Abstract
DAF-16, the C. elegans FOXO transcription factor, is an important determinant in aging and longevity. In this work, we manually curated FOXODB http://lyh.pkmu.cn/foxodb/, a database of FOXO direct targets. It now covers 208 genes. Bioinformatics analysis on 109 DAF-16 direct targets in C. elegans found interesting results. (i) DAF-16 and transcription factor PQM-1 co-regulate some targets. (ii) Seventeen targets directly regulate lifespan. (iii) Four targets are involved in lifespan extension induced by dietary restriction. And (iv) DAF-16 direct targets might play global roles in lifespan regulation.
Keywords: lifespan, C. elegans, insulin signaling, FOXO direct targets, bioinformatics, Gerotarget
INTRODUCTION
DAF-16, the C. elegans FOXO transcription factor, plays as a molecular switch in lifespan regulation [1]. When activated by reduced insulin signaling, it could extend C. elegans's lifespan by activating or inhibiting its downstream genes [2, 3]. Presumably, these downstream genes largely determine how the lifespan can be extended. Yet, little is known about their positions in the regulatory network: which are directly regulated by DAF-16, and which are indirect targets.
To identify DAF-16 targets, various high throughput techniques have been used, such as microarray [2, 3], proteomics [4], and DamID (DNA adenine methyltransferase identification) [5]. Microarray and proteomics could identify DAF-16 downstream genes, but have difficult to figure out whether they are direct or indirect targets. DamID could identify DAF-16 direct targets in theory, but may have probability to identify false positives and negatives in practice [6].
Insulin signaling is remarkably conserved in C. elegans, Drosophila melanogaster and mammals, and reduced signaling of this pathway has been shown to extend lifespan in all of these animals [7]. For FOXO and its orthologs, there are different identifiers of genes, transcripts and proteins in different species. We called them all “FOXOs” hereafter. Currently, many experimentally validated FOXOs direct targets scattered in literatures [6, 8, 9]. Collecting these known targets, and then mapping them to C. elegans through orthologous analysis would be helpful for longevity research in C. elegans.
In this work, by manually reading literatures, we collected 208 experimentally validated FOXOs direct targets. Through orthologous mapping, we eventually got 109 DAF-16 direct targets in C. elegans. To make data easily accessible, we set up FOXODB (http://lyh.pkmu.cn/foxodb/). Bioinformatics analysis on the 109 targets revealed interesting results.
RESULTS
FOXODB: a database of FOXO direct targets
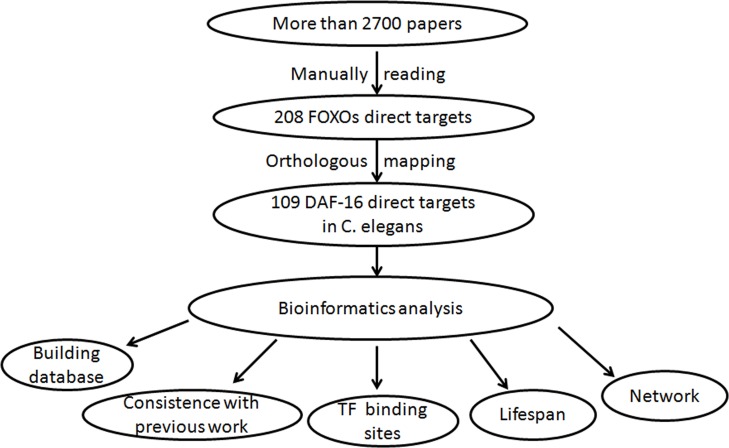
As shown in Figure 1, we searched PubMed with keywords “FOXOs” and found more than 2700 papers. We manually read the papers. When a gene was determined as a FOXOs direct target, key information was extracted and record in FOXODB (http://lyh.pkmu.cn/foxodb). Rules for collecting a gene to FOXODB were strict. (i) The gene should be differentially expressed in FOXOs (+) versus FOXOs (−); (ii) FOXOs must be able to bind to the promoter of the gene; And (iii) only traditional experimental evidence(s) was adopted. Details can be found in Materials and Methods.
Figure 1. Workflow.
We first searched PubMed with FOXOs keywords and found more than 2700 papers. Through manually reading these papers, we got 208 FOXOs direct targets. Inparanoid was used to map FOXOs targets to their orthologs in C. elegans. And 109 genes were found eventually. Bioinformatics analysis on the 109 genes were done including comparison with previous results, transcription factor binding site enrichment, lifespan regulation and network topological feature analysis. And we built a database to make all data easily accessible.
Currently, FOXODB covers 302 entries and 208 genes, including 35, 26, and 147 direct targets in C. elegans, Drosophila melanogaster and mammals, respectively. FOXODB is well designed and friendly to user (Figure 2). As in our previous works [10, 11], FOXODB was written in PHP (Hypertext Preprocessor). We believe FOXODB will be a valuable resource to the field.
Figure 2. FOXODB database.
FOXODB is a database aiming at collecting experimentally verified direct targets of FOXOs. FOXODB is well designed and friendly to user. User could easily browse, search and download the genes.
DAF-16 direct targets significantly overlaps with previous results
Inparanoid is a database specially designed for orthologue analysis [12]. We used it to map FOXODB genes to C. elegans orthologs and got 109 DAF-16 direct targets eventually. (Supplementary Table 1 and 2).
The 109 targets significantly overlapped with genes found in previous works (Supplementary Table 3). For example, Murphy et al. found 514 differential expressed genes by comparing microarrays of daf-16 (+) versus daf-16 (−) [2], and 18 (2.8 in random chance, p = 0) of them overlapped with the 109 targets. Similar, Tepper et al. found 3,396 differential expressed genes [13], and 31 (18.5 in random chance, p = 0.0019) of them overlapped with the 109 targets. Dong et al. found 86 proteins differentially expressed using proteomics [4], and 12 (0.47 under random, p = 0) of them appeared in the 109 targets. Using DamID, 65 genes were found as potential DAF-16 targets [5], and 6 (0.31 under random, p = 0) of them were in the 95 targets (14 genes were excluded from the 109 targets, since they were collected from this work. 95 = 109-14). These results showed the 109 DAF-16 direct targets were reliable.
DAF-16 and PQM-1 co-regulate some direct targets
It has been reported that DAF-16 binding element (DBE), GTAAACA or TGTTTAC, and DAF-16-associated element (DAE), TGATAAG or CTTATCA, enriched in DAF-16 regulated genes [2, 4, 13, 14]. The DBE was recognized by DAF-16, and the DAE by transcription factor PQM-1 [13]. Here, we searched both the DBE and the DAE in 1kb promoter region (relative to TSS) of the 109 targets. As a result, 30 genes contained the DBE (Supplementary Table 4), 30 genes contained the DAE (Supplementary Table 5) and 7 genes contained both (Table 1). Recent work showed that DAF-16 could even recognize DAE [15]. Taken together, these results indicated DAF-16 and PQM-1 at least co-regulate some direct targets.
Table 1. The seven DAF-16 direct targets that contain both DBE and DAE.
| GeneID | DAF-16 binding element(DBE) | DAF-16 associate element(DAE) |
|---|---|---|
| daf-2 | [GTAAACA: 108] | [CTTATCA: 85] |
| mtl-1 | [GTAAACA: 44] | [CTTATCA: 417] |
| srp-2 | [GTAAACA: 780] | [CTTATCA: 165][TGATAAG: 448] |
| rars-1 | [TGTTTAC: 925] | [CTTATCA: 697] |
| T19C9.8 | [GTAAACA: 174] | [CTTATCA: 54] |
| C25E10.8 | [GTAAACA: 231] | [CTTATCA: 935][TGATAAG: 267] |
| daf-16 | [GTAAACA: 264] | [TGATAAG: 46] |
DAF-16 binding element (DBE): GTAAACA or TGTTTAC; DAF-16 associated element (DAE): TGATAAG or CTTATCA. [GTAAACA: 108] means that GTAAACA is located 108bp relative to transcription start site.
Seventeen DAF-16 direct targets directly regulated lifespan
GenAge [16], an useful longevity research resource, covers 681 longevity genes in C. elegans. Compared with the 109 targets, 17 genes overlapped, significantly higher than 3.71 under random, p = 0 (Table 2). Of the 17 genes, 10 were obtained by orthologous mapping. This means they were for the first time known as DAF-16 direct targets that regulate lifespan.
Table 2. The 17 DAF-16 direct targets that directly regulate lifespan.
| Symbol | Longevity influence | Orthologous mapping |
|---|---|---|
| din-1 | Pro-Longevity | Yes |
| lgg-1 | Pro-Longevity | Yes |
| mdh-1 | Pro-Longevity | No |
| nnt-1* | Pro-Longevity | No |
| prdx-3 | Pro-Longevity | Yes |
| daf-16 | Pro-Longevity | Yes |
| aco-2 | Anti-Longevity | Yes |
| age-1 | Anti-Longevity | Yes |
| daf-2 | Anti-Longevity | Yes |
| daf-7 | Anti-Longevity | Yes |
| dod-17# | Anti-Longevity | No |
| gpd-2* | Anti-Longevity | No |
| ubh-4 | Anti-Longevity | No |
| pck-2* | Anti-Longevity | No |
| W09D10.3 | Anti-Longevity | Yes |
| lars-2 | Anti-Longevity | Yes |
| sod-2 | Unclear | No |
and # represent up and down regulated in daf-16(+) versus daf-16(−), respectively. Longevity genes were classified into anti-longevity and pro-longevity. Knockout or suppression of anti-longevity gene, or overexpression of pro-longevity gene resulted in lifespan extension, whereas the opposite interventions led to reduction of lifespan. Orthologous mapping describes whether the gene was gotten from orthologous mapping (Yes) or not (NO).
According to GenAge, longevity genes were classified into anti-longevity and pro-longevity [16, 17]. Knockout or suppression of anti-longevity gene, or overexpression of pro-longevity gene resulted in lifespan extension, whereas the opposite interventions led to reduction of lifespan [16, 17]. Of the 17 genes, 10 were classified as anti-longevity, 6 as pro-longevity, and one, sod-2, was not classified. We observed their expressions in Murphy et al.'s microarrays as mentioned above [2]. gpd-2, pck-2 and nnt-1 were up-regulated, and dod-17 was down-regulated in daf-16(+) vs. daf-16(-). Since gpd-2 and pck-2 were anti-longevity, their up-regulations were not expected in daf-16(+) animals. Thus, we inferred that knocking out or down either gpd-2 or pck-2 in daf-16(+) might further extend lifespan. Fortunately, this inference has been validated in previous work [4]
Four DAF-16 direct targets were involved in lifespan extension induced by dietary restriction
Many dietary restriction methods could extend C. elegans's lifespan [18]. Some of them such as eat mutation or some forms of bacterial dilution do not require DAF-16, while some other forms of bacterial dilution and peptone dilution require DAF-16 [18]. Thus, it was interesting to know whether DAF-16 direct targets were involved in lifespan extension induced by dietary restriction. GenDR, a database collecting lifespan-regulating genes related to dietary restriction, covers 48 genes in C. elegans [19]. Here, we compared them with the 109 targets and found 4 overlapping genes: age-1, hsp-12.6, daf-16 and daf-2. This was significantly higher than 0.26 under random, with p = 1.35E-4. This result supported that some dietary restriction methods required DAF-16 for lifespan extension.
DAF-16 direct targets might play global roles in lifespan regulation
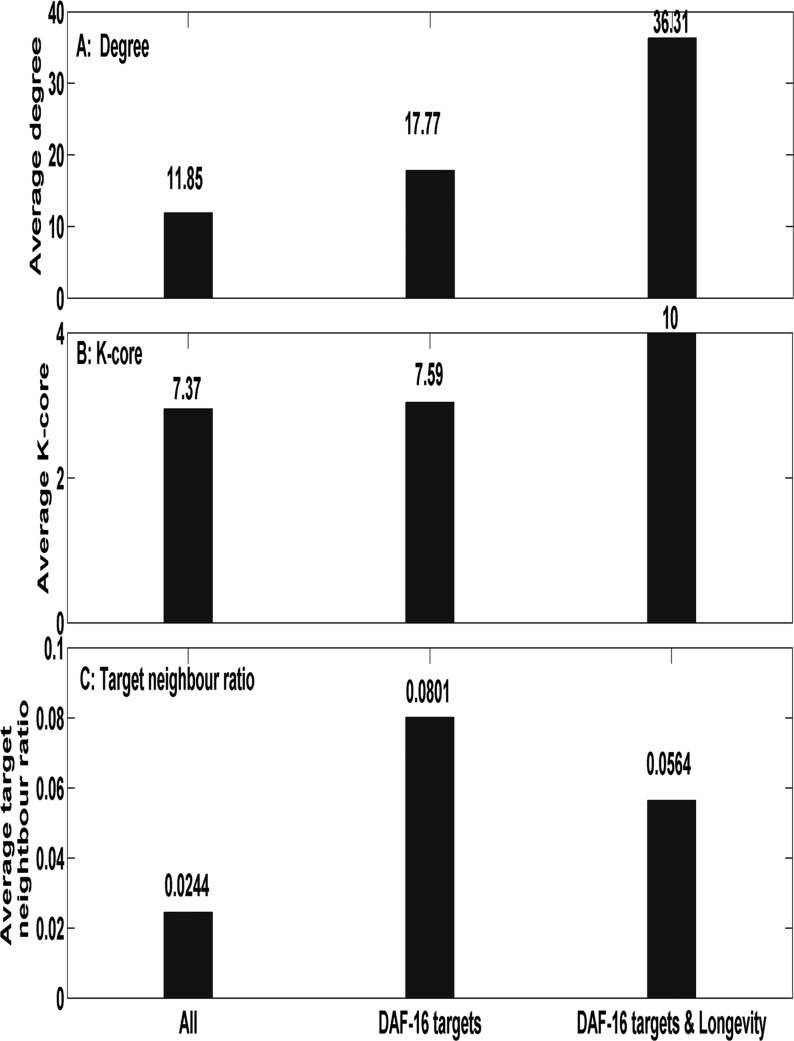
Proteins do not function in isolation but through interaction with each other. And from network view, the more interaction partners (higher degrees) one protein has, the more important the protein might be. Here, we studied the degrees of the 109 targets, and found that the average degree is 17.77, significantly higher than 11.85, the average degree for other proteins in the network (p = 0.0014, Kolmogorov-Smirnov test, KS test for short). As analyzed above, 17 targets directly regulated lifespan. The average degree for them is the highest, 36.31 (see Figure 3A). This result was consistent with our previous work, the degrees of longevity genes tend to be higher than that of non- longevity genes [20].
Figure 3. Network topological features of DAF-16 direct targets.
The network topological features of DAF-16 direct targets were shown. ‘All’ represents all proteins in the network. ‘DAF-16 targets’ represents the 109 DAF-16 direct targets in C. elegans. ‘DAF-16 targets & longevity’ represents the DAF-16 direct targets that regulate lifespan. Figure A is the average degree, Figure B is the average K-core, and Figure C is the average of target neighbor ratio.
K-core, another network index, takes into account not only the number of direct neighbors but also the placement of a protein in the network. It assumed that centrally located proteins are more important than the peripheral ones [21]. As shown in Figure 3B, the 109 targets have an average K-core 7.59, significantly higher than 7.37, the average for other proteins in the network (p = 8.4*E-4, KS test). And the 17 lifespan-regulating targets had the highest average K-core 10.
To know whether DAF-16 direct targets function through cooperation with each other, we computed for each protein the ‘target neighbor ratiO'. It is the ratio of the number of interaction partners that belong to the DAF-16 direct targets to its degree [22]. As shown in Figure 3C, DAF-16 direct targets tend to directly interact with each other (p = 2.7*E-4, KS test).
In all, these results revealed that DAF-16 direct targets tended to have more interaction neighbors, locate network center and interact with each other. This implied that DAF-16 direct targets might play global roles in lifespan regulation.
DISCUSSION
In this work, we manually curated FOXODB by reading literatures. It now covers 208 FOXOs direct targets. To our knowledge, this is the largest. 109 DAF-16 direct targets in C. elegans were found by orthologous mapping. And 17 of them directly regulated lifespan. These are also important data to the field.
We searched DAF-16 binding element (DBE) in 1kb promoter region of the 109 DAF-16 direct targets, and found 30 of them contained the DBE (GTAAACA or TGTTTAC) while the others not. It was difficult to understand why so many DAF-16 direct targets did not contain DBE. For explanation, first, different works used different DBE motifs [2-4]. It was hard to know which one was correct. We chose a strict DBE motif and thus resulted in few sequence matches. If using a loose DBE motif, more genes with DBE could be found. For example, when using DBE, RTAAAYA, R = A/G, Y = C/T, as in previous work [3], 91 of the 109 targets would contain the DBE in 1kb promoter region. Second, we only searched the 1kb promoter region. Some DBE may locate outside of the region and thus not be found.
We did the first network analysis on DAF-16 direct targets. The results showed they tended to be higher in degree, locate network center and directly interact with each other. The protein interactions used for network analysis include several kinds of interactions such as physical interaction, genetic interaction and predicted interaction. However, it's worth noting that some of the interactions might be collected from literatures. Thus, the more a gene being studied, the more likely the gene has higher degree. Though the collected interactions may be only a small part of the whole data, we still cannot exclude the possibility that this might affect the results.
MATERIALS AND METHODS
Data source
The gene sequences were downloaded from WormBase, version 220. Protein interaction network was obtained from our previous work [20]. The network was constructed by integrating different kinds of interactions including physical interactions, genetics interactions and predicted interactions, covering a total of 7, 219 proteins and 41, 132 edges [20].
Workflow
As shown in Figure 1, we searched PubMed with ‘FOXOs’ and found more than 2700 papers. We manually read the papers and found 208 FOXOs direct targets. Inparanoid is a database specially designed for orthologous analysis [12]. We used it to map the 208 targets to their orthologs in C. elegans and finally got 109 genes. Bioinformatics analysis on this list were done including comparison with previous results, transcription factor binding site enrichment, lifespan regulation and network topological feature analysis. We built a database to make all data easily accessible.
Database creation
To collect FOXOs direct targets, we searched PubMed using keywords: ‘FOXO1, FOXO3, FOXO4, FOXO6, AFX, FKHR, dFOXO and DAF-16′. Then we manually read the papers and retrieved gene symbol, species, quantitation methods, DNA binding detection methods and function descriptions et al. Strict rules were used to determine whether a gene was a FOXOs direct target or not. (i) The gene should be differentially expressed in FOXOs (+) VS. FOXOs (−). The quantitation method should be traditional such as RT-PCR or western blotting. And if high throughput quantitation method such as microarray was used, the result must be further confirmed by other technique like GFP (Green Fluorescent Protein). (ii) There must be evidence showing that FOXOs could bind to the promoter region of the gene, like ChIP (chromatin immunoprecipitation) and EMSA (Electrophoretic mobility shift assay), or mutating the FOXOs binding site could significantly change the gene expression.
Hypergeometric model
The hypergeometric model was used for calculating the significance of two gene sets with a certain number of overlapping genes. The P-value is calculated as follows:
N: Number of genes in C. elegans genome, 20,000 was used for approximation in this work.
m: Number of genes in gene set 1.
n: Number of genes in gene set 2.
k: Number of overlapping genes between the m genes and the n genes.
Kolmogorov-Smirnov test
In statistics, the two-sample Kolmogorov-Smirnov test is one of the most useful and general nonparametric methods for comparing two samples, as it is sensitive to differences in both location and shape of the empirical cumulative distribution functions of the two samples. In this work, two-sample KS test was used to compare the network topological features of DAF-16 direct targets and that of the remaining genes in C. elegans.
Network topological features
The network topological features degree and K-core were computed by using an R package igraph [23]. The definitions for them can be found in Table 3.
Table 3. Formal representation of graph measures.
| Name | Function | Descriptions |
|---|---|---|
| Degree | Ki | the number of interaction partners of node i |
| Target neighbor ratio | Kpi | K piis the number of links between node i and proteins encoded by DAF-16 direct targets |
| K-core | K | A K-core of a graph can be obtained by recursively removing all nodes with a degree less than K, until all nodes in the remaining graph have a degree at least K. |
Functions are the definitions of the topological features. Descriptions give explanations for symbols in the definitions.
SUPPLEMENTARY FIGURES AND TABLES
Footnotes
CONFLICTS OF INTEREST
All the authors declare that they have no competing interests.
GRANT SUPPORT
This work was supported by the National Natural Science Foundation of China (grant nos. 81300253).
Authors' contributions
Yan-Hui Li conceived and designed the experiments. Yan-Hui Li performed the experiments and wrote the paper. Gai-Gai Zhang collected part of the FOXOs target genes. Gai-Gai Zhang revised the paper. Both authors read and approved the final manuscript.
REFERENCES
- 1.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 2.Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 3.McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 4.Dong MQ, Venable JD, Au N, Xu T, Park SK, Cociorva D, Johnson JR, Dillin A, Yates JR., 3rd Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science. 2007;317:660–663. doi: 10.1126/science.1139952. [DOI] [PubMed] [Google Scholar]
- 5.Schuster E, McElwee JJ, Tullet JM, Doonan R, Matthijssens F, Reece-Hoyes JS, Hope IA, Vanfleteren JR, Thornton JM, Gems D. DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol Syst Biol. 2010;6:399. doi: 10.1038/msb.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy CT. The search for DAF-16/FOXO transcriptional targets: approaches and discoveries. Exp Gerontol. 2006;41:910–921. doi: 10.1016/j.exger.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 7.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 8.van der Vos KE, Coffer PJ. The extending network of FOXO transcriptional target genes. Antioxid Redox Signal. 2011;14:579–592. doi: 10.1089/ars.2010.3419. [DOI] [PubMed] [Google Scholar]
- 9.Puig O, Mattila J. Understanding Forkhead box class O function: lessons from Drosophila melanogaster. Antioxid Redox Signal. 2011;14:635–647. doi: 10.1089/ars.2010.3407. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Wang C, Miao Z, Bi X, Wu D, Jin N, Wang L, Wu H, Qian K, Li C, Zhang T, Zhang C, Yi Y, Lai H, Hu Y, Cheng L, et al. ViRBase: a resource for virus-host ncRNA-associated interactions. Nucleic Acids Res. 2015;43:D578–582. doi: 10.1093/nar/gku903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li YH, Zhang G, Cui Q. PPUS: a web server to predict PUS-specific pseudouridine sites. Bioinformatics. 2015;31:3362–3364. doi: 10.1093/bioinformatics/btv366. [DOI] [PubMed] [Google Scholar]
- 12.Ostlund G, Schmitt T, Forslund K, Kostler T, Messina DN, Roopra S, Frings O, Sonnhammer EL. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 38:D196–203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tepper RG, Ashraf J, Kaletsky R, Kleemann G, Murphy CT, Bussemaker HJ. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell. 2013;154:676–690. doi: 10.1016/j.cell.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, Johnson TE, Kim SK. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell. 2008;134:291–303. doi: 10.1016/j.cell.2008.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang P, Judy M, Lee SJ, Kenyon C. Direct and indirect gene regulation by a life-extending FOXO protein in C. elegans: roles for GATA factors and lipid gene regulators. Cell Metab. 2013;17:85–100. doi: 10.1016/j.cmet.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tacutu R, Craig T, Budovsky A, Wuttke D, Lehmann G, Taranukha D, Costa J, Fraifeld VE, de Magalhaes JP. Human Ageing Genomic Resources: integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res. 2013;41:D1027–1033. doi: 10.1093/nar/gks1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Budovsky A, Tacutu R, Yanai H, Abramovich A, Wolfson M, Fraifeld V. Common gene signature of cancer and longevity. Mech Ageing Dev. 2009;130:33–39. doi: 10.1016/j.mad.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;8:113–127. doi: 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wuttke D, Connor R, Vora C, Craig T, Li Y, Wood S, Vasieva O, Shmookler Reis R, Tang F, de Magalhaes JP. Dissecting the gene network of dietary restriction to identify evolutionarily conserved pathways and new functional genes. PLoS Genet. 2012;8:e1002834. doi: 10.1371/journal.pgen.1002834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li YH, Dong MQ, Guo Z. Systematic analysis and prediction of longevity genes in Caenorhabditis elegans. Mech Ageing Dev. 2010;131:700–709. doi: 10.1016/j.mad.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Wuchty S, Almaas E. Peeling the yeast protein network. Proteomics. 2005;5:444–449. doi: 10.1002/pmic.200400962. [DOI] [PubMed] [Google Scholar]
- 22.Xu J, Li Y. Discovering disease-genes by topological features in human protein-protein interaction network. Bioinformatics. 2006;22:2800–2805. doi: 10.1093/bioinformatics/btl467. [DOI] [PubMed] [Google Scholar]
- 23.Csardi G NT. The igraph software package for complex network research. InterJournal, Complex Systems. 2006 http://igraph.sf.net [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.