative abundance of the genera Collinsella (P = 0.02), Enterorhabdus (P = 0.02), and Gordonibacter (P = 0.02) in smokers. No statistically significant differences at the species level were observed, although smokers had lower proportions of Faecalibacterium prausnitzii (P = 0.10).
Conclusions:
Gut microbial diversity is reduced in smokers with CD compared with nonsmokers with CD. The microbial profile differs between these groups at the genus level. Future studies should evaluate whether intestinal microbes mediate the adverse effects of smoking in CD.
Crohn's disease (CD) is a type of inflammatory bowel disease (IBD) that is marked by recurrent episodes of intestinal inflammation and may lead to significant complications and disability.1,2 The pathogenesis of this chronic disorder is complex and thought to involve an interaction between host and environmental factors, in which the gut microbiota is considered to play an important role.3 Smoking is one of the strongest environmental risk factors for CD.4 It is consistently associated with an increased risk of developing CD.5 Moreover, smoking has adverse effects on the clinical course with an increased number of relapses and need for medications and surgical interventions among smokers with CD.6–8 Although deleterious effects of smoking on intestinal permeability, the immune system, and epigenetic susceptibility have been reported, the biological mechanisms by which smoking influences the pathogenesis of CD remain unclear.4
With the advent of culture-independent techniques for characterizing microbial communities, the role of the microbiota in health and disease has regained interest. Next-generation sequencing methods have provided powerful tools with regard to in-depth analysis of the microbiome.9–11 Accumulating data highlight the potential impact of environmental factors, such as antibiotic use or diet, on the gut microbiota.12–15 Accordingly, it has been suggested that intestinal microbes could be an important link between smoking and CD.4 Only few studies have specifically investigated the relationship between smoking and gut microbiota in humans.16–18 In a recent interventional study, using different techniques, significant changes were observed in the fecal microbiota of healthy individuals undergoing smoking cessation, including an increase in the relative abundance of Firmicutes (Clostridium coccoides, Eubacterium rectale, and Clostridium leptum subgroup) and Actinobacteria (high guanine and cytosine content bacteria and Bifidobacteria) and a reduction of Bacteroidetes (Prevotella spp. and Bacteroides spp.) and Proteobacteria (b- and g-subgroup).16,17 A cross-sectional study using fluorescent in situ hybridization targeting selected bacterial groups reported that smoking patients with active CD exhibited different microbial profiles, with higher Bacteroides–Prevotella, compared with nonsmoking patients with CD.18 Similar results were found in healthy smoking controls, suggesting that the association may not be due to intestinal inflammation but may reflect a direct impact of smoking on the microbiota. These studies were mostly based on the abundance of 16S ribosomal RNA (rRNA) genes. However, it has been shown that quantifying microbial species with 16S rRNA is biased and depending on the region that is being sequenced, conflicting results can be generated, whereas microbiome shotgun sequencing uses all DNA fragments found in a sample to quantify relative abundances of bacteria as well as other organisms.19 In view of these considerations and the paucity of previous data, this study aimed to compare the gut microbiota composition of smoking with nonsmoking patients with CD, using whole-metagenome sequencing.
MATERIALS AND METHODS
Study Population
Smoking and nonsmoking patients with CD were identified from 2 prospective cohort studies in France and in the Netherlands. In France, subjects with CD were recruited from 2 Parisian hospitals and the French association for IBD (Association François Aupetit), whereas in the Netherlands, consecutive patients with IBD in an academic hospital scheduled for surveillance colonoscopy were enrolled. For the present investigation, inclusion criteria were age ≥18 years and an established diagnosis of CD according to endoscopic, radiological, and/or histological features.20 Exclusion criteria were use of antibiotics and bowel cleansing for colonoscopy during the last 2 months before fecal sampling. For each smoking patient with CD, one nonsmoking patient with CD was randomly selected, carefully matched for country, gender, age, and body mass index. Disease activity was taken into account by including only patients with relatively low fecal calprotectin levels or absence of endoscopic inflammation in both groups. Matching for body mass index was only performed for patients in the French cohort.
Data Collection
In the framework of both observational studies, patients were asked to collect a stool sample at a regular basis. Concomitantly, disease activity was assessed using the Harvey–Bradshaw index and either fecal calprotectin levels (France) or endoscopic disease activity (the Netherlands).21 Current smoking status (categorized into yes or no) and daily cigarette consumption were assessed with self-reported questionnaires. The medical records of participants were reviewed for demographic data, disease classification, medications, and comorbidities.
Sample Preparation and Processing for Sequencing
Stool samples were collected at home using Sarstedt tubes (Sarstedt, Nümbrecht, Germany) filled with a preservative buffer. On reception, the tubes were stored at −80°C. Sample aliquoting, DNA isolation, and sequencing were outsourced to GATC Biotech (Konstanz, Germany). For the first batch of samples, manual DNA extraction was performed by suspending fecal samples in 250 μL of guanidine thiocyanate 0.1 M Tris (pH 7.5) and 40 μL of 10% N-lauroyl sarcosine. The suspension was then submitted to vigorous bead-beating to release DNA from microbial cells, and DNA extraction was conducted using a standard protocol.22 For the second batch of samples, a commercial extraction kit, the QIAamp Stool DNA mini kit (Qiagen, Hilden, Germany) was used. DNA concentrations were measured using Qubit fluorometric quantitation (Life Technologies, Carlsbad, California). DNA libraries were prepared following the manufacturer's instruction (Illumina, San Diego, California). Fecal DNA was sequenced on a HiSeq 2500 Illumina sequencer. The target of 40 million minimum paired-end reads was generated for each sample and sequencing read length was 100 to 125 bp.
Bioinformatics Processing
FASTQ files were processed using a customized version of MOCAT software.23 Reads were trimmed and filtered with a quality cutoff of 20. Sequence reads shorter than 45 bp, mapping to Illumina adapters, or to the human genome (version hg19) were discarded. Reads were then mapped against Enterome's proprietary CD catalog of 4 million genes. The catalog is an enrichment of a previous 3.3 million genes catalog with 700,000 additional complete genes identified from patients with CD.9 Genes were quantified within a sample using relative abundance measurements, which means that for each gene, the sum of uniquely mapped reads was divided by gene length and by the sum of all genes. Normalized gene abundances were then summarized at different taxonomic and analytical levels: phylum, genus, species, and MetaGenomics Species.24
Gene Richness and Taxonomic Diversity
The gene richness for each sample was computed from the raw abundance table after downsizing based on 11 million simulated depth sequencing and 30 repeats.11 The Shannon index was computed to assess taxonomic diversity at both the genus and species level.25
Phylogenetic Annotation
Genes were annotated using BLASTN alignment method against KEGG and RefSeq genomic databases.26–28 The gene annotation method was adapted from Li et al.29 Only the hits with a minimum of 80% of query sequence length and 65% identity were considered in the annotation process. The similarity thresholds for the phylum, genus, and species taxonomic ranges were 65%, 80%, and 95%, respectively. Genes with multiple hits deprived of any consensus (a consensus was defined as 10% of hits having the same annotation) for their taxonomic associations were annotated at a higher taxonomic range until a consensus was established.
Fecal Calprotectin Analysis
In addition to stool sample collection for metagenomic analysis, fecal samples were collected at home using Sarstedt tubes without preservation buffer in the French study. In these samples, fecal calprotectin was measured using an enzyme-linked immunosorbent assay (Bühlmann fCAL ELISA tests; Bühlmann, Schönenbuch, Switzerland) by Merieux Nutrition (Nantes, France). Range values were between 50 μg/g and 3000 μg/g.
Statistical Analysis
Clinical variables were summarized as medians with interquartile ranges (IQRs) or as frequencies with percentages. The Fisher's exact test was used to compare categorical variables between smokers and nonsmokers. The Wilcoxon's signed-rank test for paired samples was performed to compare continuous variables between the 2 groups, including metagenomic data after filtering the variables by presence. When comparing the relative abundances of phyla, genera, and species between smokers and nonsmokers, P-values were adjusted using the Benjamini–Hochberg procedure. A P-value lower than 0.05 was considered statistically significant. All statistical analyses were performed using R (version 3.2.1).
Ethical Considerations
The original cohort studies were both approved by the institutional medical ethics committees.
RESULTS
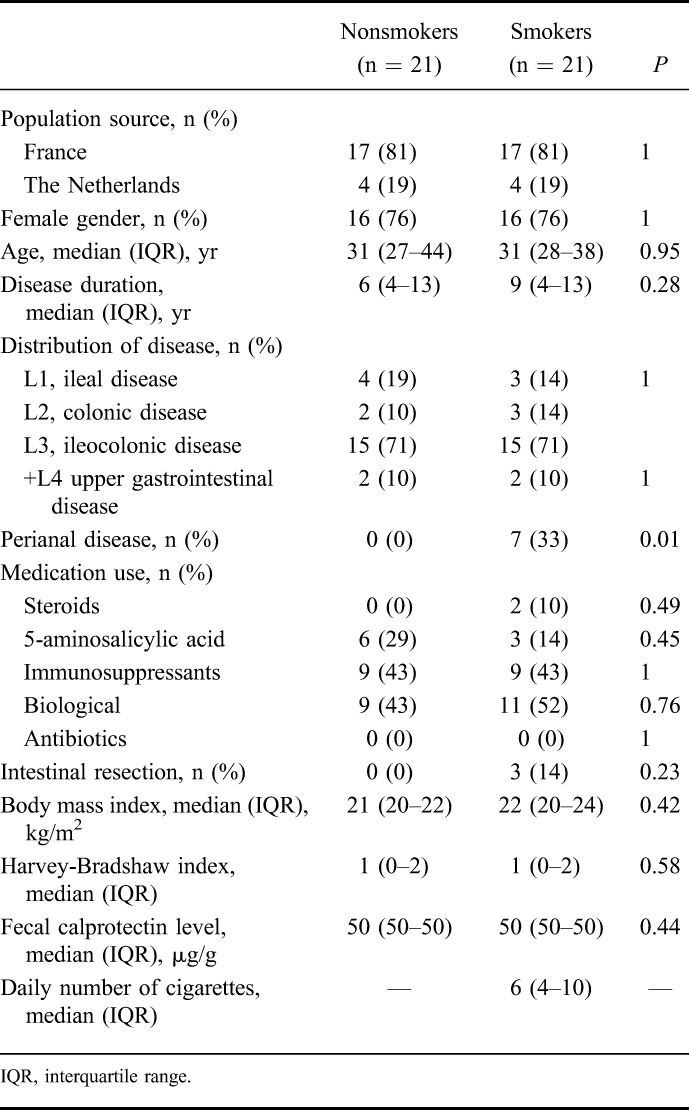
In total, 21 smoking and 21 matched nonsmoking patients with CD (76% of females) with a median age of 31 years were included (Table 1). Ileocolonic disease was the commonest disease localization in both groups (71%). Seven smoking patients had perianal disease, whereas this phenotype was not present in the nonsmoking group (P < 0.01). No other statistically significant differences in clinical characteristics between smoking and nonsmoking patients with CD were observed. The median fecal calprotectin level was 50 μg/g (IQR 50–50 μg/g) in both groups. Those patients without fecal calprotectin levels (4 smoking and 4 nonsmoking subjects) were all in endoscopic remission. The median number of daily cigarette consumption was 6 (IQR 4–10 cigarettes).
TABLE 1.
Clinical Characteristics of Nonsmoking and Smoking Crohn's Disease Patients
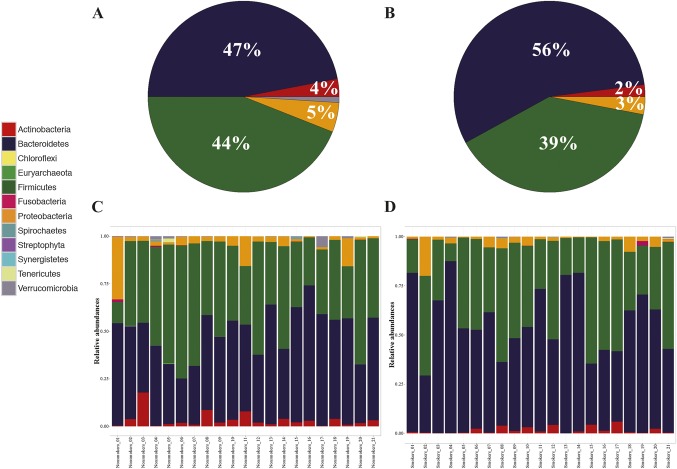
The composition of the gut microbiota of all patients was mainly characterized by the phyla Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria (Fig. 1 A–B). There were marked interindividual differences in the 2 dominant phyla Firmicutes and Bacteroidetes, but no significant differences between smokers and nonsmokers were seen at the phylum level (Fig. 1 C–D).
FIGURE 1.
Phyla composition. A and B, Pie charts of the relative abundances of the major phyla in nonsmoking and smoking patients with CD, respectively. C and D, Bar charts of the phyla composition in nonsmoking and smoking patients with CD, respectively.
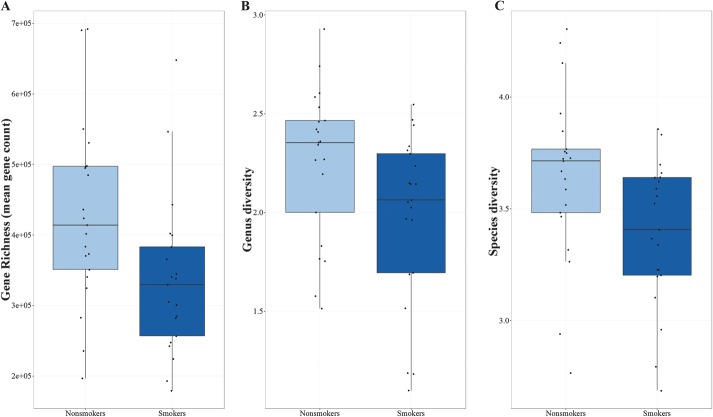
Gene richness was significantly decreased in smokers compared with nonsmokers with median gene counts of 329,600 and 414,100 (P = 0.01), respectively (Fig. 2A). The taxonomic diversity was also lower in smoking patients than in nonsmoking patients, both at the genus level (median Shannon index 2.06 versus 2.35, P < 0.01) and the species level (median Shannon index 3.41 versus 3.71, P = 0.01) (Fig. 2B–C).
FIGURE 2.
Gut microbial diversity in nonsmoking and smoking patients with CD. A, Gene richness. B and C, Shannon diversity index at the genus level and species level, respectively.
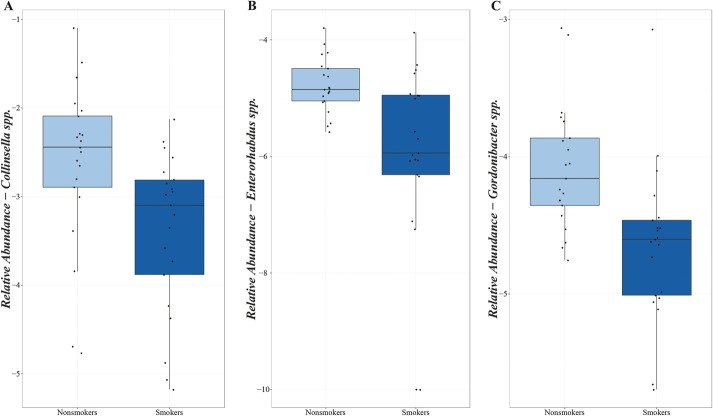
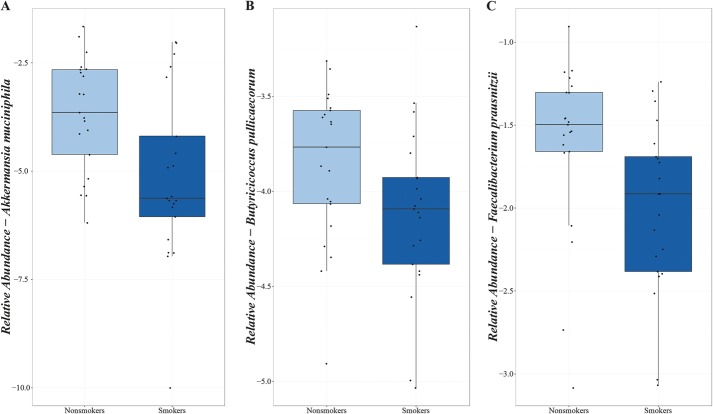
Testing for 94 genera abundant in more than 70% of the studied population, decreased proportions of the genera Collinsella (P = 0.02), Enterorhabdus (P = 0.02), and Gordonibacter (P = 0.02) were observed in smokers compared with nonsmokers (Fig. 3A–C). No statistically significant differences in the relative abundance of species and MetaGenomics Species were detected after correction for multiple testing. However, proportions of several bacterial species previously associated with CD appeared consistently reduced in smokers compared with nonsmokers, including decreased relative abundances of Akkermansia muciniphila (P = 0.15) and Butyricicoccus pullicaecorum (P = 0.26) (Fig. 4A–B). The proportion of Faecalibacterium prausnitzii was also lower in smoking patients with CD (median abundance 1.22%, IQR 0.42%–2.04%) than in nonsmoking patients with CD (median abundance 3.19%, IQR 2.19%–4.98%) (P = 0.10) (Fig. 4C).
FIGURE 3.
Relative abundances of genera in nonsmoking and smoking patients with CD (after log transformation). A, Genus Collinsella. B, Genus Enterorhabdus. C, Genus Gordonibacter.
FIGURE 4.
Relative abundances of species in nonsmoking and smoking patients with CD (after log transformation). A, Akkermansia muciniphila. B, Butyricicoccus pullicaecorum. C, Faecalibacterium prausnitzii.
DISCUSSION
Smoking has well-established detrimental effects on CD, but the biological mechanisms for this association are not clear. This study showed that gut microbial gene richness, genus diversity and species diversity were reduced in smokers with CD as compared with nonsmokers with CD. The microbial profile also differed between these groups regarding the relative abundance of bacterial taxa, including decreased proportions of the genera Collinsella, Enterorhabdus, and Gordonibacter among smoking patients. These findings indicate that an altered gut microbiota may underlie the association between smoking and CD.
Phylogenetic diversity and gene richness of the intestinal microbiome are consistently linked to human health and disease.9,11,30,31 An overall decrease in these ecological measures has been associated with a variety of disorders, including IBD and CD in particular.9,32–35 The observed reduction in gut microbial diversity among smokers in our study is in line with the general perception that this feature is related to an unhealthy state. Accordingly, a greater perturbed microbiota in smoking patients with CD could possibly more readily predispose to the development of disease activity in comparison with nonsmoking patients with CD.
The genera Collinsella, Enterorhabdus, and Gordonibacter belong to the Actinobacteria phylum, which consists of Gram-positive bacteria characterized by high guanine and cytosine contents and has previously been shown less prevalent in healthy smoking individuals.16,17 Collinsella spp. have been isolated from patients with CD and a member of this genus previously appeared less prevalent in relatives of patients with CD compared with unrelated controls.36–38 Enterorhabdus spp. have been identified from mouse models of ileitis and colitis and from the human gut.39–42 This genus was found to be associated with a genetic variant of the human leukocyte antigen complex that has been related to inflammatory diseases.41 Gordonibacter spp. have previously been isolated from the human intestinal tract, including from a patient with CD.43–45 These bacteria can produce urolithins,44,46 which are metabolites considered to have anti-inflammatory properties.47,48 Otherwise, relatively little is known about these bacterial taxa and their potential relationship with smoking or CD, highlighting the need for future studies to explore how these microbes might interact with smoking and CD.
Although not reaching statistical significance after correction for multiple testing, a difference was also observed in the relative abundance of microbes previously implicated in CD. This included a consistent decrease in proportions of A. muciniphila,49,50 B. pullicaecorum,51 and F. prausnitzii among smoking patients.52,53 As studies have shown that these bacteria can exert anti-inflammatory effects, this could further support the hypothesis of microbes mediating the adverse effects of smoking in CD.
Previous studies examining the impact of smoking on the gut microbiota are limited, especially in patients with CD. An elegant, interventional study, in which healthy smoking individuals underwent controlled smoking cessation, showed significant alterations in the fecal microbiota, including an increase in Actinobacteria (high guanine and cytosine content bacteria and Bifidobacteria) and a decrease in Bacteroidetes (Prevotella spp. and Bacteroides spp.).16,17 A higher number of Prevotella spp. and Bacteroides spp. (combined) was also found to be associated with smoking in a study reporting on the effects of smoking in patients with active CD.18 Using a whole-metagenome shotgun sequencing, we were able to assess the relative abundance of these bacterial taxa separately, finding a reduced proportion of the Bacteroides genus (unadjusted P = 0.01), but not the Prevotella genus (unadjusted P = 0.59). A few studies also documented associations for F. prausnitzii and the genus Anaerostipes with smoking in CD.18,54,55 Collectively, our data partly confirm and expand on previous studies. Although it remains to be established how smoking might affect the composition of intestinal microbes, these results emphasize that differences in the gut microbiome may be involved in the deleterious effects of smoking in CD.
A significant advantage of this study over previous studies was the metagenomic approach, which allowed us to evaluate the whole gut microbial community rather than targeting (specific) 16S rRNA sequences. Moreover, this technique offered a full quantitative assessment of the gut microbiome. To minimize the influence of disease activity, we only included patients who were either in endoscopic remission or had low fecal calprotectin levels.
This study was limited by the relatively small sample size, which might have resulted in a lack of statistical power to identify more minor but potentially biologically relevant microbial differences. However, the use of a matched design does result in a more powerful study and reduces the risk of identifying confounding variables, although data on diet or other lifestyle factors possibly associated with smoking were not at our disposal. Furthermore, the smoking status of patients was not extensively evaluated. Self-administered questionnaires may carry a risk of misclassification, but previous studies showed that self-reported data on smoking behavior were fairly accurate.56,57
To conclude, we demonstrated that gut microbial gene richness, genus diversity, and species diversity were decreased in smokers with CD as compared with nonsmokers with CD. This was accompanied by differences in the relative abundance of bacterial taxa, including reduced proportions of the genera Collinsella, Enterorhabdus, and Gordonibacter. In future, larger studies need to evaluate whether intestinal microbes mediate the adverse effects of smoking in CD.
Footnotes
The FXR-IBD consortium is financially supported by FP7 Marie Curie Actions IAPP (FXR-IBD, 611979). J. L. Opstelten was supported by an unrestricted research grant from Dr. Falk Pharma. S. W. C. van Mil was supported by the Netherlands Organization for Scientific Research (NWO) Project VIDI (917.11.365), the Utrecht University Support Grant and Wilhelmina Children's Hospital Research Fund.
The authors have no conflict of interest to disclose.
REFERENCES
- 1.Thia KT, Sandborn WJ, Harmsen WS, et al. Risk factors associated with progression to intestinal complications of Crohn's disease in a population-based cohort. Gastroenterology. 2010;139:1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Büsch K, da Silva SA, Holton M, et al. Sick leave and disability pension in inflammatory bowel disease: a systematic review. J Crohns Colitis. 2014;8:1362–1377. [DOI] [PubMed] [Google Scholar]
- 3.Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parkes GC, Whelan K, Lindsay JO. Smoking in inflammatory bowel disease: impact on disease course and insights into the aetiology of its effect. J Crohns Colitis. 2014;8:717–725. [DOI] [PubMed] [Google Scholar]
- 5.Mahid SS, Minor KS, Soto RE, et al. Smoking and inflammatory bowel disease: a meta-analysis. Mayo Clin Proc. 2006;81:1462–1471. [DOI] [PubMed] [Google Scholar]
- 6.Lindberg E, Järnerot G, Huitfeldt B. Smoking in Crohn's disease: effect on localisation and clinical course. Gut. 1992;33:779–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosnes J, Carbonnel F, Beaugerie L, et al. Effects of cigarette smoking on the long-term course of Crohn's disease. Gastroenterology. 1996;110:424–431. [DOI] [PubMed] [Google Scholar]
- 8.Nunes T, Etchevers MJ, García-Sánchez V, et al. Impact of smoking cessation on the clinical course of Crohn's disease under current therapeutic algorithms: a multicenter prospective study. Am J Gastroenterol. 2016;111:411–419. [DOI] [PubMed] [Google Scholar]
- 9.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manichanh C, Borruel N, Casellas F, et al. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599–608. [DOI] [PubMed] [Google Scholar]
- 11.Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. [DOI] [PubMed] [Google Scholar]
- 12.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(suppl 1):4554–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotillard A, Kennedy SP, Kong LC, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. [DOI] [PubMed] [Google Scholar]
- 15.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biedermann L, Zeitz J, Mwinyi J, et al. Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS One. 2013;8:e59260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biedermann L, Brülisauer K, Zeitz J, et al. Smoking cessation alters intestinal microbiota: insights from quantitative investigations on human fecal samples using FISH. Inflamm Bowel Dis. 2014;20:1496–1501. [DOI] [PubMed] [Google Scholar]
- 18.Benjamin JL, Hedin CR, Koutsoumpas A, et al. Smokers with active Crohn's disease have a clinically relevant dysbiosis of the gastrointestinal microbiota. Inflamm Bowel Dis. 2012;18:1092–1100. [DOI] [PubMed] [Google Scholar]
- 19.Ranjan R, Rani A, Metwally A, et al. Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem Biophys Res Commun. 2016;469:967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lennard-Jones JE. Classification of inflammatory bowel disease. Scand J Gastroenterol. 1989;s170:2–6; discussion 16–19. [DOI] [PubMed] [Google Scholar]
- 21.Harvey RF, Bradshaw JM. A simple index of Crohn's-disease activity. Lancet. 1980;1:514. [DOI] [PubMed] [Google Scholar]
- 22.Godon JJ, Zumstein E, Dabert P, et al. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl Environ Microbiol. 1997;63:2802–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kultima JR, Sunagawa S, Li J, et al. MOCAT: a metagenomics assembly and gene prediction toolkit. PLoS One. 2012;7:e47656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Almeida M. Caractérisation de flores microbiennes intestinale humaine et fromagère par méthode de métagénomique quantative [Characterization of human intestinal microbiota and cheese microbiota by quantitative metagenomic method]. 2013. Université Paris Sud - Paris XI [University of Paris-Sud - University of Paris XI]. Available at: https://tel.archives-ouvertes.fr/tel-01124109/document. Accessed July 2016. [Google Scholar]
- 25.Shannon CE, Weaver W. The mathematical theory of information. Bell Syst Tech J. 1949;27:359–423. [Google Scholar]
- 26.Zeller G, Tap J, Voigt AY, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014;10:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, Goto S, Sato Y, et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tatusova T, Ciufo S, Fedorov B, et al. RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res. 2014;42:D553–D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Jia H, Cai X, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32:834–841. [DOI] [PubMed] [Google Scholar]
- 30.de Vos WM, de Vos EA. Role of the intestinal microbiome in health and disease: from correlation to causation. Nutr Rev. 2012;70(suppl 1):S45–S56. [DOI] [PubMed] [Google Scholar]
- 31.Hollister EB, Gao C, Versalovic J. Compositional and functional features of the gastrointestinal microbiome and their effects on human health. Gastroenterology. 2014;146:1449–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dicksved J, Halfvarson J, Rosenquist M, et al. Molecular analysis of the gut microbiota of identical twins with Crohn's disease. ISME J. 2008;2:716–727. [DOI] [PubMed] [Google Scholar]
- 35.Kang S, Denman SE, Morrison M, et al. Dysbiosis of fecal microbiota in Crohn's disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis. 2010;16:2034–2042. [DOI] [PubMed] [Google Scholar]
- 36.Kageyama A, Benno Y, Nakase T. Phylogenetic and phenotypic evidence for the transfer of Eubacterium aerofaciens to the genus Collinsella as Collinsella aerofaciens gen. nov., comb. nov. Int J Syst Bacteriol. 1999;49:557–565. [DOI] [PubMed] [Google Scholar]
- 37.Joossens M, Huys G, Cnockaert M, et al. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut. 2011;60:631–637. [DOI] [PubMed] [Google Scholar]
- 38.Lee T, Clavel T, Smirnov K, et al. Oral versus intravenous iron replacement therapy distinctly alters the gut microbiota and metabolome in patients with IBD. Gut. [published online ahead of print February 4, 2016]. 10.1136/gutjnl-2015-309940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clavel T, Charrier C, Braune A, et al. Isolation of bacteria from the ileal mucosa of TNFdeltaARE mice and description of Enterorhabdus mucosicola gen. nov., sp. nov. Int J Syst Evol Microbiol. 2009;59:1805–1812. [DOI] [PubMed] [Google Scholar]
- 40.Clavel T, Duck W, Charrier C, et al. Enterorhabdus caecimuris sp. nov., a member of the family Coriobacteriaceae isolated from a mouse model of spontaneous colitis, and emended description of the genus Enterorhabdus Clavel, et al. 2009. Int J Syst Evol Microbiol. 2010;60:1527–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hov JR, Zhong H, Qin B, et al. The influence of the autoimmunity-associated ancestral HLA haplotype AH8.1 on the human gut microbiota: a cross-sectional study. PLoS One. 2015;10:e0133804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang J, Summanen PH, Henning SM, et al. Xylooligosaccharide supplementation alters gut bacteria in both healthy and prediabetic adults: a pilot study. Front Physiol. 2015;6:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Würdemann D, Tindall BJ, Pukall R, et al. Gordonibacter pamelaeae gen. nov., sp. nov., a new member of the Coriobacteriaceae isolated from a patient with Crohn's disease, and reclassification of Eggerthella hongkongensis Lau, et al. 2006 as Paraeggerthella hongkongensis gen. nov., comb. nov. Int J Syst Evol Microbiol. 2009;59:1405–1415. [DOI] [PubMed] [Google Scholar]
- 44.Selma MV, Tomás-Barberán FA, Beltrán D, et al. Gordonibacter urolithinfaciens sp. nov., a urolithin-producing bacterium isolated from the human gut. Int J Syst Evol Microbiol. 2014;64:2346–2352. [DOI] [PubMed] [Google Scholar]
- 45.Jin JS, Lee KC, Park IS, et al. Gordonibacter faecihominis sp. nov., isolated from human faeces. Antonie Van Leeuwenhoek. 2014;106:439–447. [DOI] [PubMed] [Google Scholar]
- 46.Selma MV, Beltrán D, García-Villalba R, et al. Description of urolithin production capacity from ellagic acid of two human intestinal Gordonibacter species. Food Funct. 2014;5:1779–1784. [DOI] [PubMed] [Google Scholar]
- 47.González-Sarrías A, Larrosa M, Tomás-Barberán FA, et al. NF-kappaB-dependent anti-inflammatory activity of urolithins, gut microbiota ellagic acid-derived metabolites, in human colonic fibroblasts. Br J Nutr. 2010;104:503–512. [DOI] [PubMed] [Google Scholar]
- 48.Larrosa M, González-Sarrías A, Yáñez-Gascón MJ, et al. Anti-inflammatory properties of a pomegranate extract and its metabolite urolithin-A in a colitis rat model and the effect of colon inflammation on phenolic metabolism. J Nutr Biochem. 2010;21:717–725. [DOI] [PubMed] [Google Scholar]
- 49.Png CW, Lindén SK, Gilshenan KS, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. [DOI] [PubMed] [Google Scholar]
- 50.Derrien M, Van Baarlen P, Hooiveld G, et al. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front Microbiol. 2011;2:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eeckhaut V, Machiels K, Perrier C, et al. Butyricicoccus pullicaecorum in inflammatory bowel disease. Gut. 2013;62:1745–1752. [DOI] [PubMed] [Google Scholar]
- 52.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sokol H, Seksik P, Furet JP, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–1189. [DOI] [PubMed] [Google Scholar]
- 54.Li E, Hamm CM, Gulati AS, et al. Inflammatory bowel diseases phenotype, C. difficile and NOD2 genotype are associated with shifts in human ileum associated microbial composition. PLoS One. 2012;7:e26284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Connor Gorber S, Schofield-Hurwitz S, Hardt J, et al. The accuracy of self-reported smoking: a systematic review of the relationship between self-reported and cotinine-assessed smoking status. Nicotine Tob Res. 2009;11:12–24. [DOI] [PubMed] [Google Scholar]
- 57.Yeager DS, Krosnick JA. The validity of self-reported nicotine product use in the 2001-2008 National Health and Nutrition Examination Survey. Med Care. 2010;48:1128–1132. [DOI] [PubMed] [Google Scholar]