Abstract
Antibiotic administration can disrupt the intestinal microbiota and down-regulate innate immune defenses, compromising colonization resistance against orally acquired bacterial pathogens. Vancomycin-resistant Enterococcus faecium (VRE), a major cause of antibiotic-resistant infections in hospitalized patients, thrives in the intestine when colonization resistance is compromised, achieving extremely high densities that can lead to bloodstream invasion and sepsis. Viral infections, by mechanisms that remain incompletely defined, can stimulate resistance against invading bacterial pathogens. We report that murine norovirus infection reduces the density of VRE in the intestinal tract of mice with antibiotic-induced loss of colonization resistance. Resiquimod (R848), a synthetic ligand for Toll-like receptor 7 (TLR-7) that stimulates antiviral innate immune defenses, restores expression of the antimicrobial peptide Reg3γ and reestablishes colonization resistance against VRE in antibiotic-treated mice. Orally administered R848 triggers TLR-7 on CD11c+ dendritic cells, inducing interleukin-23 (IL-23) expression followed by a burst of IL-22 secretion by innate lymphoid cells, leading to Reg3γ expression and restoration of colonization resistance against VRE. Our findings reveal that an orally bioavailable TLR-7 ligand that stimulates innate antiviral immune defenses in the intestine restores colonization resistance against a highly antibiotic-resistant bacterial pathogen.
INTRODUCTION
The emergence of antibiotic resistance among bacterial pathogens is outpacing the development of antibiotics that circumvent resistance mechanisms (1, 2). Enterococcus faecalis and/or Enterococcus faecium are nearly ubiquitous inhabitants of the human intestine but, in terms of frequency, minor contributors to the intestinal microbiota of antibiotic-naïve individuals (3). In the past two decades, increasing vancomycin resistance among enterococci has become a challenge in clinical settings where these bacterial strains cause systemic infections, leading the Centers for Disease Prevention and Control to categorize vancomycin-resistant E. faecium (VRE) as a serious public health threat (1, 4, 5). VRE expands to very high densities in the gastrointestinal tract of patients after antibiotic-induced disruption of microbiota-mediated colonization resistance (6, 7). Intestinal domination by bacterial species, such as VRE, precedes bacteremia and leads to poor outcomes in patients (8–11).
Commensal bacteria can directly inhibit colonization by potentially invasive pathogens by consuming limited nutrients or secreting bactericidal or bacteriostatic factors (12, 13), or indirectly mediate colonization resistance by stimulating host-derived immune defense mechanisms (12, 13). Indirect colonization resistance is driven by commensal members of the microbiota signaling through innate immune receptors such as Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain receptors to tonically stimulate immune defense pathways leading to expression of antimicrobial proteins, such as the antibacterial C-type lectin regenerating islet-derived 3γ (Reg3γ), a murine ortholog of human HIP/PAP (14, 15). Antibiotic-mediated disruption of the intestinal microbiota results in loss of direct and indirect colonization resistance, increasing host susceptibility to intestinal damage and systemic infections caused by VRE and other antibiotic-resistant pathogens (9, 16–19). Whereas reconstitution of the microbiota by fecal transplantation from antibiotic-naïve donors has cured patients of recurrent Clostridium difficile infections (20), the complexities and risks of fecal transplantation limit its therapeutic potential as a treatment for VRE elimination from gastrointestinal tract (21).
In addition to diverse populations of commensal bacterial species, the mammalian intestinal tract also hosts a broad variety of viruses that comprise the host’s microvirome (22, 23). Immune stimulation by viruses induces antibacterial defense mechanisms (24), suggesting that tonic signaling by viruses or virus-derived molecules could augment microbiota-driven colonization resistance. For example, enteric infection with murine norovirus (MNV) limits colonic inflammation after Citrobacter rodentium infection (25), and reovirus infection of the small intestines of neonatal mice induces production of interleukin-22 (IL-22), a cytokine that induces expression of Reg3γ by intestinal epithelial cells (26). However, specific viral-derived ligands that drive antibacterial defenses have not been identified, and thus, it remains unclear whether such ligands can enhance immune-mediated colonization resistance.
Here, we show that MNV infection partially restored anti-bacterial immune defense in antibiotic-treated mice and vital titers inversely correlated with the density of intestinal VRE colonization. Oral administration of resiquimod (R848), a synthetic molecule that mimics virus-derived single-stranded RNA and stimulates TLR-7 and TLR-8 (27), reestablished colonization resistance in antibiotic-treated mice and markedly reduced intestinal colonization with VRE. R848 acted through TLR-7–expressing CD11c+ dendritic cells and IL-23 to induce IL-22, leading to the induction of Reg3γ. Our studies demonstrate that imidazoquinolines used for the treatment of clinical papilloma virus infections (28–31) also enhance colonization resistance against an antibiotic-resistant bacterial pathogen.
RESULTS
Enteric viral infection reduces intestinal VRE colonization
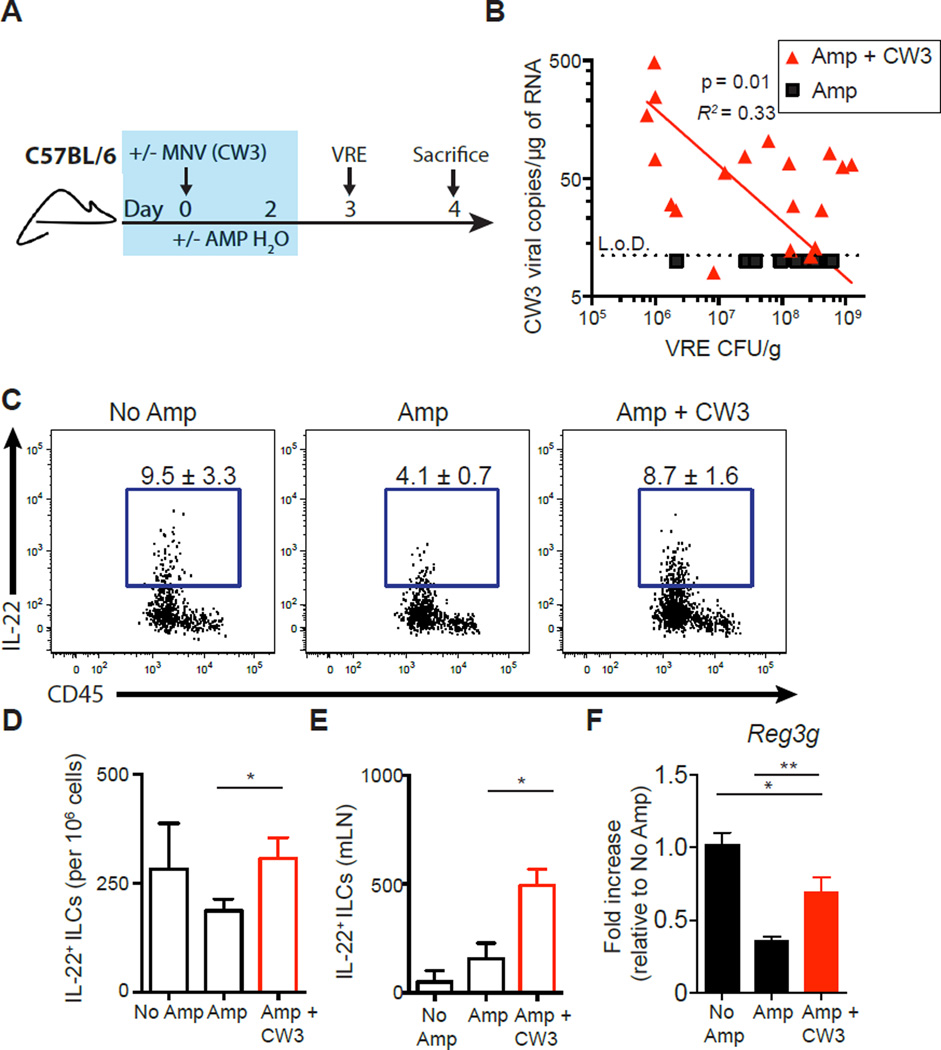
Antibiotic-mediated loss of colonization resistance enables VRE to undergo a marked expansion in the gut (6, 7). Although viral infection and consequent stimulation of the immune system can enhance defense against bacterial pathogens (24, 25), it is unclear whether enteric viruses can restrict expansion of VRE after antibiotic treatment. To address this question, ampicillin-treated C57BL/6 mice were inoculated with either MNV (MNV-CW3 strain) or vehicle control, and 3 days after viral infection, the mice were inoculated with ~106 colony-forming units (CFUs) of VRE via oral gavage (Fig. 1A). Ampicillin administration depletes indigenous intestinal bacterial communities but does not inhibit VRE growth (fig. S1A) (32). As previously reported (32), ampicillin-treated mice became densely colonized with VRE, whereas colonization resistance in ampicillin-naïve control mice prevented intestinal VRE colonization (Fig. 1B and fig. S1B). Infection of ampicillin-treated mice with MNV-CW3 modestly reduced VRE density in the ileum (fig. S1B), with the VRE burden inversely correlated with MNV-CW3 titers (Fig. 1B), although inhibition of VRE in the MNV-CW3 group did not reach statistical significance (fig. S1B). Ampicillin treatment reduced expression of IL-22 in innate lymphoid cells (ILCs) isolated from the Peyer’s patches (PPs), and MNV-CW3 infection restored IL-22+ ILCs to levels comparable to ampicillin-naïve control mice (Fig. 1, C and D, and fig. S1C). The total number of IL-22+ ILCs recovered from the mesenteric lymph nodes (mLNs) of MNV-CW3–infected mice increased compared to ampicillin-treated mice (Fig. 1E). Furthermore, MNV-CW3 infection partially restored the expression of Reg3g mRNA transcripts in the ileum, although induction of Reg3g did not reach levels observed in ampicillin-naïve mice (Fig. 1F). Partical restoration of the IL-22/Reg3g axis in ampicillin-treated mice by MNV-CW3 infection suggests that viral stimulation can recalibrate intestinal immune defenses after antibiotic-mediated loss of colonization resistance.
Fig. 1. Norovirus infection inversely correlates with VRE colonization in ampicillin-treated mice.
(A) Schematic for antibiotic treatment, norovirus (MNV-CW3) infection, and VRE infection. Blue box represents the time mice are receiving ampicillin water. (B) Comparison of detectable viral copies of MNV-CW3 in ileal tissue versus VRE CFU in the ileal content. L.o.D., limit of detection. Red line represents log-log nonlinear regression fit for Amp + CW3 (n = 19). (C to E) Cells isolated from the PPs or mLNs of non–ampicillin-treated, VRE-infected mice (No Amp), ampicillin-treated, VRE-inoculated mice (Amp), and ampicillin-treated, MNV-CW3/VRE-coinfected (Amp + CW3) mice were incubated in medium in the presence of brefeldin A (BFA) for 3 hours and assessed for IL-22 production. Flow cytometric plots gated on live, CD45+, non-T non-B, Gr-1neg CD90+ CD127+ cells. Frequency (C) and number (D) of IL-22+ ILCs in the PPs. Individual mice pooled into treatment groups. Data are sum of four independent experiments (n = 7 to 12 pooled samples per group). (E) Number of IL-22+ ILCs in the mLNs. Data are sum of three independent experiments (n = 6 to 10 mice per group). (F) Reg3g gene expression quantified by real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) and displayed as fold increase over No Amp mice and normalized to Hprt. Data are sum of four independent experiments (n = 15 to 18 mice per group) (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, t test). Data are means ± SEM.
Loss of commensal bacteria can impair the establishment of enteric viral infections (33, 34) and may explain why some ampicillin-treated mice inoculated with MNV-CW3 had low viral titers in the ileum and elevated VRE density (Fig. 1B). Indeed, it has been reported that MNV-infected, antibiotic-treated, and germ-free mice have substantially lower viral titers than conventional mice (25, 35). To avoid variability caused by poor viral infectivity, we sought to identify a specific viral ligand that could be administered to faithfully activate intestinal immune defenses and provide colonization resistance against antibiotic-resistant pathogens.
The TLR-7 ligand R848 reestablishes Reg3γ expression in antibiotic-treated mice
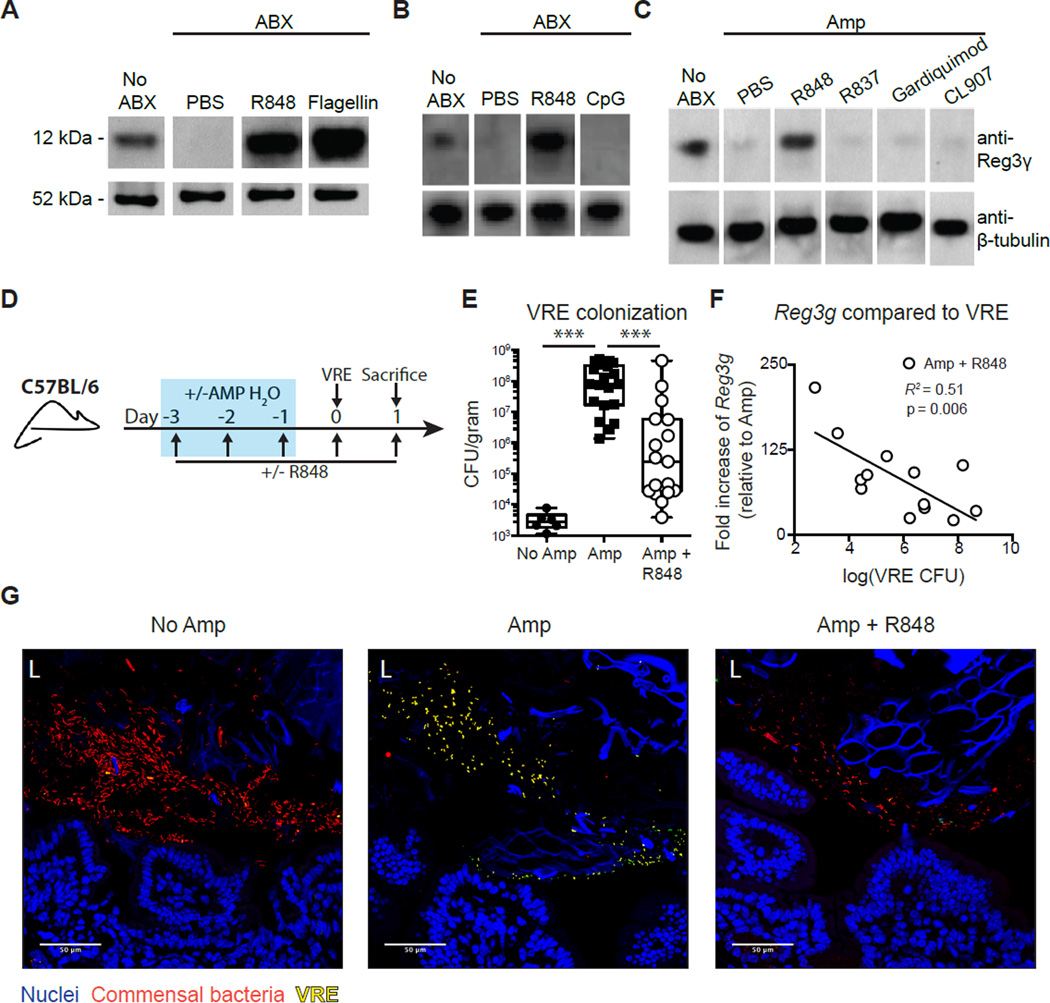
Our previous work demonstrated that administration of bacterial lipopolysaccharide (LPS) or flagellin to antibiotic-treated mice inhibits VRE expansion by stimulating the TLR-4 or TLR-5 signaling pathway, leading to the induction of Reg3γ (36, 37). TLR-7, an intracellular innate immune receptor, detects short, single-stranded RNA motifs derived from RNA viruses such as norovirus and also signals via the MyD88 adaptor protein (38). MNV-CW3 infection significantly elevated expression of Tlr7 mRNA transcripts in the ileum compared to both ampicillin-treated and ampicillin-naïve mice (fig. S1D). To investigate whether engagement of TLR-7 with virus-derived molecules could protect mice against VRE, C57BL/6 mice were treated with R848 orally, flagellin intraperitoneally, or phosphate-buffered saline (PBS) orally for 3 days while receiving antibiotic treatment. To allow direct comparisons of data with our previous studies (36, 37), a broad-spectrum antibiotic cocktail of metronidazole, neomycin, and vancomycin (ABX) was used to deplete intestinal bacterial communities. Upon verification of consistency between studies, ampicillin treatment alone was used to disrupt colonization resistance in mice. Reg3γ expression was measured in the ileum of untreated and antibiotic-treated mice after administration of R848 or flagellin. As previously described (37), antibiotic treatment reduced Reg3γ levels to nearly undetectable levels and intraperitoneal flagellin induced Reg3γ expression in the ileum of antibiotic-treated mice (Fig. 2A). R848 treatment also induced Reg3γ expression in antibiotic-treated mice to levels equal to or greater than those observed in untreated control mice (Fig. 2A and fig. S2A). In contrast, oral administration of CpG, a single-stranded DNA molecule that binds TLR-9, did not induce Reg3γ in the ileum of antibiotic-treated mice (Fig. 2B). Oral administration of other TLR-7/8 agonists, including R837, Gardiquimod, and CL907, also did not restore Reg3γ expression in the ileum (Fig. 2C), suggesting an ability of R848 to remain immunologically active after oral administration.
Fig. 2. R848 restores Reg3γ expression and colonization resistance against VRE.
(A to C) Mice were treated with ampicillin (Amp) or ABX in their drinking water for 7 days and administered daily for 3 days (days 4 to 6 of antibiotic administration) (A) R848 (50 µg) orally or flagellin (15 µg) intraperitoneally, (B) R848 (50 µg) or CpG (100 µg) orally, or (C) R848, R837, Gardiquimod, or CL907 (50 µg) orally. Protein extracts from the ileum were analyzed by Western blotting using Reg3γ-specific antiserum. Data are representative of two independent experiments (n = 3 mice per group). (D) Schematic for antibiotic treatment, R848 treatment, and VRE infection. Blue box represents the time mice are receiving ampicillin water. (E) VRE CFUs in the ileal content of non–ampicillin-treated, (No Amp), ampicillin-treated (Amp), and ampicillin + R848–treated (Amp + R848) mice 24 hours after VRE infection. Data are sum of five independent experiments (***P ≤ 0.0001, Mann-Whitney test; n = 17 to 20). (F) Comparison of Reg3g expression in the ileum of Amp + R848–treated mice versus VRE CFU in the ileal content. Reg3g expression relative to Amp mice and normalized to Hprt. Black line represents best-fit regression line for Amp + R848 mice. (G) Bacterial fluorescence in situ hybridization (FISH) staining of ileal sections from No Amp, Amp, and Amp + R848 mice. L, lumen; blue, Hoechst; red, universal 16S bacterial probe; green, VRE-specific probe.
To determine whether R848 is stimulating intestinal epithelial or bone marrow–derived cells, we generated bone marrow chimeric (BMC) mice by reconstituting lethally irradiated C57BL/6 mice with wild-type C57BL/6, Tlr7−/−, or Myd88−/− bone marrow. BMC mice were treated with antibiotics and R848 or PBS, and Reg3g mRNA expression was determined in the ileum. Whereas R848 treatment of C57BL/6 BMC mice induced expression of Reg3g mRNA, R848 administration to Myd88−/− (fig. S2B) and Tlr7−/− (fig. S2C) BMC mice did not induce Reg3g expression. These results indicate that R848 induces intestinal epithelial Reg3γ expression by stimulating the TLR-7/MyD88 pathway in bone marrow–derived, but not epithelial or stromal, cells.
The TLR-7 ligand R848 reduces VRE colonization
To determine whether R848-mediated stimulation of TLR-7 could restore colonization resistance against VRE after antibiotic treatment, we challenged untreated or ampicillin-treated mice with R848 or PBS control orally daily with VRE and quantified VRE in the ileum (Fig. 2D). R848 treatment significantly reduced VRE densities in the ileum of ampicillin-treated mice compared to ampicillin-treated mice administered PBS (Fig. 2E), consistent with the trend observed in MNV-CW3–infected mice (Fig. 1B). R848 did not inhibit VRE replication in liquid cultures, suggesting that R848 does not directly inhibit VRE growth (fig. S2D). The quantity of Reg3g mRNA transcripts in the ileum of R848- and ampicillin-treated mice inversely correlated with VRE density (Fig. 2F). Fluorescence in situ hybridization staining of ileal sections with 16S universal bacterial and enterococcus-specific probes demonstrated that ampicillin treatment reduced commensal bacteria in the ileum of both PBS- and R848-treated mice compared to ampicillin-naïve mice (Fig. 2G). Whereas the loss of colonization resistance resulted in marked expansion of VRE in ampicillin-treated mice, R848 treatment markedly reduced the ability of VRE to occupy the vacated environmental niche in the intestinal lumen of ampicillin-treated mice (Fig. 2G).
R848 rapidly induces IL-23 and IL-22 expression
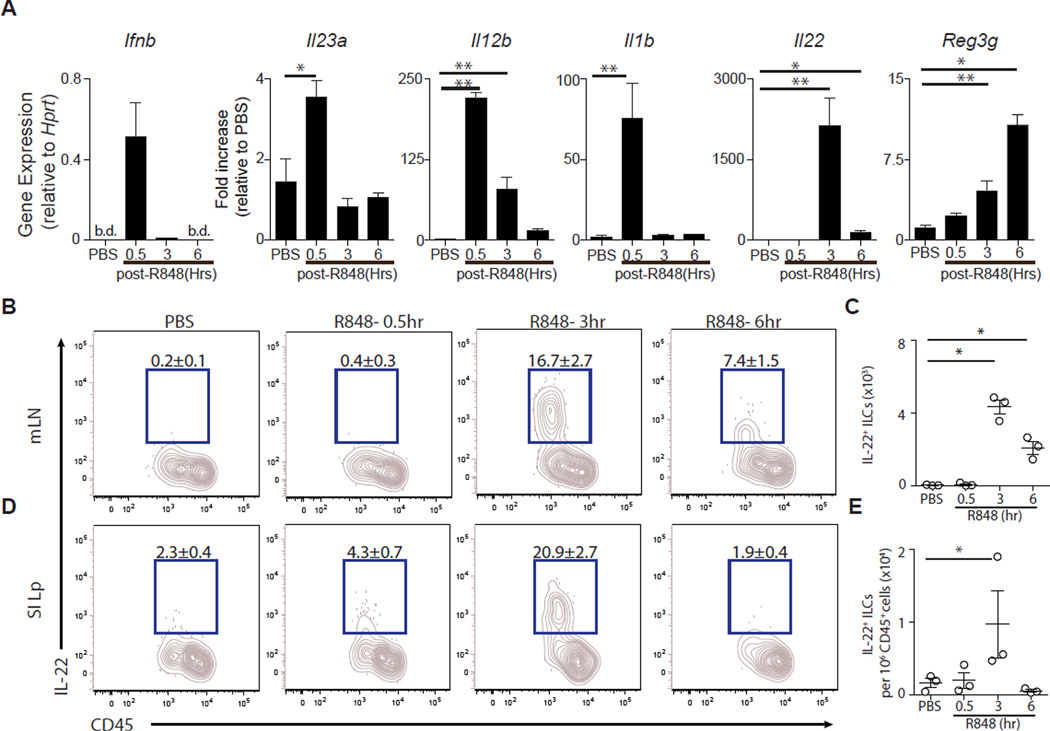
Although TLR-7 activation is best known for inducing type I interferon (IFN) and antiviral defense mechanisms (27, 39), topical administration of TLR-7 agonists to the skin induces expression of IL-23 and IL-22, two cytokines that lead to expression of Reg3γ (40, 41). We administered a single dose of R848 (50 µg orally) or PBS and measured the induction of Ifnb, Il23a(p19), Il12b(p40), Il1b, Il22, and Reg3g mRNA in the ileum by qRT-PCR 0.5, 3, and 6 hours later. Thirty minutes after intragastric administration, R848 induced transcription of Ifnb, Il23a(p19), Il12b(p40), and Il1b (Fig. 3A). Expression of Ifnb, Il23a(p19), and II1b returned to baseline levels by 3 hours, whereas Il22 transcription spiked at 3 hours (Fig. 3A). Reg3g mRNA expression continued to increase in the ileum at 3 and 6 hours after inoculation (Fig. 3A). The kinetics of gene induction is consistent with IL-23/IL-1β inducing IL-22, leading to Reg3γ expression by the intestinal epithelium (42–44). De novo expression of IL-22 protein by ILCswas analyzed using intracellular cytokine staining after ex vivo incubation without addition of exogenous stimuli and demonstrated significantly increased frequencies and numbers of IL-22+ CD90+ CD127+ ILCs in the mLNs (Fig. 3, B and C) and ileum lamina propria (Lp) (Fig. 3, D and E) after R848 administration, peaking at 3 hours. CD90+ CD127+ ILCs (figs. S3A and S4, A and B) were the dominant source of IL-22 in the mLNs (fig. S3B) and ileum Lp (fig. S3C) compared to NK1.1+ CD127neg “classical" natural killer (NK) cells, CD4+ T cells, CD8+ T cells, and γδ T cells (fig. S3A).
Fig. 3. R848 rapidly induces IL-22 production from ILCs and Reg3g in the ileum.
A single dose of R848 (50 µg) was administered orally to C57BL/6 mice. (A) Ifnb, Il23a, Il12b, Il1b, Il22, and Reg3g gene expression was assessed at 0.5, 3, and 6 hours after R848 treatment. Gene expression was quantified by qRT-PCR, normalized to Hprt, and displayed as fold increase over PBS-treated mice (3 hours after PBS treatment). Data are sum of two independent experiments (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, Mann-Whitney test; n = 3 to 7). (B to E) Single-cell suspensions isolated from the (B and C) mLNs or (D and E) ileum Lp cells of R848- or PBS-treated mice were incubated in medium in the presence of BFA and assessed for IL-22 production. Frequency (B) and number (C) of IL-22+ ILCs in the mLNs. Frequency (D) and number (E) of IL-22+ ILCs in the ileum Lp. FACS plots gated on live, CD45+, non-T non-B, Gr-1neg CD90+ CD127+ cells. Data are representative of two independent experiments (*P ≤ 0.05, Mann-Whitney test; n = 3). b.d., below detection; SI Lp, small intestinal lamina propia. Data are means ± SEM.
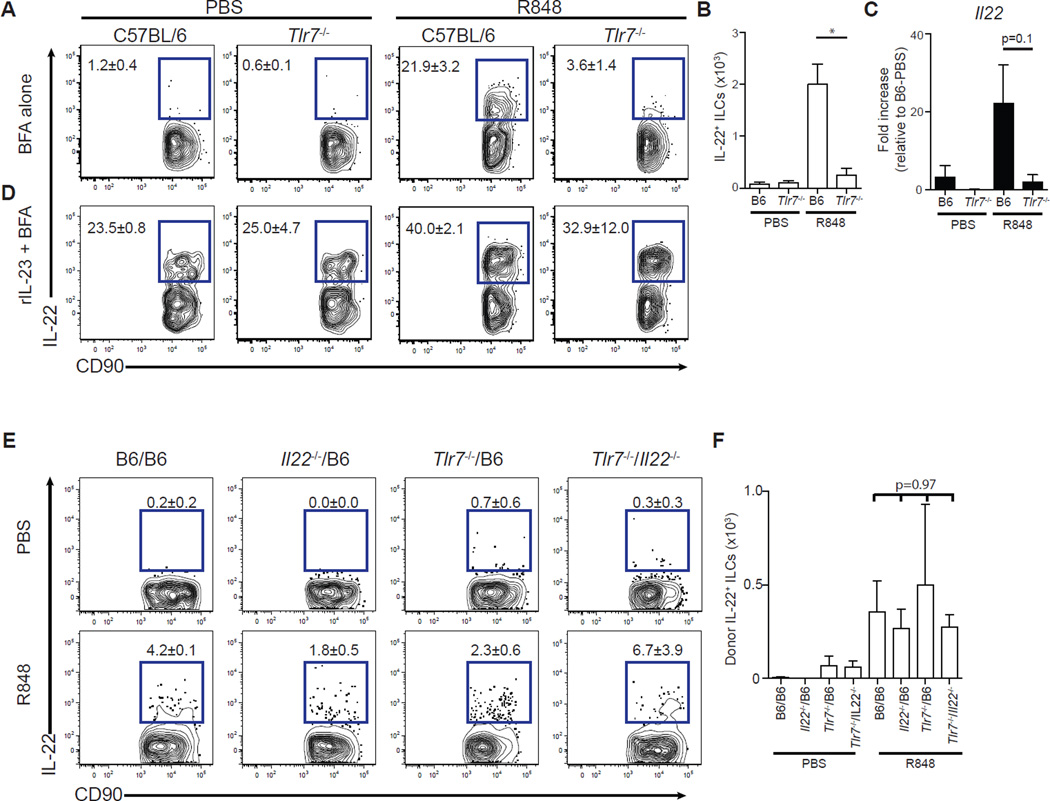
Induction of IL-22 in ILCs (Fig. 4, A and B) and up-regulation of Il22 mRNA expression in the ileum (Fig. 4C) did not occur in Tlr7−/− mice after R848 administration, demonstrating a requirement of TLR-7 for IL-22 expression. Ex vivo stimulation with recombinant IL-23, however, induced IL-22 expression in ILCs isolated from both C57BL/6 and Tlr7−/−mice (Fig. 4D and fig. S4C), demonstrating that ILCs from Tlr7−/− mice are intrinsically capable of producing IL-22 but fail to be activated after R848 treatment. Thus, similar to induction of Reg3g (fig. S2C), R848-mediated induction of IL-22 is dependent on the TLR-7 signaling pathway.
Fig. 4. R848 signals through TLR-7 but does not directly act on ILCs to drive IL-22 production.
Mice were treated with ampicillin for 3 days and administered PBS or R848 (50 µg) daily orally. (A and B) Single-cell suspensions isolated from the mLNs of C57BL/6 or Tlr7−/− mice 3 hours after final PBS or R848 treatment were incubated in medium in the presence of BFA for 3 hours and assessed for IL-22 production. Frequency (A) and number (B) of IL-22+ ILCs. FACS plots gated on live, CD45+, non-T non-B, CD11bneg, NK1.1neg CD90+ cells (*P ≤ 0.05, Mann-Whitney test; n = 3). (C) Il22 gene expression in the ileum of C57BL/6 or Tlr7−/− mice as quantified by qRT-PCR, normalized to Hprt, and displayed as fold increase over PBS-treated C57BL/6 mice (P = 0.1, Mann-Whitney test; n = 3). (D) Single-cell suspensions isolated from the mLNs of C57BL/6 or Tlr7−/− mice 3 hours after final PBS or R848 treatment were incubated in medium supplemented with recombinant IL-23 (rIL-23; 40 ng/ml) in the presence of BFA for 3 hours and assessed for IL-22 production. (E and F) Cells isolated from the mLNs of C57BL/6, Tlr7−/−/B6, Il22−/−/B6, and Tlr7−/−/Il22−/− mixed BMC mice 3 hours after final PBS or R848 treatment were incubated in medium in the presence of BFA for 3 hours and assessed for IL-22 production. Frequency (E) and number (F) of IL-22+ ILCs. FACS plots gated on donor-derived, live, CD45+, non-T non-B, Gr-1neg NK1.1neg CD90+ cells [n = 3; multiple-comparison two-way analysis of variance (ANOVA) test]. Data are means ± SEM.
The kinetics of Ifnb, Il23a(p19), Il12b(p40), and Il1b expression followed by Il22 and finally Reg3g induction suggests that R848 signaling may be perpetuated by sequential cell populations. To exclude the possibility that R848 directly stimulates IL-22 production by ILCs, we generated mixed BMC mice using Tlr7−/− and Il22−/− bone marrow cells as donor cells and lethally irradiated C57BL/6 mice as recipients. In these mixed BMC mice, none of the donor-derived ILCs has the capacity to both respond to R848 via TLR-7 and produce IL-22. Tlr7−/−/Il22−/− mixed BMC mice, along with Tlr7−/−/B6, Il22−/−/B6, and C57BL/6 BMC mice, were treated with R848, and ILCs from mLNs were assessed for IL-22 production by intracellular cytokine staining. Flow cytometric analyses were gated on donor-derived ILCs to exclude radio-resistant, host-derived ILCs (fig. S4D) (45). ILCs from all mixed BMC mice underwent comparable induction of IL-22 after R848 administration, indicating that R848 stimulates TLR-7 on a cell population that is distinct from ILCs (Fig. 4, E and F).
R848-mediated IL-22 production is independent of type I IFN and IL-1 signaling but dependent on IL-23
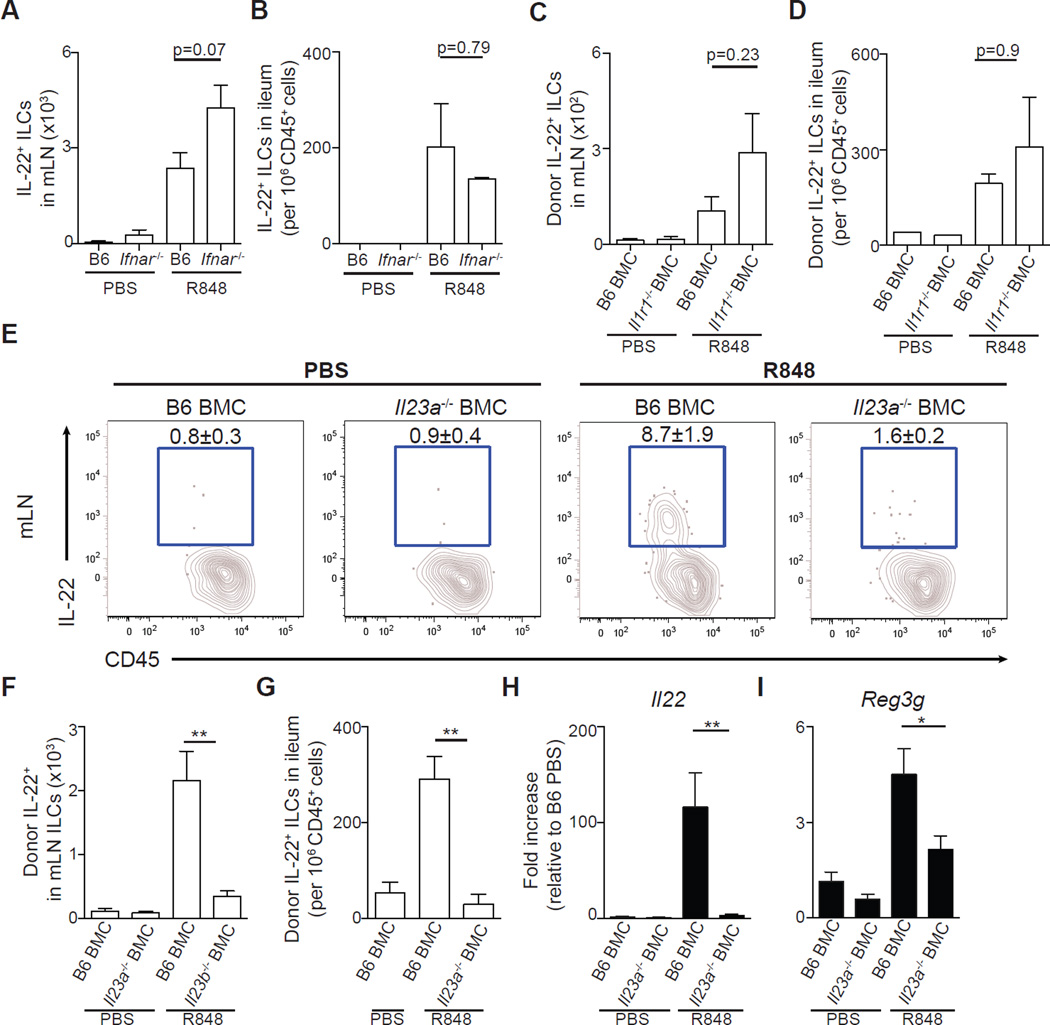
R848 treatment rapidly induced transcription of Ifnb, Il23a, Il12b, and Il1b (Fig. 3A). To assess the role of these cytokines in driving IL-22 expression, Ifnar−/− and wild-type C57BL/6 mice or Il1r1−/−, Il23a−/−, and C57BL/6 BMC mice were compared after R848 treatment. ILCs isolated from the mLNs (Fig. 5A) and ileum (Fig. 5B) of Ifnar−/− mice or the mLNs (Fig. 5C) and ileum (Fig. 5D) of Il1r1−/− BMC mice treated with R848 produced IL-22 comparable to those of control mice. Donor- and host-derived ILCs isolated from R848-treated Il1r1−/− BMC mice had similar frequencies of IL-22–producing ILCs as those isolated from control C57BL/6 BMC mice (fig. S5, A to C), demonstrating that neither extrinsic nor intrinsic IL-1 receptor signaling is necessary for R848-induced IL-22 production by ILCs. In contrast, ampicillin-treated Il23a−/− BMC mice had significantly decreased frequencies (Fig. 5E) and numbers of IL-22–producing ILCs in the mLNs (Fig. 5F and fig. S5D) and ileum Lp (Fig. 5G) compared to ampicillin-treated control C57BL/6 BMC mice after R848 administration. Induction of Il22 (Fig. 5H) and Reg3g (Fig. 5I) mRNA in the ileum was significantly impaired in Il23a−/− BMC mice after R848 treatment, demonstrating that IL-23 is necessary for R848-mediated restoration of Reg3γ expression in antibiotic-treated hosts.
Fig. 5. R848-mediated IL-22 production is independent of type I IFN and IL-1 signaling but dependent on IL-23.
(A and B) C57BL/6 and Ifnar−/− mice were administered PBS or R848 (50 µg) orally and sacrificed 90 min later. Number of IL-22+ ILCs in the (A) mLNs and (B) ileum after ex vivo incubation in medium in the presence of BFA for 3 hours. Data are representative of two independent experiments (n = 3 to 5; Mann-Whitney test). (C to I) C57BL/6, Il1r1−/−, or Il23a−/− BMC mice were treated with ampicillin for 3 days and administered PBS or R848 (50 µg) daily orally. Number of donor-derived IL-22+ ILCs in the (C) mLNs and (D) ileum of C57BL/6 or Il1r1−/− BMC mice after ex vivo incubation in medium in the presence of BFA for 3 hours (n = 3 to 4; Mann-Whitney test). Frequency (E) and number (F) of donorderived IL-22+ ILCs in the mLNs of C57BL/6 and Il23a BMC mice after ex vivo incubation in medium in the presence of BFA for 3 hours. FACS plots gated on donor-derived, live, CD45+, non-T non-B, CD11bneg CD90+ CD127+ cells. (G) Number of donor-derived IL-22+ ILCs in the ileum of PBS- or R848-treated C57BL/6 and Il23a BMC mice. Il22 (H) and Reg3g (I) gene expression in the ileum of PBS- or R848-treated C57BL/6 and Il23a−/− BMC mice as quantified by qRT-PCR and displayed as fold increase over PBS-treated C57BL/6 BMC mice and normalized to Hprt. Data are representative of three independent experiments (*P ≤ 0.05, **P ≤ 0.01, Mann-Whitney test; n = 3 to 6). Data are means ± SEM.
R848 signals via TLR-7–expressing CD11c+ dendritic cells to induce IL-22
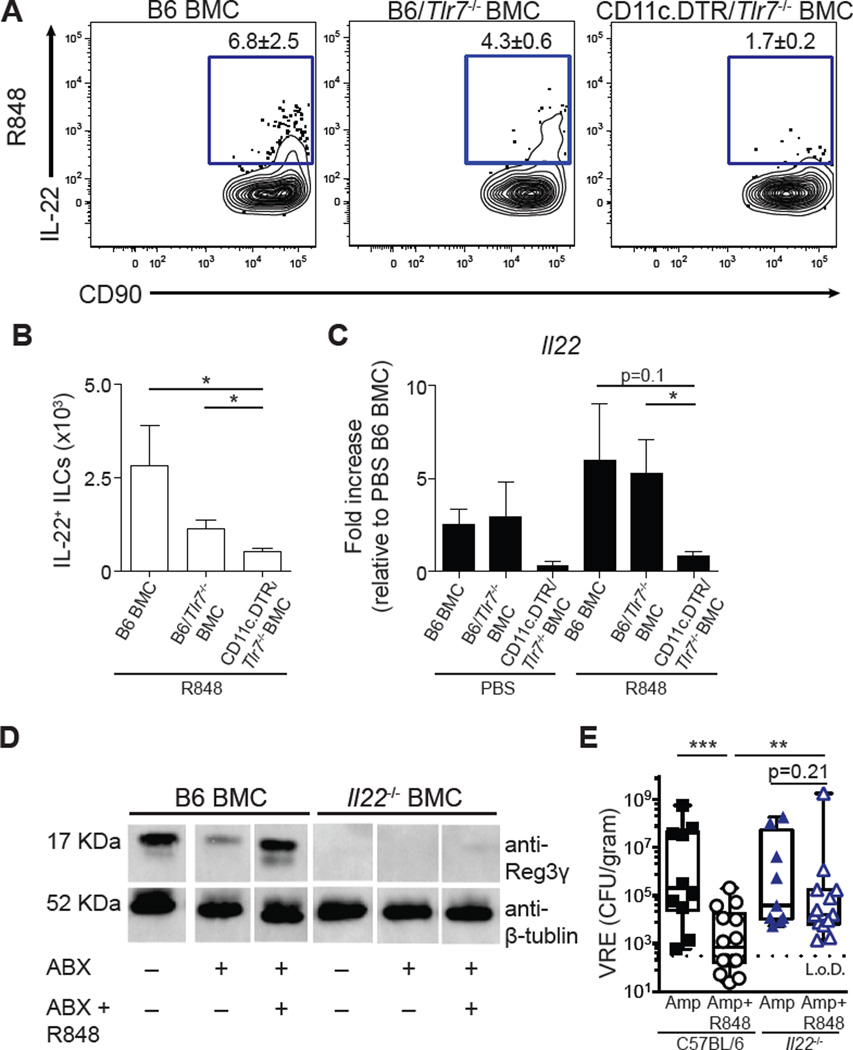
CD11c+ dendritic cells express TLR-7 (46), and activation of the TLR/MyD88 signaling pathway in dendritic cells can lead to IL-23 secretion and IL-22 production by ILCs (42, 47). To assess whether dendritic cells responded to R848 to drive IL-22 production, mixed BMC mice were made using CD11c.DTR, Tlr7−/−, and C57BL/6 mice as bone marrow donors and C57BL/6 mice as recipients. In Tlr7−/−/CD11c.DTR mixed BMC mice, diphtheria toxin (DT) treatment eliminated CD11c+ cells expressing the diphtheria toxin receptor, leaving only TLR-7–deficient CD11c+ dendritic cells intact (fig. S6D). ILCs isolated from the ileum (Fig. 6, A and B) and mLNs (fig. S6, A and B) of Tlr7−/−/CD11c.DTR mixed BMC mice had fewer IL-22+ ILCs after R848 treatment compared to both B6 BMC and Tlr7−/−/B6 mixed BMC mice. qRT-PCR analysis in the ileum confirmed that R848 failed to induce Il22 mRNA expression in Tlr7−/−/CD11c.DTR mixed BMC mice, indicating that CD11c+ dendritic cells are critical for R848-mediated induction of IL-22 (Fig. 6C).
Fig. 6. R848 signals via TLR-7–expressing CD11c+ dendritic cells to induce IL-22, which is necessary to reestablish colonization resistance against VRE.
(A and B) C57BL/6, Tlr7−/−/B6, and Tlr7−/−/CD11c.DTR mixed BMC mice were treated with ampicillin for 3 days, treated with diphtheria toxin, and administered PBS or R848 (50 µg) daily orally. Frequency (A) and number (B) of IL-22+ ILCs in the ileum of R848-treated C57BL/6, Tlr7−/−/B6, and Tlr7−/−/CD11c.DTR mixed BMC mice after ex vivo incubation in medium in the presence of BFA for 3 hours. FACS plots gated on live, CD45+, non-T non-B, Gr-1neg CD90+ CD127+ cells. Data are representative of two independent experiments (*P ≤ 0.05, **P ≤ 0.01, Mann-Whitney test; n = 3). (C) Il22 gene expression in the ileum of Tlr7−/−/B6 and Tlr7−/−/CD11c.DTR mixed BMC mice as quantified by qRT-PCR and displayed as fold increase over PBS-treated B6 BMC mice and normalized to Hprt. Data are representative of two independent experiments (*P ≤ 0.05, **P ≤ 0.01, Mann-Whitney test; n = 3). Data are means ± SEM. (D) ABX-treated or untreated C57BL/6 or Il22−/− BMC mice were administered PBS or R848 (50 µg) daily orally. Protein extracts from the ileum were analyzed by Western blotting using the Reg3γ-specific antiserum. Data are representative of two independent experiments. Each lane is for a representative mouse from the indicated group. (E) C57BL/6 and Il22−/− were treated with ampicillin for 3 days and administered PBS or R848 (50 µg) daily orally. Mice were inoculated with VRE, and CFUs in the ileal content were assessed 24 hours after infection. Data are sum of three independent experiments (**P ≤ 0.01, Mann-Whitney test; n = 10 to 14).
IL-22 is critical for R848-mediated Reg3γ expression and protection against VRE colonization
TLR-7–expressing CD11c+ dendritic cells and IL-23 are essential for R848-mediated induction of IL-22 (Figs. 5 and 6, A to C). Antibiotic-treated Il22−/− BMC (Fig. 6D) and Il12b(p40)−/− BMC mice (fig. S6C) failed to increase Reg3γ expression in the ileum after R848 treatment. Next, Il22−/− and C57BL/6 mice were treated with ampicillin and R848 or PBS for 3 days followed by inoculation with VRE. R848-treated C57BL/6 mice had reduced VRE density in the ileum compared to PBS-treated C57BL/6 control mice (Fig. 6E). R848 treatment of Il22−/− mice, however, did not reduce VRE density compared to PBS-treated Il22−/− mice (Fig. 6E). Thus, IL-22 is essential for R848-mediated colonization resistance against VRE. These results demonstrate that stimulation of TLR-7 with an orally bioavailable drug reestablishes intestinal innate immune defenses, thereby enhancing colonization resistance against a highly antibiotic-resistant pathogen.
DISCUSSION
VRE has emerged as one of the most common causes of bloodstream infection in patients undergoing bone marrow transplantation, and recent studies indicate that systemic infection is preceded by dense intestinal colonization that results from antibiotic-induced loss of colonization resistance (8, 9, 11). Although intestinal domination represents an important step toward VRE dissemination into the bloodstream, it also increases the risk of VRE transmission to other hospitalized patients. Thus, reducing the density of intestinal VRE colonization would reduce the incidence of systemic VRE infections in colonized patients and also the rate of VRE transmission between patients. Enteric viruses shape both the composition of intestinal bacteria communities and the host’s immune system (25, 48). We report that MNV infection partially restores basal immune defenses in the intestine, and norovirus titers inversely correlate with intestinal VRE colonization. Development of an infectious virus as a therapeutic agent is clinically impractical; therefore, we sought to identify a pharmacological agent that could mimic the immunostimulatory properties of viruses and reestablish immune-mediated mechanisms of colonization resistance. Our study demonstrates that R848, a synthetic molecule that induces antiviral immune defenses, can markedly reduce the density of intestinal VRE colonization by signaling through TLR-7 and activating IL-22 expression by ILCs to enhance innate antibacterial defense mechanisms.
Colonization resistance is remarkably effective at eliminating exogenously acquired bacterial species from the gastrointestinal tract. Antibiotics are often necessary in hospital settings; however, their impact on intestinal commensal bacterial communities can impair direct and indirect colonization resistance defense mechanisms. Studies performed roughly 60 years ago demonstrated that a single dose of antibiotic could render mice nearly a million-fold more susceptible to infection by Salmonella typhimurium and implicated obligate anaerobic commensal bacteria as essential mediators of colonization resistance (49). Therapeutics capable of restoring the host’s natural defenses could be used to supplement antibiotic treatment to reduce the risk of infection with antibiotic-resistant bacteria in patients. Whereas antibiotic treatment markedly reduces the expression of Reg3γ in the small intestine of mice, oral administration of LPS to stimulate TLR-4 or systemic administration of flagellin to stimulate TLR-5 can restore murine Reg3γ expression and enhance resistance to dense colonization by VRE (36, 37). Our findings demonstrate that targeted activation of ILCs is a potential therapeutic intervention strategy against intestinal bacterial pathogens.
Whereas reestablishing colonization resistance after antibiotic-mediated disruption of the microbiota may benefit patients, potential risks also require consideration. The severity of microbiota injury can be variable and likely depends on the antibiotic resistance of the patient’s baseline microbiota, the antimicrobial spectrum, the route/duration of treatment, and the clearance mechanisms of the administered antibiotic. Recovery of the intestinal microbiota likely also varies between patients, in part depending on the composition of the posttreatment microbiota and re-acquisition of microbes from the environment. Thus, in some settings, compromise of colonization resistance may be transient, whereas in other settings, damage to the microbiota may be prolonged. Although fecal transplantation has successfully treated recurrent C. difficile infections (20), the inability to fully define the composition of feces raises concerns in patients with compromised immune defenses. Furthermore, recently discovered associations of fecal microbiota composition with a range of metabolic and inflammatory diseases raise concerns about the long-term impact of fecal transplantation from heterologous donors. Although administration of defined bacterial populations to patients after antibiotic treatment may eventually facilitate the reestablishment of colonization resistance, the optimal bacterial species and consortia have yet to be defined. Thus, a combinational approach of using pharmacological induction of colonization resistance by stimulating innate immune receptors during and after antibiotic treatment represents a potential approach to reduce the risk of dense colonization by antibiotic-resistant pathogens such as VRE.
The risks of innate immune receptor stimulation are not negligible and include potentially inducing acute or chronic inflammation and enhancing existing inflammatory diseases. In the case of R848 administration, induction of IL-23 and IL-22 in the small intestinal Lp and reduction of intestinal VRE colonization must be balanced against the potentially adverse effects of increased type I IFN expression. For example, a phase 2a clinical trial reported that oral administration of R848 could reduce viral titers of chronic hepatitis C virus but only at doses associated with a high frequency of adverse events in patients (50). Induction of Reg3γ was inversely correlated with VRE density in R848-treated mice, and Reg3γ can directly target Gram-positive bacteria such as VRE (14, 36). However, our studies do not exclude the potential role for other antimicrobial peptide induced by TLR-7 activation in limiting VRE expansion. Although further studies will be required to determine whether the adverse effects of innate immune activation can be suppressed while preserving the beneficial effects, the data presented in this study suggest that controlled amplification of the TLR-7/IL-22 pathway will permit restoration of immune-mediated colonization resistance and may limit infection by intestinal pathogens in antibiotic-exposed, susceptible individuals.
MATERIALS AND METHODS
Study design
The goal of this study was to investigate the ability of viral infection or agonist that stimulates receptors associated with antiviral innate immune defense to restore antibiotic-induced colonization resistance against VRE. A series of BMC mice were made to identify the molecular mechanism of action of R848-mediated inhibition of VRE colonization. All MNV-infected mice were included in the analyses to avoid bias despite reduced capability of MNV to infect ampicillin-treated mice (25, 35). Group sizes were limited in size (n = 3 to 5 per R848-treated group and n = 1 to 3 per PBS-treated group) to ensure rapid processing of tissue and accurate assessment of in vivo immune activity. For in vivo experiments, the number of mice is stated in the figure legends. The mice were randomized to different groups, but the experimenters were not blinded to group identity.
Mice and generation of bone marrow chimeras
Wild-type maximum barrier C57BL/6, Tlr7−/−, Il1r1−/−, Il12b−/−, and CD11c.DTR mice were purchased from The Jackson Laboratory. All knockout mouse strains were derived on a C57BL/6 background. Ifnar−/− mice were purchased from B&K Universal and bred in-house onto a C57BL/6 background. Il22−/− mice were provided by R. Flavell (Yale University, New Haven, CT). Il23a−/− bone marrow cells were provided by D. Littman (New York University, New York, NY). All mice were bred and maintained under specific pathogen–free conditions at the Memorial Sloan Kettering Research Animal Resource Center. Sex- and age-matched controls were used in all experiments according to institutional guidelines for animal care. All animal procedures were approved by the Institutional Animal Care and Use Committee of the Memorial Sloan Kettering Cancer Center. Four- to 6-week-old congenically marked C57BL/6 mice were lethally irradiated with 950 cGy using a 137Cs source. One day following irradiation, mice received an intravenous injection of 2 × 106 bone marrow cells harvested from the femurs and tibias of wild-type C57BL/6, Tlr7−/−, Il1r1−/−, Ifnar−/−, Il12b−/−, Il22−/−, or CD11c.DTR mice. Mixed BMC mice were generated with a 1:1 mixture of 2.0 × 106 bone marrow cells from the femurs and tibias of C57BL/6, Tlr7−/−, Il22−/−,or CD11c.DTR mice. The mice were allowed to rest for at least 6 weeks after irradiation to allow for reconstitution of the hematopoietic compartment before being used for experiments.
Antibiotic treatment, MNV and VRE infection, and R848 treatment
Mice were cohoused for 3 weeks before antibiotic treatment to allow for equilibration of the microbiota. Antibiotic administration consisted of metronidazole (0.5 g/liter; Sigma), neomycin (0.5 g/liter; Sigma), and vancomycin (1 g/liter; Novaplus) mixed in drinking water for 7 days (37) or a targeted regiment of ampicillin (0.25 g/liter; Fisher Scientific) for 72 hours and then replaced with regular water for the duration of the experiment. Twenty-four hours after cessation of antibiotic water, mice were administered 2 × 106 CFUs of VRE, purchased from the American Type Culture Collection (700221), via oral gavage. VRE was grown at 37°C in Brain Heart Infusion broth to early stationary phase and diluted in PBS to 107 CFUs. For norovirus infections, MNV (CW3 strain) was grown and isolated from RAW264.7 cells (51), and mice were administered 106 plaque-forming units of MNV-CW3 by oral gavage. For R848 (InvivoGen) treatment, mice were administered either a single 50-µg dose via oral gavage or 50 µg of R848 daily starting the day of antibiotic treatment and continuing until the day of mouse sacrifice. CpG (ODN 1585, 100 µg), R837 (50 µg), Gardiquimod (50 µg), CL907 (50 µg), and poly(dT) (InvivoGen) were administered via oral gavage daily for 3 days. For diphtheria toxin treatment, mice received 15 ng of diphtheria toxin intraperitoneally, in conjunction with R848 or PBS treatment at days −5, −3, and −1 before sacrifice.
Quantification of VRE burden and MNV-CW3 titers
Intestinal contents from the ileum were weighed and resuspended in 1 ml of PBS. Tenfold dilutions were plated on BBL Enterococcosel agar (Becton Dickinson) plates [supplemented with vancomycin (8 µg/ml; Novaplus) and streptomycin (100 µg/ml; Fisher)] and incubated for 48 hours at 37°C for the specific detection of VRE.
MNV-CW3 quantification was assessed using TaqMan qPCR analysis. RNA was isolated from the ileal tissue using mechanical homogenization and TRIzol isolation (Invitrogen) according to the manufacturer’s instructions. Complementary DNA (cDNA) was generated using the QuantiTect Reverse Transcription Kit (Qiagen). The forward and reverse primers used were CACGCCACCGATCTGTTGTG and GCGCTGCGCCATCACTC, respectively. The MNV probe was CGCTTTGGAACAATG. RT-PCR was carried out using TaqMan PCR Master Mix (Applied Biosystems) in 10-µl reaction mixture volumes according to the manufacturer’s instructions, and reactions were run on an RT-PCR system (StepOne Plus; Applied Biosystems). All samples were assessed in triplicate, and the results were standardized to total RNA per sample.
Isolation of cells from the mLNs and intestine
Lymphocytes were isolated from the mLNs by mechanical disruption through 100-µm cell strainers. Single-cell suspensions were obtained from the ileum Lp by longitudinally cutting the ileum and then washing out content in PBS. Intestinal tissues were incubated at 37°C under gentle agitation in stripping buffer [PBS, 5 mM EDTA, 1 mM dithiothreitol, 4% fetal calf serum, and penicillin/streptomycin (10 µg/ml)] for 10 min to remove epithelial cells and then for another 20 min for the IEL. Supernatants containing the IEL fraction were passed through 100-µm cell strainers and resuspended in 40% Percoll. The remaining tissue was digested with collagenase IV (1.5 mg/ ml; 500 U/ml) and deoxyribonuclease (20 µg/ml) in complete medium [Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, penicillin/streptomycin (10 µg/ml), gentamicin (50 µg/ml), 10 mM Hepes, 0.5 mM β-mercaptoethanol, and l-glutamine (20 µg/ml)] for 30 min at 37°C under gentle agitation. Supernatants containing the Lp fraction were passed through a 100-µm cell strainers and resuspended in 40% Percoll. Samples were then centrifuged for 20 min at 600g to obtain IEL and Lp cell fractions.
Ex vivo cytokine detection and flow cytometry
For direct ex vivo cytokine detection, cells isolated from the mLNs or ileum were cultured in a 96-well plate in complete medium and BFA alone (GolgiPlug, eBioscience) for 3 hours at 37°C. After incubation, cells were surface-stained in FACS buffer (PBS, 2% bovine serum albumin, 0.2 mg of sodium azide, 2 mM EDTA) using a standard flow cytometric staining protocol with fluorescently conjugated antibodies specific to CD3ε, CD4, CD5, CD8α, CD19, CD45.1, CD45.2 CD90.1, CD90.2, CD127, Gr-1 (clone RB85), CD11b (Invitrogen), γδTCR (eBioscience), and NK1.1 (BioLegend). After staining for surface antigens, cells were stained for intracellular cytokines using intracellular cytokine fixation buffer (eBioscience) and fluorescently conjugated antibodies specific for IL-22 (clone 1H8PWSR, eBioscience). Cell viability was assessed with LIVE/DEAD Aqua stain (Invitrogen). Samples were collected by an LSR II flow cytometer (Becton Dickinson). All flow cytometry data were analyzed by FlowJo version 9.7 (Tree Star).
Tissue isolation, RT-PCR, and Western blot analysis
RNA was isolated from ileum using mechanical homogenization and TRIzol isolation (Invitrogen) according to the manufacturer’s instructions. cDNA was generated using the QuantiTect Reverse Transcription Kit (Qiagen). RT-PCR was performed on cDNA using TaqMan primers and probes in combination with TaqMan PCR Master Mix (Applied Biosystems), and reactions were run on an RT-PCR system (StepOne Plus; Applied Biosystems). Gene expression is displayed as fold increase over wild-type control mice, unless otherwise stated, and normalized to Hprt.
The extracted protein samples were run for the presence of Reg3γ and β-tubulin protein as described in (37). Briefly, identical amounts of protein were loaded on a 4 to 12% SDS-PAGE (polyacrylamide gel electrophoresis) gel and transferred to a nitrocellulose membrane, and rabbit polyclonal Reg3γ-specific antiserum or mouse anti–β-tubulin antibodies (Santa Cruz Biotechnology) were used to detect Reg3γ and the loading control protein. Original blot images are shown in Fig. S7. Bands were quantified using the ImageJ software, and the relative density values are listed in table S1.
Fluorescence in situ hybridization
The FISH method was adapted from previous publications (52, 53). Briefly, tissue sections were deparaffinized with xylene (twice, 10 min each) and rehydrated through an ethanol gradient (95%, 10 min; 90%, 10 min) to water. Sections were incubated with a universal bacterial probe directed against the 16S ribosomal RNA gene (EUB338: [Cy3]-GCTGCCTCCCGTAGGAGT-[AmC7~Q+Cy3es]) and an Enterococcus-specific probe (Enfm93: [AminoC6+Alexa488]-GCCACTCCTCTTTTTCCGG-[AmC7~Q+Alexa488]) at 50°C for 3 hours. Probes were diluted to 5 ng/µl in 0.9 M NaCl, 20 mM tris-HCl (pH 7.2), and 0.1% SDS before use. Sections were later washed twice in 0.9 M NaCl and 20 mM tris-HCl (pH 7.2) (wash buffer) for 10 min and counterstained with Hoechst (1:3000 in wash buffer) for nuclear staining (32, 54). Images were acquired with a Leica TCS SP5-II upright confocal microscope using a 40× oil immersion lens (numerical aperture, 1.4; HCX PL APO) as a series of short Z-stacks. Maximum intensity projection processing of Z-stacks was done in Fiji (ImageJ) software.
Statistical analysis
Results represent means ± SEM. Statistical significance was determined by the unpaired one-sided Mann-Whitney test unless otherwise stated. Statistical analyses were performed using GraphPad Prism software version 6.0. Exact P values are listed in table S1.
Supplementary Material
Acknowledgments
We thank members of the Pamer laboratory for helpful discussions and critical reading of the manuscript. We thank the Molecular Cytology Facility at Memorial Sloan Kettering Cancer Center (Core grant P30 CA008748) for histology sectioning, staining, and imaging.
Funding: Supported by the U.S. NIH (RO1 AI042135 and AI095706 to E.G.P.), the Irvington Institute Postdoctoral Fellowship of the Cancer Research Institute (M.C.A.), and the Swiss National Science Foundation Early Postdoc.Mobility Fellowship (S.B.).
Footnotes
www.sciencetranslationalmedicine.org/cgi/content/full/8/327/327ra25/DC1
Materials and Methods
Fig. S1. Norovirus infection is associated with reduced VRE colonization.
Fig. S2. R848 restores Reg3γ expression via TLR-7/MyD88 signaling on hematopoietic cells.
Fig. S3. ILCs are the dominant source of IL-22 after R848 treatment.
Fig. S4. Gating strategy for Figs. 3 and 4.
Fig. S5. R848 stimulates IL-22 expression in wild-type and Il1r1−/− ILCs.
Fig. S6. R848-driven induction of IL-22 and Reg3γ is dependent on TLR-7–expressing CD11c+ dendritic cells and IL-23 signaling.
Fig. S7. Full Western blot images from Figs. 2 (A to C) and 6 D and figs. S2A and S5D.
Table S1. Source data and exact P values.
Author contributions: M.C.A., C.G.B., and E.G.P. designed the experiments and wrote the manuscript. M.C.A. and C.G.B. performed experiments and analysis of data. B.S. and R.A.C. assisted in bacterial culturing, RNA tissue isolation, and RT-PCRs. S.B. and J.W.K. assisted in tissue harvesting and processing. D.A. and L.C.O. provided viral stocks and assisted with MNV-related experimental design, execution, and analysis.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Antibiotic Resistance Threats in the United States 2013. Atlanta, GA: Centers for Disease Control and Prevention; 2014. [Google Scholar]
- 2.Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM, Bartlett JG, Edwards J., Jr Infectious Diseases Society of America, The epidemic of antibiotic-resistant infections: A call to action for the medical community from the Infectious Diseases Society of America. Clin. Infect. Dis. 2008;46:155–164. doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- 3.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arias CA, Murray BE. Emergence and management of drug-resistant enterococcal infections. Expert Rev. Anti Infect. Ther. 2008;6:637–655. doi: 10.1586/14787210.6.5.637. [DOI] [PubMed] [Google Scholar]
- 5.Arias CA, Murray BE. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 2012;10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donskey CJ, Chowdhry TK, Hecker MT, Hoyen CK, Hanrahan JA, Hujer AM, Hutton-Thomas RA, Whalen CC, Bonomo RA, Rice LB. Effect of antibiotic therapy on the density of vancomycin-resistant enterococci in the stool of colonized patients. N Engl. J. Med. 2000;343:1925–1932. doi: 10.1056/NEJM200012283432604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MRM, Kamboj M, Pamer EG. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Invest. 2010;120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taur Y, Jenq RR, Perales M-A, Littmann ER, Morjaria S, Ling L, No D, Gobourne A, Viale A, Dahi PB, Ponce DM, Barker JN, Giralt S, van den Brink M, Pamer EG. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124:1174–1182. doi: 10.1182/blood-2014-02-554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, Lee YJ, Dubin KA, Socci ND, Viale A, Perales M-A, Jenq RR, van den Brink MRM, Pamer EG. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin. Infect. Dis. 2012;55:905–914. doi: 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magauran CE, Salgado CD. Challenges and advances in infection control of hematopoietic stem cell transplant recipients. Infect. Disord. Drug Targets. 2011;11:18–26. doi: 10.2174/187152611794407764. [DOI] [PubMed] [Google Scholar]
- 11.Weinstock DM, Conlon M, Iovino C, Aubrey T, Gudiol C, Riedel E, Young JW, Kiehn TE, Zuccotti G. Colonization, bloodstream infection, and mortality caused by vancomycin-resistant enterococcus early after allogeneic hematopoietic stem cell transplant. Biol. Blood Marrow Transplant. 2007;13:615–621. doi: 10.1016/j.bbmt.2007.01.078. [DOI] [PubMed] [Google Scholar]
- 12.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abt MC, Pamer EG. Commensal bacteria mediated defenses against pathogens. Curr. Opin. Immunol. 2014;29:16–22. doi: 10.1016/j.coi.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nuñez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 16.Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: New developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 2009;7:526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 17.Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, Russell SL, Vallance BA, Finlay BB. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect. Immun. 2011;79:1536–1545. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira RBR, Gill N, Willing BP, Antunes LCM, Russell SL, Croxen MA, Finlay BB. The intestinal microbiota plays a role in Salmonella-induced colitis independent of pathogen colonization. PLOS One. 2011;6:e20338. doi: 10.1371/journal.pone.0020338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohnhoff M, Miller CP, Martin WR. Resistance of the mouse’s intestinal tract to experimental Salmonella infection. I. Factors which interfere with the initiation of infection by oral inoculation. J. Exp. Med. 1964;120:805–816. doi: 10.1084/jem.120.5.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JFWM, Tijssen JGP, Speelman P, Dijkgraaf MGW, Keller JJ. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl. J. Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 21.Pamer EG. Fecal microbiota transplantation: Effectiveness, complexities, and lingering concerns. Mucosal Immunol. 2014;7:210–214. doi: 10.1038/mi.2013.117. [DOI] [PubMed] [Google Scholar]
- 22.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Virgin HW. The virome in mammalian physiology and disease. Cell. 2014;157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS, Miller VL, Virgin HW., IV Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- 25.Kernbauer E, Ding Y, Cadwell K. An enteric virus can replace the beneficial function of commensal bacteria. Nature. 2014;516:94–98. doi: 10.1038/nature13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernández PP, Mahlakõiv T, Yang I, Schwierzeck V, Nguyen N, Guendel F, Gronke K, Ryffel B, Hölscher C, Dumoutier L, Renauld J-C, Suerbaum S, Staeheli P, Diefenbach A. Interferon-λ and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat. Immunol. 2015;16:698–707. doi: 10.1038/ni.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, Horiuchi T, Tomizawa H, Takeda K, Akira S. Small anti-viral compounds activate immune cells via the TLR7 MyD88–dependent signaling pathway. Nat. Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 28.Dockrell DH, Kinghorn GR. Imiquimod and resiquimod as novel immunomodulators. J. Antimicrob. Chemother. 2001;48:751–755. doi: 10.1093/jac/48.6.751. [DOI] [PubMed] [Google Scholar]
- 29.Edwards L, Ferenczy A, Eron L, Baker D, Owens ML, Fox TL, Hougham AJ, Schmitt KA HPV Study Group. Self-administered topical 5% imiquimod cream for external anogenital warts. Arch. Dermatol. 1998;134:25–30. doi: 10.1001/archderm.134.1.25. [DOI] [PubMed] [Google Scholar]
- 30.Beutner KR, Tyring SK, Trofatter KF, Jr, Douglas JM, Jr, Spruance S, Owens ML, Fox TL, Hougham AJ, Schmitt KA. Imiquimod, a patient-applied immune-response modifier for treatment of external genital warts. Antimicrob. Agents Chemother. 1998;42:789–794. doi: 10.1128/aac.42.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beutner KR, Spruance SL, Hougham AJ, Fox TL, Owens ML, Douglas JM., Jr Treatment of genital warts with an immune-response modifier (imiquimod) J. Am. Acad. Dermatol. 1998;38:230–239. doi: 10.1016/s0190-9622(98)70243-9. [DOI] [PubMed] [Google Scholar]
- 32.Caballero S, Carter R, Ke X, Sušac B, Leiner IM, Kim GJ, Miller L, Ling L, Manova K, Pamer EG. Distinct but spatially overlapping intestinal niches for vancomycin-resistant Enterococcus faecium and carbapenem-resistant Klebsiella pneumoniae. PLOS Pathog. 2015;11:e1005132. doi: 10.1371/journal.ppat.1005132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, Chervonsky AV, Golovkina TV. Successful transmission of a retrovirus depends on the commensal microbiota. Science. 2011;334:245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, Dermody TS, Pfeiffer JK. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baldridge MT, Nice TJ, McCune BT, Yokoyama CC, Kambal A, Wheadon M, Diamond MS, Ivanova Y, Artyomov M, Virgin HW. Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science. 2015;347:266–269. doi: 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kinnebrew MA, Ubeda C, Zenewicz LA, Smith N, Flavell RA, Pamer EG. Bacterial flagellin stimulates toll-like receptor 5–dependent defense against vancomycin-resistant Enterococcus infection. J. Infect. Dis. 2010;201:534–543. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawai T, Akira S. Toll-like receptor and RIG-1-like receptor signaling. Ann. N. Y. Acad. Sci. 2008;1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- 39.Gibson SJ, Lindh JM, Riter TR, Gleason RM, Rogers LM, Fuller AE, Oesterich JL, Gorden KB, Qiu X, McKane SW, Noelle RJ, Miller RL, Kedl RM, Fitzgerald-Bocarsly P, Tomai MA, Vasilakos JP. Plasmacytoid dendritic cells produce cytokines and mature in response to the TLR7 agonists, imiquimod and resiquimod. Cell. Immunol. 2002;218:74–86. doi: 10.1016/s0008-8749(02)00517-8. [DOI] [PubMed] [Google Scholar]
- 40.Van Belle AB, de Heusch M, Lemaire MM, Hendrickx E, Warnier G, Dunussi-Joannopoulos K, Fouser LA, Renauld J-C, Dumoutier L. IL-22 is required for imiquimod-induced psoriasiform skin inflammation in mice. J. Immunol. 2012;188:462–469. doi: 10.4049/jimmunol.1102224. [DOI] [PubMed] [Google Scholar]
- 41.van der Fits L, Mourits S, Voerman JSA, Kant M, Boon L, Laman JD, Cornelissen F, Mus A-M, Florencia E, Prens EP, Lubberts E. Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis. J. Immunol. 2009;182:5836–5845. doi: 10.4049/jimmunol.0802999. [DOI] [PubMed] [Google Scholar]
- 42.Kinnebrew MA, Buffie CG, Diehl GE, Zenewicz LA, Leiner I, Hohl TM, Flavell RA, Littman DR, Pamer EG. Interleukin 23 production by intestinal CD103+CD11b+ dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36:276–287. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 44.Sutton CE, Lalor SJ, Sweeney CM, Brereton CF, Lavelle EC, Mills KHG. Interleukin-1 and IL-23 induce innate IL-17 production from γδ T cells, amplifying Th17 responses and autoimmunity. Immunity. 2009;31:331–341. doi: 10.1016/j.immuni.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 45.Hanash AM, Dudakov JA, Hua G, O’Connor MH, Young LF, Singer NV, West ML, Jenq RR, Holland AM, Kappel LW, Ghosh A, Tsai JJ, Rao UK, Yim NL, Smith OM, Velardi E, Hawryluk EB, Murphy GF, Liu C, Fouser LA, Kolesnick R, Blazar BR, van den Brink MRM. Interleukin-22 protects intestinal stem cells from immune-mediated tissue damage and regulates sensitivity to graft versus host disease. Immunity. 2012;37:339–350. doi: 10.1016/j.immuni.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Milling S, Yrlid U, Cerovic V, MacPherson G. Subsets of migrating intestinal dendritic cells. Immunol. Rev. 2010;234:259–267. doi: 10.1111/j.0105-2896.2009.00866.x. [DOI] [PubMed] [Google Scholar]
- 47.Roses RE, Xu S, Xu M, Koldovsky U, Koski G, Czerniecki BJ. Differential production of IL-23 and IL-12 by myeloid-derived dendritic cells in response to TLR agonists. J. Immunol. 2008;181:5120–5127. doi: 10.4049/jimmunol.181.7.5120. [DOI] [PubMed] [Google Scholar]
- 48.Duerkop BA, Clements CV, Rollins D, Rodrigues JLM, Hooper LV. A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc. Natl. Acad. Sci. U.S.A. 2012;109:17621–17626. doi: 10.1073/pnas.1206136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hentges DJ, Freter R. In vivo and in vitro antagonism of intestinal bacteria against Shigella flexneri. I. Correlation between various tests. J. Infect. Dis. 1962;110:30–37. doi: 10.1093/infdis/110.1.30. [DOI] [PubMed] [Google Scholar]
- 50.Pockros PJ, Guyader D, Patton H, Tong MJ, Wright T, McHutchison JG, Meng T-C. Oral resiquimod in chronic HCV infection: Safety and efficacy in 2 placebo-controlled, double-blind phase IIa studies. J. Hepatol. 2007;47:174–182. doi: 10.1016/j.jhep.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 51.Tomov VT, Osborne LC, Dolfi DV, Sonnenberg GF, Monticelli LA, Mansfield K, Virgin HW, Artis D, Wherry EJ. Persistent enteric murine norovirus infection is associated with functionally suboptimal virus-specific CD8 T cell responses. J. Virol. 2013;87:7015–7031. doi: 10.1128/JVI.03389-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swidsinski A, Loening-Baucke V, Lochs H, Hale LP. Spatial organization of bacterial flora in normal and inflamed intestine: A fluorescence in situ hybridization study in mice. World J. Gastroenterol. 2005;11:1131–1140. doi: 10.3748/wjg.v11.i8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waar K, Degener JE, van Luyn MJ, Harmsen HJM. Fluorescent in situ hybridization with specific DNA probes offers adequate detection of Enterococcus faecalis and Enterococcus faecium in clinical samples. J. Med. Microbiol. 2005;54:937–944. doi: 10.1099/jmm.0.46022-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.