Summary
Secretory immunoglobulin A (SIgA) antibodies play an important role in protecting the mucosal surfaces against pathogens and maintaining homeostasis with the commensal microbiota. Because a substantial portion of the gut microbiota is coated with SIgA, we hypothesized that microbiota–SIgA complexes are important for the maintenance of gut homeostasis. Here we investigated the relationship between microbiota–SIgA complexes and inflammatory epithelial cell responses. We used a multi‐cellular three‐dimensional (3D) organotypical model of the human intestinal mucosa composed of an intestinal epithelial cell line and primary human lymphocytes/monocytes, endothelial cells and fibroblasts. We also used human SIgA from human colostrum, and a prominent bacterial member of the first colonizers, Escherichia coli, as a surrogate commensal. We found that free and microbiota‐complexed SIgA triggered different epithelial responses. While free SIgA up‐regulated mucus production, expression of polymeric immunoglobulin receptor (pIgR) and secretion of interleukin‐8 and tumoir necrosis factor‐α, microbiota‐complexed SIgA mitigated these responses. These results suggest that free and complexed SIgA have different functions as immunoregulatory agents in the gut and that an imbalance between the two may affect gut homeostasis.
Keywords: gut, human, mucosal immunity, SIgA
Introduction
The intestinal microbiota has a key role in influencing the normal development of the mucosal immune system 1 and is instrumental in the metabolism of food and drugs 2, 3. Microbiota is also in constant contact with secretory immunoglobulin A (SIgA), the predominant immunoglobulin in mucosal compartments 4. Indeed, the production of SIgA depends upon microbiota colonization of the gastrointestinal tract. It has been demonstrated that germ‐free mice exhibit a marked reduction of SIgA in the gut 5, 6, 7. In addition, a substantial proportion (24–74%) of the microbiota is coated with SIgA 8, 9, 10. Of note, the enzymatic resistance of SIgA to protease‐containing secretions such as those present in the gut is enhanced when the antibody is complexed with antigen 11. Moreover, binding of SIgA to microbiota promotes a controlled sampling of microbial‐derived antigens by microfold cells (M cells) within the epithelial layer 10, 12.
SIgA has also been shown to help reduce pathogen‐mediated proinflammatory responses by the gut epithelium 13. For example, using a rabbit model of ligated ileal loops, investigators have shown that SIgA neutralization of Shigella flexneri can down‐regulate inflammatory responses 14. However, little is known regarding how free and microbiota‐complexed SIgA regulate these epithelial cell responses. Herein, we used a multi‐cellular three‐dimensional (3D) model of the human intestinal mucosa to evaluate the early events triggered in epithelial cells following interactions with free and microbiota‐complexed SIgA. This 3D model is comprised of human intestinal epithelial cells, lymphocytes/monocytes, endothelial cells and fibroblasts 15, 16. In this 3D model, the epithelial cell line behaves as a multi‐potent progenitor cell that gives rise to functional and highly differentiated cells from multiple lineages (i.e. absorptive enterocyte, goblet and M cells) 15. Epithelial cells in our 3D model grow as a confluent monolayer surrounding the extracellular matrix (ECM), with their luminal surface facing outwards 15, 16. We also used a single prominent bacterial member of the first colonizers, Escherichia coli, as a surrogate commensal and human SIgA from human colostrum. It is well known that colostrum contains SIgA specific to microbiota (including E. coli) which have once passed through the mother's gut (namely the ‘enteric–mammary link’) 17.
Here we report that the majority of E. coli present in the supernatants from the 3D model were coated with SIgA and that this interaction was instrumental in changing the epithelial cell immune responses when compared to those elicited by free SIgA. While free SIgA up‐regulated mucus production, expression of polymeric immunoglobulin receptor (pIgR) and secretion of interleukin (IL)−8 and tumour necrosis factor (TNF)‐α, microbiota‐complexed SIgA mitigated these responses. These results suggest that free and complexed SIgA might have different immunoregulatory properties in the gut and that an imbalance between the two may affect gut homeostasis.
Methods
3D model cells and culture media
The 3D model system was comprised of intestinal epithelial cell line HCT‐8 cells [CCL‐244; American Type Culture Collection (ATCC), Manassas, VA, USA] and primary human lymphocytes/monocytes, endothelial cells (HUVEC cells, CRL‐1459; ATCC) and fibroblasts (CCD‐18Co cells, CRL‐1459; ATCC) cultured under microgravity conditions. Cell cultivation and the set‐up of the 3D model were performed as described previously 15, 16. Briefly, fibroblasts and endothelial cells were embedded in a collagen‐I matrix (Invitrogen, Carlsbad, CA, USA), enriched with additional gut basement membrane proteins 18 and added to rotating wall vessels (RWV) (Synthecon, Houston, TX, USA). The collagen mixture was composed of Dulbecco's modified Eagle's medium (DMEM) (Invitrogen), supplemented with 50 µg/ml gentamicin, 2 mM L‐glutamine and 10% heat‐inactivated fetal bovine serum plus 3 mg/ml bovine collagen‐I (Invitrogen), 10 μg/ml laminin (Sigma, St Louis, MO, USA), 40 μg/ml collagen IV (Sigma), 10 μg/ml fibronectin (BD Pharmingen, San Jose, CA, USA), 2 μg/ml heparin sulphate proteoglycan (Sigma) and 15 mM NaOH (to reach the physiological pH). Epithelial cells were then added to the vessels.
Lymphocytes/monocytes isolated from healthy volunteers were added to the 3D model culture at days 4 and 9 (± 1 day) 19. Briefly, after the Ficoll density gradient centrifugation step, lymphocytes and monocytes were collected, washed and added immediately to the cultures without stimulation, or cryopreserved in liquid N2. It is important to highlight that peripheral blood mononuclear cells (PBMC) consist largely of lymphocytes and monocytes, but also contain a small proportion of dendritic cells and other low‐frequency cell subsets. Isolated lymphocytes/monocytes were added to the 3D model at the same frequency (2 × 107/vessel) and timing, as described previously 15, 16. The experiments in this paper were performed with 15–17‐day‐old 3D models.
Ethics statement
All blood specimens were collected from volunteers who participated in the University of Maryland Institutional Review Board approved protocol (number HP‐00040025) that authorized the collection of blood specimens from healthy volunteers for the studies included in this paper. This protocol was conducted in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. The purpose of this study was explained to the volunteers, who gave informed, signed consent before the blood draw. PBMC were isolated from the blood by density gradient centrifugation and cryopreserved in liquid N2 following standard techniques 15, 19.
E. coli infection and SIgA treatment
3D model constructs were stimulated by incubating them for up to 6 h at 37°C in RPMI (without antibiotics) in the presence of SIgA (100 μg/ml; MP Biomedicals Life Science, Santa Ana, CA, USA) and/or E. coli strain HS (HS). Epithelial cells were exposed to a multiplicity of infection (MOI) ranging from 500 : 1 to 1500 : 1. Based on our previous flow cytometry analyses, using collagenase‐dissociated cells from the 3D model, we estimate that 3D organoids, like those used to perform the experiments in this paper, will contain approximately 20 × 106 HCT‐8 cells. SIgA were purified commercially from pooled human colostrum using multi‐step procedures, including salt fractionation, gel filtration, ion‐exchange chromatography and immunoabsorption. The product was then dialyzed into 0·01 M sodium phosphate, 0·07 M sodium chloride, pH 7·3, filtered through a 0·22 µm filter, vialled and lyophilized. No traces of immunoglobulin (Ig)G, IgM or non‐immunoglobulin proteins were detected in this SIgA preparation. Exposure to purified human serum IgA (100 μg/ml; MP Biomedicals Life Science, Santa Ana, CA, USA) or purified human serum IgG (100 μg/ml, ThermoFisher, Waltham, MA, USA) were used as controls. In co‐culture experiments, HS and either SIgA or serum IgG or IgA were added concomitantly to the 3D models. In some experiments, neutralizing goat anti‐human pIgR polyclonal antibody (8 μg/ml; R&D, Minneapolis, MN, USA) was added 1 h before the addition of HS and SIgA to the 3D culture system. No preservative was added to any of the Ig used to treat the 3D model constructs (i.e. SIgA, serum IgA, IgG or anti‐pIgR).
Preparation of 3D model constructs for histology and immunohistochemistry
The following primary anti‐human antibodies were used for immunohistochemistry: (1) goat anti‐polymeric immunoglobulin receptor (pIgR) polyclonal antibody (1 : 50; R&D), (2) mouse anti‐MUC2 monoclonal antibodies (mAb) (clone CCP58, 1 : 100; Invitrogen, Camarillo, CA, USA) and (3) rabbit protein‐A purified anti‐E. coli–IgG polyclonal antibody (1 : 1000; Abcam, Cambridge, MA, USA). Of note, a mixture of E. coli strains (O157:H7, O20, O125, O55, O111 and K12), intact plus lysed, non‐denatured was used for the preparation of the polyclonal anti‐E. coli antibody. These antibodies are known to react readily to E. coli strain HS 20. The 3D model constructs were removed from the vessels and fixed by immersion in 5% paraformaldehyde (overnight incubation) or methanol Carnoy (4 h incubation) at room temperature. After fixation, the 3D model constructs were embedded in paraffin‐blocks, and 5 μm sections cut and dried in an oven at 37ºC overnight. The sections were then washed consecutively two times for 5 min in Histo‐Clear (National Diagnostics, Atlanta, GA, USA), in 100% ethanol, 95% ethanol and 75% ethanol and finally rehydrated in phosphate‐buffered saline (PBS) for 10 min. For histological staining, tissue sections were stained with haematoxylin and eosin (H&E) and examined under a light microscope for morphological evaluations.
For immunochemical staining procedures, tissue sections were rinsed in water and the antigen retrieved by autoclaving samples (120ºC, 30 s) (Pascal chamber; Dako, Carpinteria, CA, USA) in sodium‐citrate buffer (Invitrogen). After washing in distilled H2O for 5 min, the sections were treated with PBS, 3% H2O2 for 15 min at room temperature. Tissue sections were then washed in PBS/0·5% Tween‐20 and blocked with blocking buffer [PBS/0·5% Tween‐20/5% bovine serum albumin (BSA)]. After 1 h, an excess of blocking buffer was blotted and the sections incubated with blocking buffer‐diluted primary antibody for 1 h at room temperature. After incubating two times in PBS/0·5% Tween‐20 for 5 min, detection of the primary antibody was performed by incubating the sections with anti‐mouse, ‐rabbit or ‐goat horseradish peroxidase‐labelled antibodies (Vector, Burlingame, CA, USA) for 30 min at room temperature. Immunostaining was visualized using diaminobenzidine (DAB) peroxidase‐chromogen reaction (ImmPACT DAB kit; Vector). Sections were counterstained with Mayer's haematoxylin (30 s), dehydrated, mounted using Histomount (Invitrogen) and visualized with a Nikon E800 microscope using spot software. To assess antigen markers quantitatively, Image J software (http://rsbweb.nih.gov/ij) was used to analyse histological sections of 3D model constructs. Briefly, images were converted into 16‐bit images and the threshold adjusted until it showed only the area of interest. This process resulted in the creation of a binary version of the image with only two‐pixel intensities: black = 0 and white = 255. The total area and number of pixels were then calculated. For each section, at least, five different fields were evaluated.
Preparation of supernatants for flow cytometry analysis
The amount of free and E. coli‐complexed SIgA in cell culture supernatants was determined by flow cytometry. Supernatants were harvested 3 h after addition of the stimuli (e.g. HS and SIgA) to the cultures and kept at −20°C until assayed. In these studies, uninfected cells (medium only) or cultures with IgG were used as controls. Briefly, supernatants were collected into 4·0 ml tubes, washed by centrifugation in PBS and stained with biotinylated mouse anti‐IgA1/IgA2 mAb (BD) and rabbit anti‐E. coli–IgG polyclonal antibody (Abcam). After an incubation of 45 min at 37°C, samples were washed twice with PBS and incubated for an additional 45 min at 37°C with streptavidin‐Pacific Orange (Invitrogen). After washing and fixation, specimens were analysed by flow cytometry on a custom LSR‐II instrument (BD Biosciences). Data were analysed with WinList version 7·0 (Verity Software House, Topsham, ME, USA).
Mucus and cytokine production
Levels of mucus in culture supernatants were measured by using a commercial mucin 2 (MUC2) enzyme‐linked immunosorbent assay (ELISA) (MyBioSource, San Diego, CA, USA). Levels of IL‐8 were measured by using a commercial IL‐8 ELISA (eBioscience, San Diego, CA, USA) or meso scale discovery (MSD, Gaithersburg, MD, USA) multiplexed assay. MSD was also used to measure levels of IL‐1β and TNF‐α. Supernatants were harvested up to 6 h after addition of the stimuli (e.g. HS and SIgA) to the cultures and kept at −20°C until assayed. In these studies, uninfected cells (medium only) or cultures with IgG were used as controls. ELISA and MSD were carried out following the manufacturer's instructions. The level of sensitivity of MUC2 and IL‐8 ELISAs were 0·3 ng/ml and 2·0 pg/ml, respectively. The levels of sensitivity for the various cytokines measured by MSD ranged from 0·3 to 2·5 pg/ml.
Western blot
The following primary anti‐human antibodies were used for Western blot analysis: (1) mouse anti‐MUC2 mAb (clone CCP58, 1 : 250; Imgenex, San Diego, CA, USA), (2) pIgR polyclonal antibody (1 : 500; R&D) and (3) rabbit anti‐glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) polyclonal antibody (1 : 000; Imgenex) (used as loading control). Whole cell lysates were prepared by adding the 3D model constructs into ready‐to‐use radioimmunoprecipitation assay (RIPA) buffer [50 mM Tris (pH 7·4), 150 mM NaCl, 1% Igepal (vol/vol), 1% sodium deoxycholate (wt/vol), 5 mM iodoacetamide, 0·1% sodium dodecyl sulphate (wt/vol)] (Sigma) containing a cocktail of protease and phosphatase inhibitors (Sigma). 3D model constructs were then mixed briefly using a vortex and incubated on ice for 30 min. After incubation, and to further disrupt the 3D model tissues, they were passed through a 21‐gauge needle and incubated for another 30 min on ice. The lysate was then clarified by centrifugation at 10 000 g for 15 min at 4°C. The supernatants were transferred to new microfuge tubes and the pellets discarded. The protein concentration of the cell lysates was determined by using a Thermo Scientific Pierce Micro BCA Protein Assay kit (Thermo Scientific, Rockford, IL, USA). Cell lysates (7 μg) were mixed with Laemmli buffer (Bio‐Rad, Hercules, CA, USA) containing 5% β‐mercaptoethanol (vol/vol), boiled for 5 min, separated on 4 and 20% sodium dodecyl sulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) gels and transferred to nitrocellulose membranes (Amersham Pharmacia Biotech, Amersham, UK). The membranes were blocked subsequently with 5% (wt/vol) non‐fat dry milk in PBS containing 0·1% Tween 20 (wt/vol) for 2 h at room temperature (RT), before exposure to primary antibodies overnight at 4°C (anti‐MUC2, ‐pIgR; or ‐GAPDH). The blots were then washed and incubated with a biotin‐labelled horse anti‐mouse, anti‐goat or anti‐rabbit IgG (1 : 600; Vector) for 1 h at RT. After washing, the blots were incubated with horseradish peroxidase (HRP) streptavidin (1 : 6000; Vector) for 1 h at RT. Signals were detected by chemiluminescence using Pierce ECL Plus Western Blotting Substrate kit (Thermo Scientific) using the manufacturer's instructions. Band intensity was normalized to GAPDH and quantitated by densitometry using Image J software.
Quantitative polymerase chain reaction (qPCR)
Isolation of total cellular RNA was performed using the RNeasy Mini Isolation Kit (Qiagen, Valencia, CA, USA) and RNA concentrations measured on a NanoDrop 1000 spectrophotometer. Thus, 1·0 μg of RNA was treated with DNase I (Qiagen) and reverse‐transcribed using RT2 first‐strand Reverse Transcription Kit (Qiagen). Amplified material was detected using RT2 SYBR® Green qPCR Mastermix (Qiagen). Real‐time quantitative PCR was performed on an ABI 7900HT thermocycler (Applied Biosystems, Foster City, CA, USA), with cycling conditions of 95°C for 10 min, 40 cycles of 95°C for 15 s and 60°C for 1 min. Analysis of results was performed using the GeneGlobe Data Analysis Center web‐based software (Qiagen). Relative gene expression data were calculated against GAPDH, HPRT1 and RPLP0 housekeeping genes and relative to the media only (negative control) by using the comparative 2(−delta delta Ct) method.
Statistical analysis
All statistical tests were performed using Prism software (version 5·02; GraphPad Software, La Jolla, CA, USA). Comparisons between groups were performed using one‐way analysis of variance (anova) tests. Newman–Keuls multiple comparison tests were used as post‐hoc tests. P‐values < 0·05 were considered significant.
Results
Assessment of E. coli–SIgA immune complex formation in the ‘lumen’‐like compartment of the 3D organotypical model
To demonstrate that our 3D organotypical model is a suitable tool for investigating the early events of host interaction between microbiota and SIgA, we first sought to examine the level of expression of IgA receptors on the surface of epithelial cells. Enterocytes express a receptor able to bind polymeric IgA, i.e. the classical polymeric immunoglobulin receptor (pIgR) 21. This receptor is responsible for the transcytosis of IgA from the lamina propria to the apical membrane of the epithelial cells 22. In the apical membrane a large part of the pIgR is cleaved, giving rise to a compound named secretory component (SC) that together with IgA is released into the intestinal lumen 22. The expression of the pIgR was evaluated in 3D model constructs cultured with E. coli or SIgA, or with E. coli and SIgA. Cultures with media alone (none) or with IgG were used as controls. We used a commercially purified SIgA that was derived from pooled human colostrum. HS, isolated originally from a healthy human volunteer 23, was used as a surrogate commensal. HS strongly colonize the human gastrointestinal tract [1010 colony‐forming units (CFU)/g faeces] with no symptoms of disease 24. After 6 h, 3D model constructs were collected and evaluated by immunochemistry for the detection of the pIgR. As has been shown for human intestine epithelium 21, we found that under basal conditions (none), the epithelial cells from the 3D model constructs exhibited pIgR expression comparable to those observed in the human intestine (Fig. 1a,b) 25. Interestingly, pIgR expression was increased in cultures in the presence of SIgA compared to both controls [i.e. media alone (none) and cultures with HS only] (Fig. 1a). Moreover, concomitant exposure to HS and SIgA induced a decrease in pIgR expression compared with cultures with SIgA only (Fig. 1a). These results were confirmed further by quantitative reverse transcription (qRT)–PCR and Western blot (Fig. 1c,d,e). Thus, these observations suggest the existence of a mechanism modulating pIgR expression that is dependent upon the presence of either SIgA alone or combined with HS.
Figure 1.
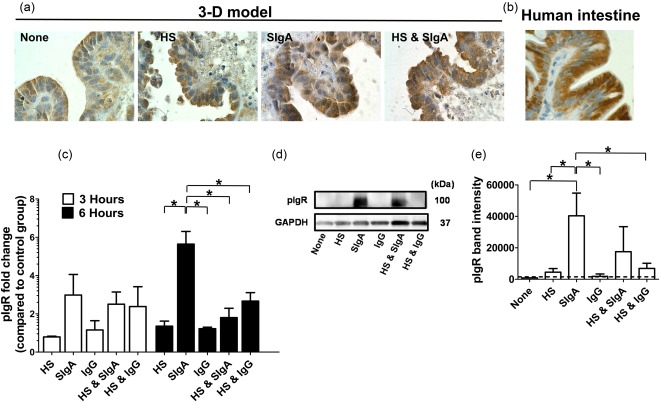
Expression of polymeric immunoglobulin receptor on epithelial cells from the 3‐dimensional (3D) organotypical model. 3D model constructs were cultured under different experimental conditions: (a) cultures with Escherichia coli strain HS (HS) or secretory immunoglobulin A (SIgA), (b) co‐cultures with HS and SIgA and (c) control groups of untreated cells (none) or culture with immunoglobulin (Ig)G. (a) After 6 h, 3D model constructs were collected and stained immunochemically for polymeric immunoglobulin receptor (pIgR). (b) Paraffin tissue section of human small intestine used as positive control. Dark brown indicates pIgR reactivity. Photographs are displayed as ×100 magnification. (c) Transcriptional analysis of pIgR regulation by quantitative reverse transcription–polymerase chain reaction (qRT–PCR). Values represent means ± standard error (s.e.) of three independent experiments with two replicates each. (d) pIgR expression determined in Western blots. (e) Values represent means ± s.e. of three independent Western blot experiments. Band density was normalized to glyceraldehyde 3‐phosphate dehydrogenase (GAPDH). The dashed lines represent the pIgR levels of the cells exposed to media only. *P‐values < 0·05.
We next sought to investigate whether HS‐SIgA immune complexes were indeed formed in the 3D model system. In the human gut, 24–74% of the microbiota is coated with SIgA 8, 9. Moreover, intestinal commensals, including E. coli, have been shown to be coated with SIgA from colostrum 26. This coating can be mediated by a mixture of specific and non‐specific (‘natural’) SIgA 5, 13. To determine whether a similar phenomenon is observed in the 3D model system, supernatants containing E. coli were collected from cultures with HS as well as cultures with HS and either SIgA, monomeric IgA or monomeric IgG, and stained using antibodies to E. coli antigens and IgA1/IgA2. Formation of immune complexes, defined as double staining for E. coli antigens and IgA1/IgA2, were analysed by flow cytometry. As shown in a representative experiment, we found that in supernatants collected from cultures with HS and SIgA the majority of the HS present were coated with SIgA (Fig. 2a,b). Of note, only negligible levels of immune complexes were observed in the control cultures, i.e. HS in the absence of SIgA or HS in presence of either human serum IgA or IgG (Fig. 2a,b). We next quantified bacterial attachment to the epithelial brush border in the presence or absence of human SIgA. The localization of HS was examined by immunohistochemistry using anti‐E. coli antibodies. We observed that in cultures with HS (i.e. no SIgA), the bacteria were rarely detected attached to the brush border (Fig. 2c, upper panel). In contrast, in cultures with HS and SIgA, HS was more abundant and detected easily (Fig. 2c, lower panel). Quantitative data showed that the presence of SIgA resulted in a fivefold increase in HS attachment to the brush border compared with cultures in the absence of SIgA (Fig. 2d). These results are in agreement with previous observations showing that SIgA can facilitate biofilm formation 26, 27, 28.
Figure 2.
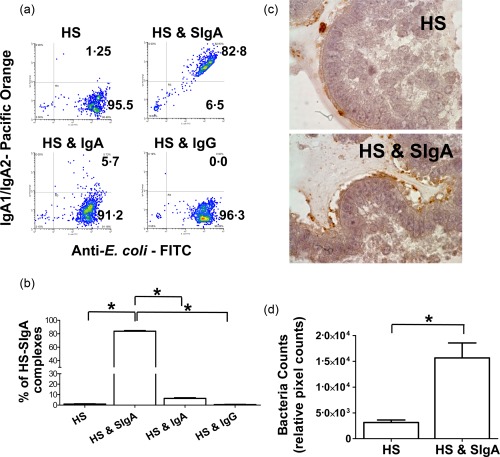
Formation of Escherichia coli–secretory immunoglobulin A (SIgA) immune complexes. (a) The presence of HS–SIgA immune complexes was detected by flow cytometry using 3‐dimensional (3D) model supernatants collected 3 h after stimulation. The following experimental conditions were evaluated: HS only or HS in the presence of either SIgA, serum IgA or serum IgG. (b) Shown is the overall % of HS–SIgA immune complexes. (c) Immunochemical staining for HS in cultures with HS only or in co‐culture with HS and SIgA. Photographs are displayed as ×100 magnification. (d) The bacteria counts on paraffin‐embedded sections are expressed as the relative pixel counts using image j software. The bars represent means ± standard error (s.e.). The data shown are representative of one of three experiments with two replicates each. *P‐values < 0·05.
Anti‐inflammatory effects of HS–SIgA immune complexes
Because previous studies have shown that SIgA can inhibit pathogen‐induced epithelial cell secretion of proinflammatory cytokines 14, we investigated whether SIgA could regulate inflammatory epithelial cell responses in the 3D model. We examined the secretion of IL‐1β, IL‐8 and TNF‐α in the 3D model supernatants 6 h after addition of the aforementioned stimuli. When compared with negative controls, exposure to HS alone resulted in increased IL‐1β, IL‐8 and TNF‐α secretion (Fig. 3a). Albeit at different levels, the exposure to SIgA alone also resulted in increases in IL‐8 and TNF‐α, but not IL‐1β secretion compared with negative controls. Interestingly, we observed that concomitant exposure to HS and SIgA induced a decrease in IL‐8 and TNF‐α but not IL‐1β secretion when compared to cultures with SIgA (Fig. 3a). Moreover, the addition of neutralizing antibodies specific to polymeric IgA receptors, the conventional pIgR, induced higher IL‐8 production in cultures with HS and SIgA (Fig. 3a). As controls for these responses, we examined IL‐8 secretion in cultures 3 and 6 h after addition of serum IgG only or serum IgG and HS. No increases in IL‐8 release were observed in cultures treated with IgG only (Fig. 3b). However, a synergetic effect was observed in cultures with HS and IgG. We observed that IL‐8 secretion in cultures with HS and IgG were significantly higher than HS alone (Fig. 3b). In conclusion, we hypothesize that the anti‐inflammatory activity observed in HS‐SIgA cultures was due, at least in part, to SIgA and involved the pIgR on epithelial cells.
Figure 3.

Escherichia coli–secretory immunoglobulin A (SIgA) immune complexes and the induction of proinflammatory cytokine production. (a) Supernatants were collected 6 h after stimulation with SIgA or E. coli strain HS (HS) or SIgA and HS in presence or absence of neutralizing anti‐polymeric immunoglobulin receptor (pIgR) antibodies. Cultures with media only were used as negative control. Levels of interleukin (IL)−1β, IL‐8 and tumour necrosis factor (TNF)‐α were measured by using a meso scale discovery (MSD) multiplexed assay. The data are representative of an average of eight independent experiments. (b) Supernatants were collected 3 and 6 h after stimulation with SIgA or HS, or SIgA and HS. Cultures with media only or immunoglobulin (Ig)G were used as controls. Levels of interleukin (IL)−8 were measured by enzyme‐linked immunosorbent assay (ELISA). The dashed line represents the cytokine levels of cells exposed to HS alone at 3 h after infection. Two independent experiments were performed with similar results. The data are representative of one experiment with two replicates. *P‐values < 0·05.
Mucus production
As described previously 15, epithelial cells in our organotypical model can differentiate into mucus‐secreting goblet cells (Fig. 4a). In fact, the mucus layer and SIgA operate together to confine bacteria to the intestinal lumen 29. Thus, it is possible that concomitant mechanisms by the mucus layer and SIgA can act in concert leading to the responses described above. To test this possibility, after 6 h of stimulation 3D model constructs were collected and evaluated by immunochemistry for the detection MUC2 mucin, the large glycoprotein stored in goblet cells. We observed that the epithelial cells exhibited MUC2 expression comparable to that observed in the human small intestine (Fig. 4a). Interestingly, as for pIgR, MUC2 expression was increased in cultures with SIgA compared to unstimulated (none) and cultures with HS only (Fig. 4a). Moreover, concomitant exposure to HS and SIgA induced a decrease in MUC2 expression compared with cultures with SIgA only (Fig. 4a). To evaluate these findings further, transcriptional analyses of MUC2 regulation were performed by qRT–PCR and Western blot assays. In both assays we observed that cultures with SIgA only had higher expression of MUC2 than cultures with HS or HS and SIgA (Fig. 4b,c,d). To assess the functionality of goblet cells we also measured the levels of MUC2 in the supernatants by ELISA. In agreement with the results described above, cultures in the presence of SIgA only had higher levels of MUC2 in the supernatants than cultures with HS or cultures with HS and SIgA (Fig. 4e). Moreover, the levels of MUC2 were similar between cultures with HS and cultures with HS plus SIgA. These results suggest that the MUC2 decreases triggered by the microbiota–SIgA immune complexes were directed mainly towards countering the effects of free SIgA. These results also demonstrate a correlation between the expression of pIgR and MUC2. The comparable expression pattern between MUC2 and pIgR shows an interconnection between these two markers.
Figure 4.
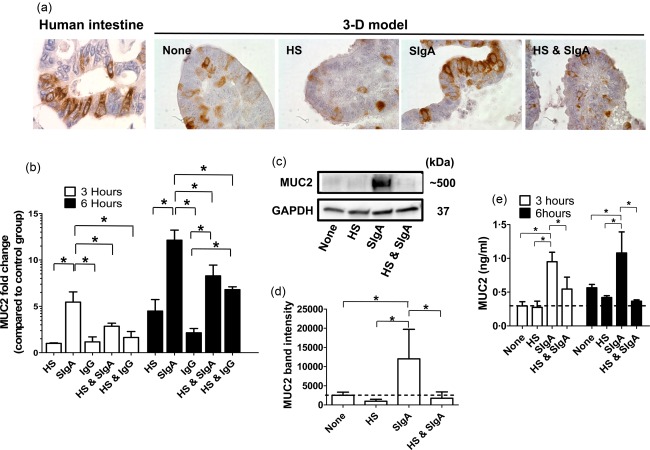
Effect of Escherichia coli–secretory immunoglobulin A (SIgA) immune complexes on mucus secretion. Three‐dimensional (3D) model constructs were collected after stimulation with HS or SIgA or E. coli strain HS (HS) and SIgA. Cultures with media (none) only or with immunoglobulin (Ig)G were used as controls. (a) Immunochemistry staining for mucin‐2 (MUC2) levels. Human small intestine (control) (left panel) and 3D model are shown at higher magnification (×100). (b) After 3 (open bars) and 6 h (closed bars) of stimulation, transcriptional analysis of MUC2 regulation was performed by quantitative reverse transcription–polymerase chain reaction (qRT–PCR). Values represent means ± standard error (s.e.) of three independent experiments with two replicates each. (c) After 6 h of stimulation, MUC2 protein expression was also detected by Western blot. (d) Values represent means ± s.e. of four independent Western blot experiments. Band density was normalized to glyceraldehyde 3‐phosphate dehydrogenase (GAPDH). (e) After 3 (open bars) and 6 h (closed bars) of stimulation, secretion of MUC2 into the supernatants was measured by enzyme‐linked immunosorbent assay (ELISA). The dashed lines represent the MUC2 levels of the cells exposed to media only (none). *P‐values < 0·05.
Discussion
Understanding of the homeostatic function of SIgA in the gut is critical to elucidate its role in health and disease. Mucosal SIgA comprises the bulk of antibodies in the human gut. An average of 3 g of SIgA is secreted daily into the intestinal tract 22. SIgA is capable of recognizing bacteria by different modes: ‘natural’ polyreactive (or cross‐reactive) and specific IgA 11. The SIgAs are also able to bind microbiota and promote a controlled antigen sampling of microbial antigens by M cells within the epithelial layer 12. Indeed, in the human gut, 24–74% of the microbiota is coated with SIgA 8, 9. SIgA also helps to reduce pathogen‐mediated proinflammatory responses by the epithelium 14. Despite the importance of these phenomena, little is known regarding how free and microbiota‐complexed SIgA regulate these epithelial cell responses.
Herein we report that, similar to in‐vivo studies 8, 9, a substantial proportion of E. coli present in the supernatants from 3D models were coated with SIgA. Like other groups, we speculated that this coating might be beneficial for the host 11. Bacterial coating by SIgA might provide containment of the microbiota as well as increased antigen sampling by antigen‐presenting cells in the gut microenvironment 11.
We also demonstrate that, as for pathogens, the formation of microbiota–SIgA complexes has a profound effect on inflammatory epithelial cell responses. We observed that microbiota‐complexed SIgA exerts anti‐inflammatory effects by down‐regulating the production of proinflammatory cytokines (e.g. IL‐8) rather than triggering them, as is the case in cultures with free SIgA. Furthermore, the levels of IL‐8 were similar between cultures with HS and cultures with HS plus SIgA, confirming that the anti‐inflammatory effects of the microbiota‐SIgA immune complexes are directed mainly towards countering the effects of free SIgA. These anti‐inflammatory effects were not present in cultures with E. coli and IgG. In contrast, IL‐8 secretion in cultures with HS and IgG were significantly higher than in cultures with either HS or IgG. We speculate that these increases were related to immune complex‐mediated inflammation (e.g. HS–IgG immune complexes), as described previously in other models 30. Further experiments are needed to confirm this hypothesis. We believe strongly that IL‐8 was produced mainly by epithelial cells, as incubation time of up to 6 h will not lead to stimulation and subsequent release of cytokines by the ECM‐embedded fibroblasts and endothelial cells into supernatants. The coefficient of diffusion of biomolecules in fibroblast‐containing contracted collagen gels is largely decreased compared with the solution (∼3 days for the cells in the vicinity) 31. However, one cannot exclude the participation, to a smaller extent, of other cell types such as macrophages in the secretion of these cytokines. Macrophages, when exposed to inflammatory stimuli, can secrete cytokines such as IL‐8 32. Indeed, by using immunochemical staining for macrophages (e.g. anti‐CD163 antibodies), we have observed the presence of monocyte‐differentiated macrophages beneath the epithelial layer of our 3D model (Supporting information, Fig. S1). Moreover, neutralization of pIgR enhanced production of proinflammatory cytokines by epithelial cells, suggesting a link between SIgA, pIgR and cytokine production. These results are in agreement with previous work showing that pIgR‐deficient mice are more prone to intestinal inflammation than wild‐type mice 33. Moreover, it has been shown that the expression of pIgR is up‐regulated by proinflammatory signalling genes such as IL‐8 34. Further investigations will be required to clarify whether this phenomenon is a function of (a) the level of SIgA coating per bacteria (functional affinity) and/or (b) the ability of SIgA to discriminate between the species/strain of commensal bacteria that stimulate inflammatory responses from those that do not. Indeed, previous studies using a mouse model of inflammatory bowel disease (IBD) have shown that high IgA coating, in contrast to low IgA coating, bind selectively to specific members of the mouse and human intestinal microbiota leading to exacerbation of intestinal inflammation 35. Previous studies have also shown that by using a Caco‐2 cell monolayer model to mimic the intestinal epithelium, the concomitant exposure to SIgA and either Lactobacillus rhamnosus or Bifidobacterium lactis probiotic strains resulted in enhanced levels of production of pIgR compared with cultures with probiotics or SIgA alone 36. The discrepancy between the latter studies and our observations is likely to be a result of the lack of mucus production in the Caco‐2 cell monolayer model. Here, we found comparable expression pattern between MUC2 and pIgR showing an interconnection between these two markers. This discrepancy also reinforces further the importance of having a multi‐cellular 3D system including fibroblasts, endothelial cells and immune cells to enable epithelial cell differentiation and function (i.e. goblet cells). In particular, the importance of cell–cell interactions in influencing intestinal cell expansion, survival and differentiation is well documented 37, 38, 39, 40, 41, 42.
Our hypothesis is that the microbiota‐SIgA complexes that are associated with the mucus provide bacteria containment preventing them from interacting directly with the host (e.g. neutralization, exclusion) leading to a decrease in IL‐8‐dependent pIgR expression. This hypothesis is supported by the observation that Salmonella O‐antigen‐specific SIgA prevents the interaction of the pathogen with the intestinal mucosa 43. Conversely, or alternatively, it is possible that SIgA‐bound E. coli may induce expression of different specific pattern recognition receptors and/or proinflammatory intracellular signalling than E. coli alone.
In conclusion, the above results open new avenues for future studies using systems involving a more complex microbiota to uncover signalling pathways affected by free SIgA versus microbiota‐complexed SIgA. These studies will be important in the establishment of effective therapeutic approaches for the treatment of inflammatory intestinal diseases; they may also provide important information on the specific nature of these interactions.
Author contributions
R. S.‐G. designed the study, performed the experiments, analysed the data and wrote the manuscript; F. S. performed the experiments and analysed the data; A. F. and M. B. S. contributed to the design and analysis of the data and wrote the manuscript.
Disclosure
The authors declare that the 3D model of the intestinal mucosa is covered by a US patent (number: US 9,200,258B2).
Supporting information
Additional Supporting information may be found in the online version of this article at the publisher's web‐site:
Fig. S1. Macrophages detection in 20 day‐cultured cells from the 3‐D model.
Acknowledgements
We are indebted to Dr Alan Cross for valuable critiques and Mrs Haiyan Chen, Regina Harley and Catherine Storrer for excellent technical assistance. The content is solely the responsibility of the authors and does not necessarily represent the official views of the sponsor, National Institute of Allergy And Infectious Diseases or the National Institutes of Health. This work was supported, in part, by NIAID, NIH, DHHS federal research grants R01 AI036525 and U19 AI082655 (CCHI) to MBS (RSG pilot project) and by NIDDK, NIH grant DK048373 to AF.
References
- 1. O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep 2006; 7:688–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Claus SP, Ellero SL, Berger B et al Colonization‐induced host–gut microbial metabolic interaction. MBio 2011; 2:e00271–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nicholson JK, Holmes E, Kinross J et al Host–gut microbiota metabolic interactions. Science 2012; 336:1262–7. [DOI] [PubMed] [Google Scholar]
- 4. Slack E, Balmer ML, Fritz JH, Hapfelmeier S. Functional flexibility of intestinal IgA – broadening the fine line. Front Immunol 2012; 3:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell‐independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 2000; 288:2222–6. [DOI] [PubMed] [Google Scholar]
- 6. Shroff KE, Cebra JJ. Development of mucosal humoral immune responses in germ‐free (GF) mice. Adv Exp Med Biol 1995; 371A:441–6. [DOI] [PubMed] [Google Scholar]
- 7. Cebra JJ, Periwal SB, Lee G, Lee F, Shroff KE. Development and maintenance of the gut‐associated lymphoid tissue (GALT): the roles of enteric bacteria and viruses. Dev Immunol 1998; 6:13–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tsuruta T, Inoue R, Nojima I, Tsukahara T, Hara H, Yajima T. The amount of secreted IgA may not determine the secretory IgA coating ratio of gastrointestinal bacteria. FEMS Immunol Med Microbiol 2009; 56:185–9. [DOI] [PubMed] [Google Scholar]
- 9. van der Waaij LA, Harmsen HJ, Madjipour M et al Bacterial population analysis of human colon and terminal ileum biopsies with 16S rRNA‐based fluorescent probes: commensal bacteria live in suspension and have no direct contact with epithelial cells. Inflamm Bowel Dis 2005; 11:865–71. [DOI] [PubMed] [Google Scholar]
- 10. Rol N, Favre L, Benyacoub J, Corthesy B. The role of secretory immunoglobulin A in the natural sensing of commensal bacteria by mouse Peyer's patch dendritic cells. J Biol Chem 2012; 287:40074–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brandtzaeg P. Secretory IgA: designed for anti‐microbial defense. Front Immunol 2013; 4:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Benckert J, Schmolka N, Kreschel C et al The majority of intestinal IgA+ and IgG+ plasmablasts in the human gut are antigen‐specific. J Clin Invest 2011; 121:1946–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mantis NJ, Rol N, Corthesy B. Secretory IgA's complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol 2011; 4:603–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boullier S, Tanguy M, Kadaoui KA et al Secretory IgA‐mediated neutralization of Shigella flexneri prevents intestinal tissue destruction by down‐regulating inflammatory circuits. J Immunol 2009; 183:5879–85. [DOI] [PubMed] [Google Scholar]
- 15. Salerno‐Goncalves R, Fasano A, Sztein MB. Engineering of a multicellular organotypic model of the human intestinal mucosa. Gastroenterology 2011; 141:e18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Salerno‐Goncalves R, Fasano A, Sztein MB. Development of a multicellular three‐dimensional organotypic model of the human intestinal mucosa grown under microgravity. Jove 2015; in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Corthesy B. Roundtrip ticket for secretory IgA: role in mucosal homeostasis? J Immunol 2007; 178:27–32. [DOI] [PubMed] [Google Scholar]
- 18. Eastburn DJ, Mostov KE. Laying the foundation for epithelia: insights into polarized basement membrane deposition. EMBO Rep 2010; 11:329–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Salerno‐Goncalves R, Pasetti MF, Sztein MB. Characterization of CD8(+) effector T cell responses in volunteers immunized with Salmonella enterica serovar typhi strain Ty21a typhoid vaccine. J Immunol 2002; 169:2196–203. [DOI] [PubMed] [Google Scholar]
- 20. Salerno‐Goncalves R, Rezwan T, Sztein MB. B cells modulate mucosal associated invariant T cell immune responses. Front Immunol 2014; 4:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mostov KE. Transepithelial transport of immunoglobulins. Annu Rev Immunol 1994; 12:63–84. [DOI] [PubMed] [Google Scholar]
- 22. Johansen FE, Kaetzel CS. Regulation of the polymeric immunoglobulin receptor and IgA transport: new advances in environmental factors that stimulate pIgR expression and its role in mucosal immunity. Mucosal Immunol 2011; 4:598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Formal SB, Dammin GJ, Labrec EH, Schneider H. Experimental Shigella infections: characteristics of a fatal infection produced in guinea pigs. J Bacteriol 1958; 75:604–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Levine MM, Bergquist EJ, Nalin DR et al Escherichia coli strains that cause diarrhoea but do not produce heat‐labile or heat‐stable enterotoxins and are non‐invasive. Lancet 1978; 1:1119–22. [DOI] [PubMed] [Google Scholar]
- 25. Uhlen M, Oksvold P, Fagerberg L et al Towards a knowledge‐based Human Protein Atlas. Nat Biotechnol 2010; 28:1248–50. [DOI] [PubMed] [Google Scholar]
- 26. Bollinger RR, Everett ML, Palestrant D, Love SD, Lin SS, Parker W. Human secretory immunoglobulin A may contribute to biofilm formation in the gut. Immunology 2003; 109:580–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bollinger RR, Everett ML, Wahl SD, Lee YH, Orndorff PE, Parker W. Secretory IgA and mucin‐mediated biofilm formation by environmental strains of Escherichia coli: role of type 1 pili. Mol Immunol 2006; 43:378–87. [DOI] [PubMed] [Google Scholar]
- 28. Palestrant D, Holzknecht ZE, Collins BH, Parker W, Miller SE, Bollinger RR. Microbial biofilms in the gut: visualization by electron microscopy and by acridine orange staining. Ultrastruct Pathol 2004; 28:23–7. [PubMed] [Google Scholar]
- 29. Hooper LV. Do symbiotic bacteria subvert host immunity? Nat Rev Microbiol 2009; 7:367–74. [DOI] [PubMed] [Google Scholar]
- 30. Li X, Kimberly RP. Targeting the Fc receptor in autoimmune disease. Expert Opin Ther Targets 2014; 18:335–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kihara T, Ito J, Miyake J. Measurement of biomolecular diffusion in extracellular matrix condensed by fibroblasts using fluorescence correlation spectroscopy. PLOS ONE 2013; 8:e82382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arango Duque G, Descoteaux A. Macrophage cytokines: involvement in immunity and infectious diseases. Front Immunol 2014; 5:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reikvam DH, Derrien M, Islam R et al Epithelial–microbial crosstalk in polymeric Ig receptor deficient mice. Eur J Immunol 2012; 42:2959–70. [DOI] [PubMed] [Google Scholar]
- 34. Bruno ME, Frantz AL, Rogier EW, Johansen FE, Kaetzel CS. Regulation of the polymeric immunoglobulin receptor by the classical and alternative NF‐kappaB pathways in intestinal epithelial cells. Mucosal Immunol 2011; 4:468–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Palm NW, de Zoete MR, Cullen TW et al Immunoglobulin a coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014; 158:1000–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mathias A, Duc M, Favre L, Benyacoub J, Blum S, Corthesy B. Potentiation of polarized intestinal Caco‐2 cell responsiveness to probiotics complexed with secretory IgA. J Biol Chem 2010; 285:33906–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Iliev ID, Spadoni I, Mileti E et al Human intestinal epithelial cells promote the differentiation of tolerogenic dendritic cells. Gut 2009; 58:1481–9. [DOI] [PubMed] [Google Scholar]
- 38. Kerneis S, Bogdanova A, Kraehenbuhl JP, Pringault E. Conversion by Peyer's patch lymphocytes of human enterocytes into M cells that transport bacteria. Science 1997; 277:949–52. [DOI] [PubMed] [Google Scholar]
- 39. Sato T, van Es JH, Snippert HJ et al Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011; 469:415–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lei NY, Jabaji Z, Wang J et al Intestinal subepithelial myofibroblasts support the growth of intestinal epithelial stem cells. PLOS ONE 2014; 9:e84651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mabbott NA, Donaldson DS, Ohno H, Williams IR, Mahajan A. Microfold (M) cells: important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol 2013; 6:666–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Voog J, Jones DL. Stem cells and the niche: a dynamic duo. Cell Stem Cell 2010; 6:103–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Endt K, Stecher B, Chaffron S et al The microbiota mediates pathogen clearance from the gut lumen after non‐typhoidal Salmonella diarrhea. PLOS Pathog 2010; 6:e1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting information may be found in the online version of this article at the publisher's web‐site:
Fig. S1. Macrophages detection in 20 day‐cultured cells from the 3‐D model.


