Abstract
Fibroblasts can be transdifferentiated directly into other somatic cells such as cardiomyocytes, hematopoietic cells, and neurons. An advantage of somatic cell differentiation without first generating induced pluripotent stem cells (iPSCs) is that it avoids contamination of the differentiated cells with residual iPSCs, which may cause teratoma. However, since primary fibroblasts from biopsy undergo senescence during repeated culture, it may be difficult to grow transdifferentiated cells in sufficient numbers for future therapeutic purposes. To circumvent this problem, we reversibly immortalized primary fibroblasts by using the piggyBac transposon to deliver the human telomerase reverse transcriptase (hTERT) gene hTERT plus SV40 Large T. Both approaches enabled fibroblasts to grow continuously without senescence, and neither caused teratoma formation in immunodeficient mice. However, fibroblasts immortalized with hTERT plus SV40 large T antigen accumulated chromosomal rearrangements, whereas fibroblasts immortalized with hTERT retained the normal karyotype. To transdifferentiate hTERT-immortalized fibroblasts into other somatic lineage cells, we transiently transfected them with episomal OCT4 and cultured them under neural cell growth condition with transposase to remove the transposon. Tripotent neural progenitor cells were seamlessly and efficiently generated. Thus, reversible immortalization of primary fibroblasts with hTERT will allow potential autologous cell-based therapeutics that bypass and simulate iPSC generation.
Introduction
Somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) with simple and powerful techniques. iPSCs may provide a wide range of terminal cell lineages for cell therapy and drug screening. However, for potential iPSC therapy, the somatic cells must first be reprogrammed into iPSCs and then differentiated into the appropriate cell type. This approach is time and labor intensive, and residual iPSCs may persist after differentiation and cause tumors after transplantation. These disadvantages can be avoided by transdifferentiation, in which one cell type is converted into another by ectopic expression of specific genes; for example, fibroblasts can be differentiated directly into neurons, cardiomyocytes, or beta cells [1–5]. However, primary somatic cells cannot be expanded indefinitely in vitro and thus cannot be produced in sufficient quantities for clinical transplantation, as a high cell density will be needed to establish functional coupling and signaling.
An alternative is to transiently immortalize somatic cells before transdifferentiation so that enough cells can be expanded for therapy. With this approach, immortalization genes such as those encoding human telomerase reverse transcriptase (hTERT), SV40 large T antigen (SV40LT), and Bmi-1 are expressed alone or in combination in primary cells and then removed typically with the Cre-loxP system [6–9]. However, a 34-bp fragment is left, which may disrupt the functions of other genes [6,8,10]. Using a piggyBac (PB) transposon to integrate and excise selectable marker genes leaves no residual sequence behind [11].
In this study, we used the PB transposon to reversibly immortalize primary fibroblasts and expand them in quantities sufficient for possible cell therapy. Fibroblasts transformed by hTERT or hTERT plus SV40LT proliferated for the duration of our studies at 140 days. Cells transformed with hTERT+SV40LT had chromosomal abnormalities and proliferated twice as fast as those transformed with hTERT, which retained the normal karyotype. Removal of hTERT by transfection with transposase abolished their proliferative potential. When fibroblasts immortalized with hTERT were transfected with episomal Oct4 and PB transposase, the transposon insert was seamlessly removed, and the cells could be directly converted into neural cells.
Materials and Methods
The Institutional Review Board of the California University, San Francisco (UCSF) School of Medicine and the UCSF Stem Cell Research Oversight Committee approved this study. The animal protocols were approved by the UCSF Animal Research Committee. A total of 20 NOD/SCID mice, ages 8 weeks were used for this study.
Plasmid construction
All restriction enzymes and Phusion high-fidelity DNA polymerase were from New England Biolabs. The plasmids 5′-PTK-3′ and hyperactive piggyBac transposase (PBase) were kind gifts from Allan Bradley (Wellcome Trust Sanger Institute, Hinxton, United Kingdom). Fragments containing the CMV early enhancer/chicken β actin (CAG) promoter were released from pCAGGS (kindly provided by Dr. Jun-ichi Miyazaki, Osaka University, Japan) by SalI and EcoRI digestion and ligated into SalI and EcoRI sites in pBluescript-SK (termed pSKCAG). A 2.5-kb fragment of hTERT was released from pBabe-hygro-hTERT with EcoRI and BamHI and ligated into pSKCAG (termed pSKCAGhT1).
The coding fragments of hTERT (hT2), enhanced green fluorescent protein (EGFP), and SV40LT were amplified from pBabe-hygro-hTERT, pEGFP, pEGFP-N3 (Invitrogen), respectively, and pSG5-LT with primers containing 2A self-cleaving peptide sequences (Supplementary Table S2) and subcloned into pGEM-T for sequencing. CAG fragments were released from the SalI and BamH I sites of pSKCAGhT and ligated into the XhoI and MfeI sites of 5′-PTK-3′ with hT2, which was released from pGEM-T-hT2 by BamH1 and EcoRI digestion. This vector was designated pT1. To generate the pT2 vector, the coding fragments of hT2, SV40LT, and EGFP were released from their individual T vectors and ligated together into the AscI and NotI sites of 5′-PTK-3′ with CAGhT1. pBabe-hygro-hTERT and pSG5-LT plasmids were from Addgene.
Cell culture
The fibroblasts we used in this study were from one donor given to us anonymously. and maintained in the fibroblast medium consisting of Dulbecco's modified Eagle's medium (DMEM) (Gibco), 10% fetal bovine serum (Hyclone), 1 mM l-glutamine (Gibco), 1% Pen-strep solution, and 1% nonessential amino acids (Gibco). Primary fibroblasts transduced with 2 μg of PB-based donor and 4 μg of PBase; after 10 days of puromycin selection, colonies were picked for expansion and analysis.
Generating tripotent neural progenitor cells from immortalized fibroblasts
hTERT fibroblasts (4 × 105) were transfected with 4 μg of episomal human Oct4 and 2 μg of PBx (Transposagen Bio) using Lonza primary fibroblasts transfection kits. Transfected cells were cultured in a reprogramming medium consisting of DMEM/F12, 10% knockout serum replacement (Gibco), 1 mM l-glutamine (Gibco), 1% Pen-strep solution, and 1% nonessential amino acids (Gibco) for 3 days and resuspended for culture in DMEM/F12 containing 20 μg of epidermal growth factor, 20 μg of basic fibroblast growth factor, 1× N2 medium, 1× B27 medium, 0.5 μM FIAU, 1% Pen-strep solution, and 1% nonessential amino acids for 10 days. Neural spheres were digested into single cells using 0.25% Trypsin, seeded on Matrigel, and cultured in DMEM/F12. These neural progenitor cells (NPCs) were then differentiated into three neural cell lineages as described [12].
Statistical analysis
All experiments were performed at least twice, and all samples were tested in triplicate. Data from each group are expressed as mean ± standard error. The Student's t-test was used for statistical analysis.
Results and Discussion
Conditional immortalization of fibroblasts with PB transposon
Traditionally, cells have been immortalized by expressing hTERT, Bmi, SV40LT, or oncogenes such as HPVE6/E7 or c-MYC. Ectopic expression of hTERT has been used to immortalize somatic cells without altering the normal karyotype [9]. The ability of SV40LT to immortalize somatic cells is largely dependent on its ability to complex with p53 [14]. Thus, SV40LT has become a popular gene to immortalize primary cells; the immortalized cells are generally not tumorigenic [14,17].
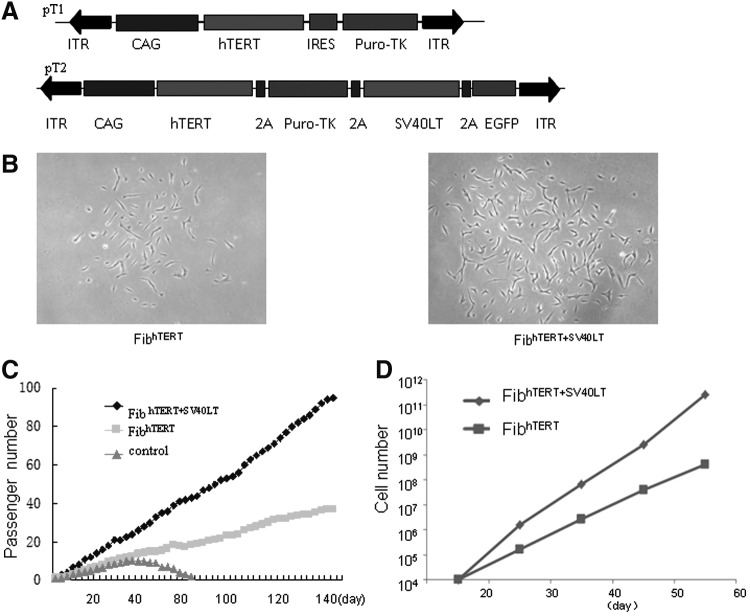
We inserted hTERT (driven by CAG promoter) and the puromycin-TK hybrid gene (Puro-TK), joined by an internal ribosome entry site, between the inverted terminal repeats of the construct pT1. In pT2, we inserted both these genes and SV40LT and EGFP; all four genes were joined by sequences encoding 2A peptides (Fig. 1A). The TTAA sequences on both ends of the inverted terminal repeats allow PB-mediated integration and seamless excision of the constructs from the integration sites [11]. Each construct was transfected in the presence of PBase into foreskin fibroblasts. To monitor transfection efficiency, 293T cells were transfected with pT2. Expression of EGFP was strong in 293T cells and weak in primary fibroblasts (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/scd). The cells were then cultured with puromycin for 2 weeks to select resistant colonies (Fig. 1B). PB integration was confirmed by using primers in the donor plasmid for polymerase chain reaction (PCR) amplification (Supplementary Fig. S2). Expression of the immortalized genes was confirmed by quantitative real-time PCR and western blot analysis (Supplementary Fig. S3A–C). Also, the immortalized cell lines from these two constructs had a strong telomerase activity, as shown by the TRAP (telomeric repeat amplification protocol) assay (Supplementary Fig. S3D). The primary fibroblasts grew slowly at first and became senescent after 30 days of culture. The pT1- and the pT2-derived fibroblasts both maintained a steady growth rate over 140 days of observation (Fig. 1C). Two individual cell lines immortalized by pT1 and pT2 vector were analyzed in the expansion phase. Both of them exhibited steady growth without senescence and their numbers increased by 105 to 106 in 50 days, respectively (Fig. 1D).
FIG. 1.
Generation of immortalized fibroblasts by PB-mediated gene transfer. (A) Structure of the two plasmid constructs for immortalization. pT1 contains hTERT, and pT2 contains hTERT, SV40LT, and EGFP; both have puro-TK for selection. (B) Colony morphology of fibroblasts reversibly immortalized with pT1 and pT2. (C) Growth curves of parental fibroblasts and cells immortalized with pT1 and pT2. Parental fibroblasts grew slowly and became senescent at passage 10, while pT1- and pT2-immortalized fibroblasts continued to proliferate, the latter at twice the rate of the former. (D) Number of immortalized fibroblast cell lines derived from pT1 and pT2 transfection from day 20 to 60 of culture. These cell lines grew exponentially for several months. EGFP, enhanced green fluorescent protein; hTERT, human telomerase reverse transcriptase; PB, piggyBac.
Karyotype, soft agar colony assay, and in vivo tumorigenicity
As shown by karyotype analysis at passage 55, fibroblasts immortalized with hTERT had normal diploid chromosomes, but fibroblasts immortalized with hTERT+SV40LT had abnormal karyotypes, including complex chromosome losses and multiple (>20) overlapped clonal abnormalities (Supplementary Fig. S4A). These chromosome defects were not surprising, as large T antigen often causes genomic instability and malignant transformation. Neither type of immortalized fibroblasts formed colonies in soft agar during 3 weeks of culture. In contrast, 293T cells did form colonies (Supplementary Fig. S4B). Three months after subcutaneous injection into NOD/SCID mice, neither hTERT nor hTERT/SV40LT-transduced cell lines gave rise to tumors, even though the latter had grossly abnormal karyotypes. As control, tumors were found in mice injected with 293T cells (Supplementary Table S1).
These findings show that fibroblasts can be immortalized by both constructs. However, the cell phenotypes were distinctly different. Cells transduced with SV40LT+hTERT proliferated rapidly, and led to round cells, aggregation, and absence of contact inhibition and anchorage-independency in the soft agar assay. In contrast, hTERT-transduced cells proliferated much more slowly and exhibited contact inhibition and anchorage-dependency. For further studies, we used only fibroblasthTERT with normal karyotypes.
Determination of integration sites mediated by PB transposon
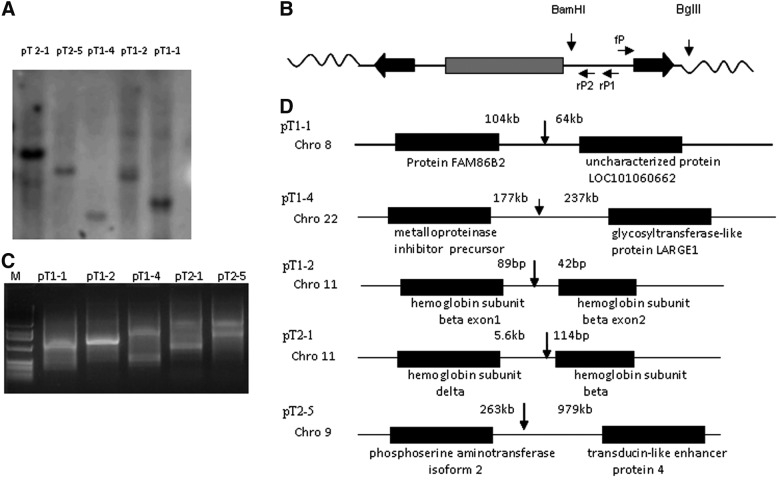
PB transposons can integrate into multiple chromosomal sites. This property has been used for gene discovery, as insertion in or near some genes disrupts their functions [18]. However, for seamless alteration of genes, cells with a single integration site are preferable because it would be difficult to excise all the insertions in cells with multiple integration sites. We used Southern blot to determine the number of PB integrations in the cell lines (Fig. 2A) and inverse PCR to determine the chromosomal location of the integration in cell lines with a single integration site (Fig. 2B). The PCR products were separated on agarose gels, the fragments isolated for sequencing (Fig. 2C), and BLAST searches used to align the insertion site to the NCBI human genomic DNA database. Analysis of the PB-mediated integration site bearing “TTAA” indicated that there were five integration sites, 2 on chromosome 11 and one each on chromosomes 8, 9, and 22 (Fig. 2D); four were in intergenic locations and one was in an intron. The 2 integration sites on chromosome 11 were at the β-globin cluster, one in the intron of the β-globin gene, and the other in the region between the δ- and the β-globin genes. Perhaps the PB we used preferentially recognizes and integrates in this area. For deimmortalization, we selected the immortalized fibroblasts with an integration site at chromosome 9, as that site is farthest from neighboring genes.
FIG. 2.
Determination of integration sites in immortalized cell lines by inverse PCR. (A) Southern blot analysis of transgenic cassettes in several immortalized lines with a puromycin probe. pT1-2 and pT2-1 denote different clones derived from transfection by pT1 and pT2, respectively. (B) Strategy for identifying integration sites using inverse PCR. The forward primer fP and two reverse primers rP1 and rP2 were designed and listed in the Supplementary Table S1. (C) Inverse PCR. Genomic DNA (1 μg) from immortalized cells was digested with BamH1 and BglII digestion, ligated, and amplified with primers fP and rP1; the products were further enriched by amplification with primers fP and rP2 and cloned into pGEM-T. (D) Five chromosomal DNA locations flanking the inserted PB transposon were identified by sequencing the PCR products. Four of the integration sites were intergenic, and one was in the intron of the β-globin gene. The distances from the next coding sequences on either side are noted. PCR, polymerase chain reaction.
Reversal of immortalization of fibroblasts after PBase transfection
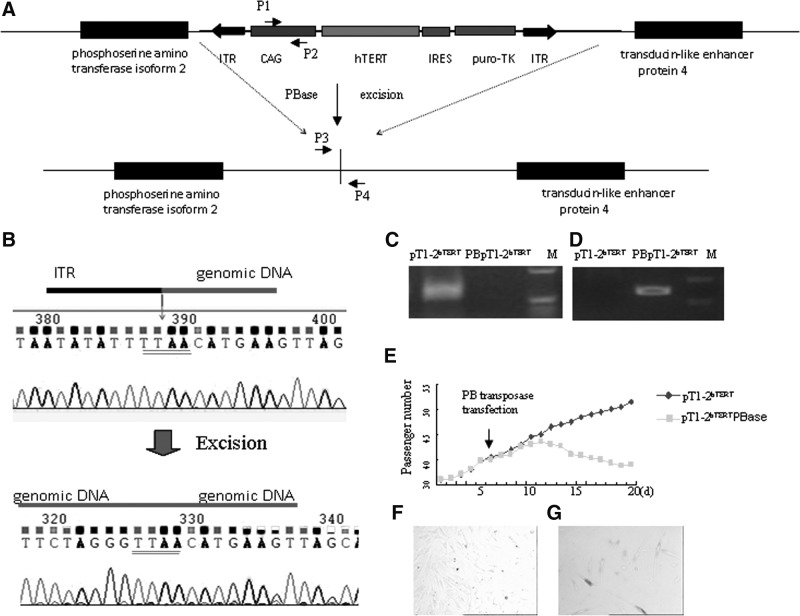
We sought to evaluate whether the immortalization-related phenotype of the fibroblasts could be reversed after removing the transgene integrated at the TTAA sites between two inverse terminals repeats. One hTERT-transduced cell line was expanded after passage 35, transfected with PBase, and negatively selected with 0.5 μM FIAU for 7 days. Analysis of half of the surviving cells by PCR with primers designed to amplify the CAG in the cassette or at the vector–genome junction confirmed that the selection cassette had been removed (Fig. 3A–D). Sequencing of the PCR product with primers flanking the integration site showed seamless replacement of the vector–genome junction with the normal genomic sequence (Fig. 3B). The remainders of the surviving cells were analyzed for proliferation rate. PBase-transduced FIAU-resistant fibroblasts with the PB insert removed had significantly decreased proliferation and quickly became senescent showing positive SA-β gal expression in days (Fig. 3E–G). Thus, the immortalized fibroblasts had a long-term high proliferative activity that was reversible.
FIG. 3.
Reversal of cell immortalization after excision of PB with transposase. (A) Schematic representation of the excision of the PB insert. (B) Sequencing before and after removal of the PB cassette. Top sequence shows the junction of the PB ITR and genomic sequence using primer P1/P2 before excision. Bottom shows the genomic sequence after excision amplified by P3-P4. Note the TTAA sequence for insertion and seamless removal characteristic of the PB system. (C) Removal of PB transposon cassette was confirmed by absence of CAG sequence amplification using the primer1 and primer2 (P1 and P2). The cell line pT1-2hTERT was analyzed side by side with deimmortalized pT1-2hTERT, as control. (D) Removal confirmed by positive amplification with prime 3 and 4 (P3 and P4). The presence of the cassette would preclude amplification under the PCR condition used. (E) Growth curve of deimmortalized pT1-2hTERT. The long-term and enhanced proliferation was reversed after hTERT was removed. Immortalized (F) and deimmortalized fibroblasts (G) were demonstrated positive with the senescence-associated beta-galactosidase (SA-βgal) activity.
Direct conversion of immortalized fibroblasts to PB-free NPCs
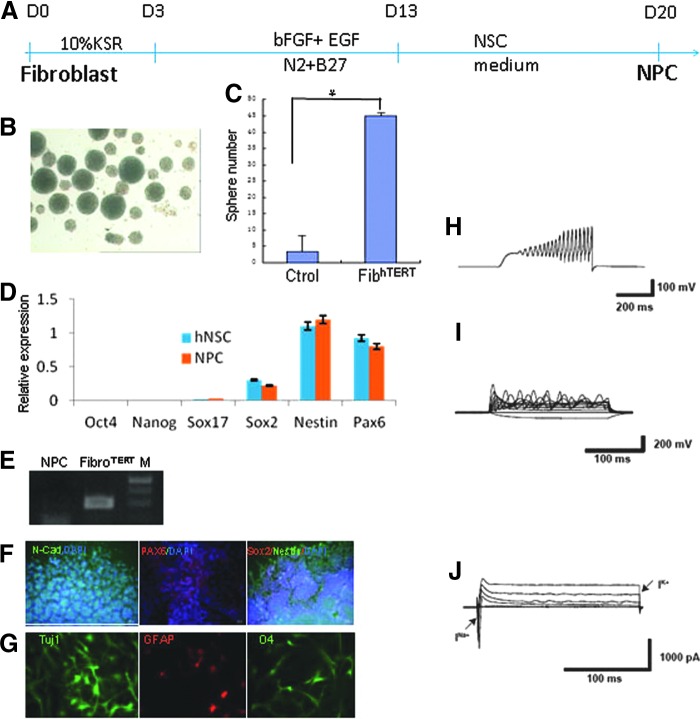
Fibroblasts have been converted into NPCs with a single transcription factor, human Oct4, delivered by lentivirus [12]. To determine whether hTERT-immortalized fibroblasts can be transdifferentiated into NPCs, we transfected hTERT fibroblasts (passage 20) and control fibroblasts (passage 10) with an episomal plasmid encoding human Oct4 and a plasmid encoding PBase, cultured the cells under neural transdifferentiation conditions for 10 days (Fig. 4A), and transferred them to the NPC medium. After 1 week, neural sphere-like cells were detected (Fig. 4B). The immortalized cells generated 10-fold more spheres than the controls (Fig. 4C). After the neural spheres were dissociated and cultured on the Matrigel for 1 week, the cells stained positively for the NPC markers N-cadherin, PAX6, and nestin (Fig. 4D, F, G). PCR analysis of cells cultured with FIAU (negative selection) confirmed the removal of the hTERT cassette (Fig. 4E).
FIG. 4.
Direct generation of NPC from immortalized fibroblast. (A) Schematic representation of generation of NPC from immortalized fibroblast. (B) Phase contrast image of neural sphere converted from immortalized fibroblast. (C) Comparison of the sphere number from immortalized group (P20) and control group (P10; data represent mean ± SEM, N = 3; *P < 0.05). (D) Quantitative RT-PCR analysis of pluripotent markers (Nanog and Oct4), endoderm marker (Sox17), and the neural stem cell-specific markers (Sox2, Nestin, and Pax6). Data represent mean ± SEM, N = 3. (E) Removal of PB transposon cassette was confirmed by absence of CAG sequence amplification using the primers P1 and P2 shown in Figure 3. As control, the fibroblasthTERT was analyzed. (F) Characterization of fibroblast-derived NPC by immunocytochemistry by staining the markers of N-Cad, Nestin, and PAX6. (G) Immunostaining of astrocyte (GFAP), neuron cell (β-Tubulin), and oligodendrocyte (O4) in further differentiated neural lineage cells. Neurophysiology of 13 neurons was recorded and 9 of them (69.2%) showed action potential firing (H), fast sodium currents (I), and outward potassium currents (J). NPC, neural progenitor cell.
To further differentiate the NPCs, we removed the growth factors and used culture protocols reported to differentiate NPC into different lineages. After 3 weeks of differentiation, neurons, astrocytes, and oligodendrocytes were detected (Fig. 4G). To characterize the neurons, patch clamp recording was used to examine their electrophysiological properties. Totally 13 neurons were recorded and 9 of them (69.2%) showed action potential firing, fast sodium currents, and outward potassium currents (Fig. 4H–J). Those NPCs cultured for only 1 week were used as controls. For control cells, none of them (n = 6) showed action potential firing or sodium currents.
In summary, we reversibly immortalized primary fibroblasts by using the PB transposon to deliver hTERT and as an example, directly transdifferentiated them into neural cells. This strategy will make it possible to generate large numbers of primary cells from small clinical biopsies for gene editing or making other modifications for therapeutic purposes. The cells can then be directly transdifferentiated into other somatic cells, thereby bypassing the potentially tumorigenic pluripotent state.
Supplementary Material
Acknowledgments
We thank Changsheng Lin at the Gladstone Institutes for kindly providing cDNAs from hNSC. This work was supp by NIH grant P01-DK088760.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S, Lipton SA, Zhang K, and Ding S. (2011). Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci U S A 108:7838–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang H, Cao N, Spencer CI, Nie B, Ma T, Xu T, Zhang Y, Wang X, Srivastava D, and Ding S. (2014). Small molecules enable cardiac reprogramming of mouse fibroblasts with a single factor, Oct4. Cell Rep 6:951–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li K, Zhu S, Russ HA, Xu S, Xu T, Zhang Y, Ma T, Hebrok M, and Ding S. (2014). Small molecules facilitate the reprogramming of mouse fibroblasts into pancreatic lineages. Cell Stem Cell 14:228–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li WL, Li K, Wei WG, and Ding S. (2013). Chemical approaches to stem cell biology and therapeutics. Cell Stem Cell 3:270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu T, Zhang M, Laurent T, Xie M, and Ding S. (2013). Concise review: chemical approaches for modulating lineage-specific stem cells and progenitors. Stem Cells Transl Med 2:355–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Westerman KA, and Leboulch P. (1996). Reversible immortalization of mammalian cells mediated by retroviral transfer and site-specific recombination. Proc Natl Acad Sci U S A 93:8971–8976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salmon P, Oberholzer J, Occhiodoro T, Morel P, Lou J, and Trono D. (2000). Reversible immortalization of human primary cells by lentivector-mediated transfer of specific genes. Mol Ther 2:404–414 [DOI] [PubMed] [Google Scholar]
- 8.Kowolik CM1, Liang S, Yu Y, and Yee JK. (2004). Cre-mediated reversible immortalization of human renal proximal tubular epithelial cells. Oncogene 23:5950–5957 [DOI] [PubMed] [Google Scholar]
- 9.Zhang ML, Tong XJ, Fu XH, Zhou BO, Wang J, Liao XH, Li QJ, Shen N, Ding J, and Zhou JQ. (2010). Yeast telomerase subunit Est1p has guanine quadruplex-promoting activity that is required for telomere elongation. Nat Struct Mol Biol 17:202–209 [DOI] [PubMed] [Google Scholar]
- 10.Xie F, Ma Q, Jiang S, Ren Z, Wang J, Huang S, Zeng F, and Zeng Y. (2012). Adjusting the attB site in donor plasmid improves the efficiency of ΦC31 integrase system. DNA Cell Biol 31:1335–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie F, Ye L, Chang JC, Beyer AI, Wang J, Muench MO, and Kan YW. (2014). Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res 24:1526–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell R, Szabo E, Shapovalova Z, Aslostovar L, Makondo K, and Bhatia M. (2014). Molecular evidence for OCT4-induced plasticity in adult human fibroblasts required for direct cell fate conversion to lineage specific progenitors. Stem Cells 32:2178–2187 [DOI] [PubMed] [Google Scholar]
- 13.Saito M, Handa K, Kiyono T, Hattori S, Yokoi T, Tsubakimoto T, Harada H, Noguchi T, Toyoda M, Sato S, and Teranaka T. (2005). Immortalization of cementoblast progenitor cells with Bmi-1 and TERT. J Bone Miner Res 20:50–57 [DOI] [PubMed] [Google Scholar]
- 14.Zhu JY, Abate M, Rice PW, and Cole CN. (1991). The ability of simian virus 40 large T antigen to immortalize primary mouse embryo fibroblasts cosegregates with its ability to bind to p53. J Virol 65:6872–6880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hawley-Nelson P1, Vousden KH, Hubbert NL, Lowy DR, and Schiller JT. (1989). HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J 8:3905–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gil J, Kerai P, Lleonart M, Bernard D, Cigudosa JC, Peters G, Carnero A, and Beach D. (2005). Immortalization of primary human prostate epithelial cells by c-Myc. Cancer Res 65:2179–2185 [DOI] [PubMed] [Google Scholar]
- 17.Ozer HL, Banga SS, Dasgupta T, Houghton J, Hubbard K, Jha KK, Kim SH, Lenahan M, Pang Z, Pardinas JR, and Patsalis PC. (1996). SV40-mediated immortalization of human fibroblasts. Exp Gerontol 31:303–310 [DOI] [PubMed] [Google Scholar]
- 18.Wang W, Bradley A, and Huang Y. (2009). A piggyBac transposon-based genome wide library of insertionally mutated Blm-deficient murine ES cells. Genome Res 19:667–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.