Abstract
Hematopoietic stem cells (HSCs) replenish all types of blood cells. It is debating whether HSCs in adults solely originate from the aorta-gonad-mesonephros (AGM) region, more specifically, the dorsal aorta, during embryogenesis. Here, we report that somite hematopoiesis, a previously unwitnessed hematopoiesis, can generate definitive HSCs (dHSCs) in zebrafish. By transgenic lineage tracing, we found that a subset of cells within the forming somites emigrate ventromedially and mix with lateral plate mesoderm-derived primitive hematopoietic cells before the blood circulation starts. These somite-derived hematopoietic precursors and stem cells (sHPSCs) subsequently enter the circulation and colonize the kidney of larvae and adults. RNA-seq analysis reveals that sHPSCs express hematopoietic genes with sustained expression of many muscle/skeletal genes. Embryonic sHPSCs transplanted into wild-type embryos expand during growth and survive for life time with differentiation into various hematopoietic lineages, indicating self-renewal and multipotency features. Therefore, the embryonic origin of dHSCs in adults is not restricted to the AGM.
Keywords: primitive hematopoiesis, definitive hematopoiesis, stem cells, blood, somite, embryos, zebrafish
Introduction
The first wave of hematopoiesis in mammals takes place in the yolk sac to form blood islands, in which inside cells differentiate into primitive nucleated erythrocytes and myeloid cells (Silver and Palis, 1997; Jagannathan-Bogdan and Zon, 2013). In the zebrafish, the primitive hematopoiesis occurs at early somite stages in the intermediate cell mass (ICM) and the rostral blood island (RBI) (Detrich et al., 1995; Warga et al., 2009). The ICM cells originate from the posterior lateral plate mesoderm (LPM) and differentiate into primitive erythrocytes, neutrophils, and thrombocytes; and the RBI cells are derived from the lateral mesoderm of the head and differentiate into macrophages. It is widely believed that primitive hematopoietic cells are only required for embryonic development and will be replaced by definitive hematopoietic stem cells (dHSCs) (Orkin and Zon, 2008; Tavian et al., 2010; Jagannathan-Bogdan and Zon, 2013).
Across vertebrate species, dHSCs are believed to be generated in the aorta-gonad-mesonephros (AGM) region and are mostly differentiated from endothelial cells in the ventral wall of the dorsal aorta (Dieterlen-Lievre, 1975; Cormier and Dieterlen-Lievre, 1988; Godin et al., 1993; Medvinsky et al., 1993; Medvinsky and Dzierzak, 1996; Tavian et al., 1996; Taoudi and Medvinsky, 2007; Zovein et al., 2008; Jin et al., 2009; Bertrand et al., 2010; Kissa and Herbomel, 2010). The dHSCs in mammals first migrate to and repopulate in the fetal liver and then home to the bone marrow (Medvinsky et al., 2011; Jagannathan-Bogdan and Zon, 2013), while the dHSCs in fish migrate first to the caudal hematopoietic tissue and then to their definitive anatomical sites in the kidney and thymus (Traver et al., 2003; Murayama et al., 2006; Orkin and Zon, 2008; Bertrand et al., 2010; Kissa and Herbomel, 2010). The important characteristics of dHSCs include self-renew ability and multipotency to differentiate into all types of blood cells.
Somites are formed in pairs from the paraxial mesoderm in an anterior to posterior sequence (Maroto et al., 2012). Cells in different parts of the nascent somites differentiate into different fates: those in the dorsal part (dermamyotome) generate skeletal muscle and dermis and those in the ventromedial compartment (sclerotome) form the vertebrae, the ribs, and the tendon (Maroto et al., 2012). It is reported that sclerotome cells may also differentiate into endothelial and vascular smooth muscle lineages of the dorsal aorta (Morin-Kensicki and Eisen, 1997; Pouget et al., 2006, 2008). However, it is unknown whether somitic cells directly commit to the hematopoietic lineages during embryonic development.
In this study, we discover that somite is an embryonic hematogenic tissue in zebrafish. A subset of cells in the forming somites migrate ventromedially into the ICM before the start of blood circulation and later on circulate together with the LPM-derived primitive hematopoietic cells. The occurrence of somite hematopoiesis is independent of LPM primitive hematopoiesis and vasculature. The somite-derived hematopoietic cells, which retain high-level expression of muscle and skeletal genes, can colonize the kidney and last for life time with self-renewal ability and multipotency.
Results
Transgenic embryos and fish with GFP-labeled somites have GFP+ blood cells
In several transgenic lines expressing GFP in somites and muscles during embryogenesis, we unexpectedly observed circulating GFP+ blood cells. In Tg(ripply1:gfp) embryos, GFP expression, which is driven by the promoter of the somite-specific gene ripply1 (Kawamura et al., 2005), appears restricted to the whole somite and the notochord (Supplementary Figure S1). In embryos of the gene trap line Tg(rbfox1l:gfp), GFP expression is expressed under the control of the endogenous rbfoxl1 locus in somites and in the heart primordium (Supplementary Figure S2) (Gallagher et al., 2011). In Tg(fmyhc2:gfp) line, the Tol2-based enhancer trap element is inserted into the intergenic region between the fast muscle heavy chain genes fmyhc2.1/myhz2 and fmyhc2.2/myhc4 (homologous to mammalian Myh4), which results in GFP expression mainly in somites and muscles resembling both fmyhc2.1 and fmyhc2.2 (Supplementary Figure S3) (Maves et al., 2007). Flow cytometry analysis indicated that Tg(ripply1:gfp), Tg(rbfox1l:gfp), and Tg(fmyhc2:gfp) embryos at 28 h postfertilization (hpf) had 78.3%, 1.08%, and 42.13% of GFP+ blood cells, respectively (Supplementary Figures S1G, S2E, and S3F). The GFP+ blood cells could be clearly seen in the heart chamber of transgenic embryos at 36 hpf (Supplementary Figures S1E, S2D, and S3E, Movies S1 and S2). The Tg(ripply1:gfp) and Tg(fmyhc2:gfp) adult fish retain GFP expression (Supplementary Figures S1F and S3D) and contain GFP+ blood cells (Supplementary Figures S1G and S3F). Based on these initial observations, we hypothesized that hematopoietic cells may start to express some somitic genes at a particular time point, or more likely, cells of somites, belonging to the paraxial mesoderm derivatives, directly differentiate into hematopoietic progenitors.
Somitic cells directly differentiate into hematopoietic cells
To trace the lineages of somitic cells, we generated a stable transgenic line Tg(foxc1b:EOS) using the foxc1b promoter and the photoconvertible fluorescent protein EOS (Wiedenmann et al., 2004). The expression of eos mRNA is initiated in the dorsal blastodermal margin in the transgenic embryos around oblong-sphere stages (3.7−4 hpf) (Supplementary Figure S4B), which is similar to the expression of endogenous foxc1b (Supplementary Figure S4A). During early somitogenesis, eos mRNA level is high in the unsegmental paraxial mesoderm and progressively decreases in the maturing somites (Figure 1A, Supplementary Figure S4C and F). Double in situ hybridization indicated that the expression domain of eos mRNA is well separated from the LPM marked by scl/tal1 and gata1a expression (Figure 1C and C’, Supplementary Figure S4C and F). Due to longer half-life of EOS protein compared to that of eos mRNA, its green fluorescence remains strong in somites and derivatives until 48 hpf (Figure 1B, Supplementary Figure S4D, G−K). Flow cytometry analysis revealed that 22.8% of circulating blood cells in Tg(foxc1b:EOS) embryos at 28 hpf were EOS+ (Figure 1D). By confocal time-lapse microscopy, we found that some green fluorescent somitic cells migrated ventromedially into the ICM region from 22 to 30 hpf, which looked morphologically indistinguishable from neighboring proerythroblasts in the ICM (Supplementary Figure S5 and Movie S3).
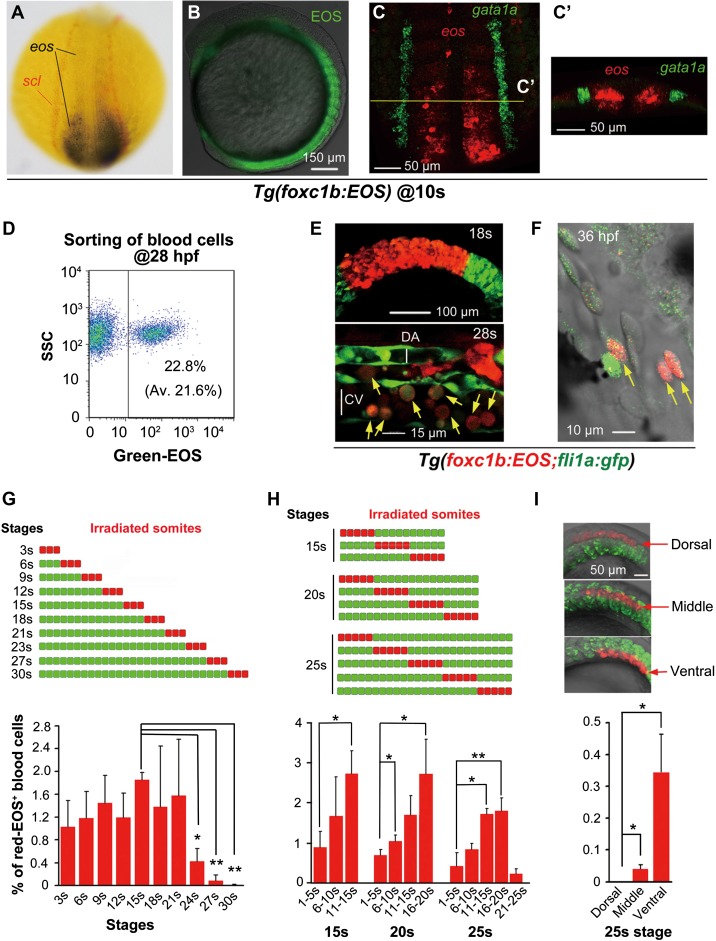
Figure 1.
Stage- and position-dependent hematogenic activity of somites. (A) Double in situ hybridization patterns of scl (red) and eos (black/blue) in a dorsally viewed Tg(foxc1b:EOS) embryo at the 10s stage. (B) EOS protein fluorescence in somites and paraxial mesoderm in a laterally viewed Tg(foxc1b:EOS) embryo at the 10s stage. (C and C’) Double fluorescence in situ hybridization patterns of eos (red) and gata1a (green) in a Tg(foxc1b:EOS) embryo at the 10s stage. The confocal image of trunk region was dorsally viewed (C) with an optical cross section showed in C’. (D) A representative FACS result of green-EOS+ blood cells from 10 Tg(foxc1b:EOS) embryos. The average from three independent experiments was shown in parenthesis. (E and F) green-EOS in five pairs of somites in Tg(foxc1b:EOS;fli1a:gfp) embryos was converted to red-EOS by irradiation at the 18s stage (top) and the resulted red-EOS+ cells (indicated by arrows) were found in the ICM at the 28s stage (bottom) (E) and in the heart (F). DA and CV represent the forming dorsal aorta and cardinal vein. (G−I) In Tg(foxc1b:EOS) embryos, three nascent somites at different stages (G), a group of 5 somites along the anteroposterior axis at different stages (H), or different portions of five nascent somites along the dorsoventral axis (I) were irradiated (top) and red-EOS+ blood cells were analyzed by flow cytometry at 30−36 hpf. Data were averaged from three independent experiments with 5−10 embryos each. Statistical significance: *P < 0.05; **P < 0.01.
EOS protein in specific somites or a group of somitic cells in Tg(foxc1b:EOS) could be efficiently photoactivated by near UV light (405 nm), allowing fast switch from green (green-EOS) to red fluorescence (red-EOS) (Supplementary Figure S6), which made it possible to track the lineages of spatiotemporally photoactivated red-EOS+ cells. After irradiating the middle region of somites, we observed migration of photoactivated somitic cells towards the midline, which might include cells that were differentiating into hematopoietic and sclerotomal fates (Supplementary Movie S4). Then, we irradiated somites of Tg(foxc1b:EOS;fli1a:gfp) double-transgenic embryos at the 18-somite (18s) stage (18 hpf), and subsequently found red-EOS+, round-shaped hematopoietic cells in the forming lumen of the dorsal aorta and the cardinal vein at the 28s stage (23 hpf) (Figure 1E). Occasionally, a few endothelial cells were red-EOS+ (Supplementary Figure S7). At ~36−40 hpf, the red-EOS+ hematopoietic cells were circulating through the heart (Figure 1F and Supplementary Movie S5). Given that the aorta-derived dHSCs start to merge at 30 hpf (Bertrand et al., 2008, 2010; Kissa et al., 2008; Kissa and Herbomel, 2010), those red-EOS+ hematopoietic cells are derived from somites well before dHSCs emergence, thus belonging to a previously unidentified type of hematopoietic cells.
Young somites contain a subset of hematogenic cells
To determine hematogenic competence of somitic cells at different stages, we photoactivated three pairs of nascent somites from 3s (11 hpf) to 30s (24 hpf) stages in Tg(foxc1b:EOS) embryos and then calculated the proportion of the red-EOS+ hematopoietic cells taken from the heart at 30−36 hpf by flow cytometry. Results indicated that nascent somites after the 21s (19.5 hpf) stage showed a marked reduction of hematogenic competence (Figure 1G). Then, we investigated hematogenic competence of somites at different positions along the anteroposterior axis at fixed stages, i.e. 15s (16.5 hpf), 20s (19 hpf), and 25s (21.5 hpf) stages. We found that somites located more posteriorly (newly formed) generally produced more hematopoietic cells, which suggests a gradual loss of hematopoietic competence of somitic cells during differentiation (Figure 1H). To know the compartmental origin of somite-derived blood cells along the dorsoventral axis, the dorsal (above the notochord), middle (parallel to the notochord), and ventral (below the notochord) portions of posterior somites were irradiated laterally at the 25s stage, respectively, and the resulted red-EOS+ hematopoietic cells were analyzed at 30−36 hpf. Results revealed that at this stage, ventral somitic cells had high hematogenic activity and middle somitic cells had low hematogenic activity, while dorsal somitic cells rarely differentiated into hematopoietic cells (Figure 1I). However, by irradiating the dorsal portion of all somites at the 15s stage (16.5 hpf), we found 0.365% of red-EOS+ hematopoietic cells in the circulation at 30 hpf (Figure 2A–D). Nevertheless, these data collectively suggest that the hematogenic activity of newly formed somites gradually decreases along the ventrodorsal axis. It is worth noting that lateral irradiation of the dorsal portion of somites would definitely not flash the LPM-derived hematopoietic cells, and hence any photoconverted circulating blood cells in this case should be the derivatives of somites rather than the LPM. Besides, we downward irradiated a single somite (eighth) on one side of Tg(foxc1b:EOS;fli1a:gfp) embryos at the 20s stage where the ICM has formed in the midline and thus would not be irradiated and subsequently found red-EOS+ hematopoietic cells within the forming vessel (e.g. Figure 2E–M). Again, this observation supports the idea that red-EOS+ hematopoietic cells are derived from somitic cells instead of the LPM cells.
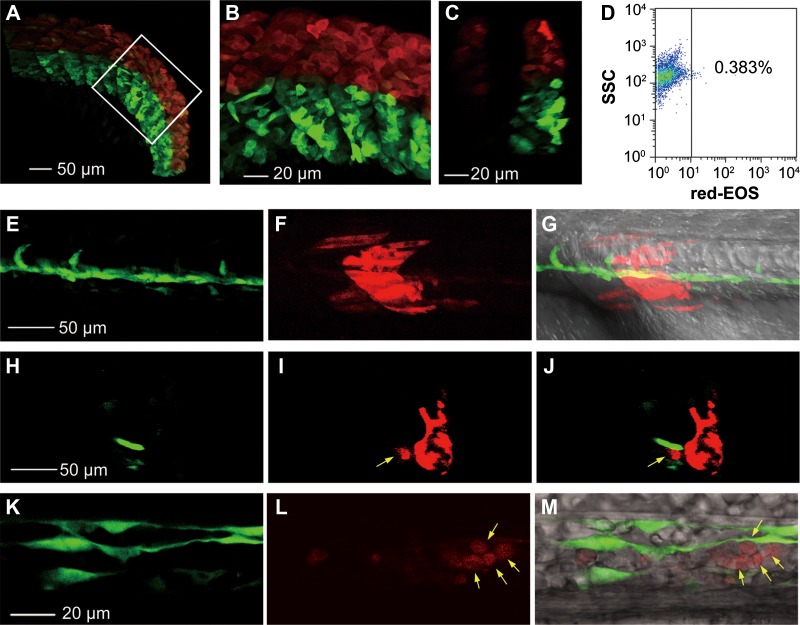
Figure 2.
Photoconverted hematopoietic cells are derived from somites but not LPM-derived blood precursors. (A−D) Dorsal somitic cells have a certain hematogenic activity at the 15s stage. The dorsal portion (above the notochord) of all somites in Tg(foxc1b:EOS) embryos was irradiated to avoid flashing the LPM-derived hematopoietic precursors. The posterior trunk region was shown (A) with the boxed area enlarged (B) and an optical cross section shown in C. The blood cells taken from 10 irradiated embryos in group at 30 hpf were analyzed by flow cytometry for red-EOS+ cells (D). (E−M) A downward irradiated single somite generates hematopoietic cells. The eighth somite on one side of a Tg(foxc1b:EOS;fli1a:gfp) embryo at the 20s stage was irradiated from the dorsal downward and the embryo was observed laterally (anterior to the left) by confocal microscopy at the 28s stage. (E−G) Trunk region focusing on the notochord. (H−J) An optical cross section from (E to G) with a photoconverted blood cell (indicated by an arrow) within the vessel. (K−M) An enlarged ICM region. Photoconverted blood cells were indicated by arrows.
A subset of ventral somitic cells start to differentiate into sclerotomal cells ~2−3 h after a somite is formed (Morin-Kensicki and Eisen, 1997). We suspected that hematogenic somitic cells may include sclerotomal progenitors. To test this hypothesis, we generated a Tg(twist1a:EOS) transgenic line using the promoter of twist1a, an early sclerotomal marker (Yeo et al., 2007). As revealed by double in situ hybridization, the scl-labeled LPM was lateral to the expression domain (in the paraxial mesoderm) of the reporter eos mRNA in Tg(twist1a:EOS) embryos at 5s (11.7 hpf) and 10s (14 hpf) stages (Supplementary Figure S8A−D). The fluorescence of EOS protein in transgenic embryos was detected in presumptive sclerotomes at as early as the 6s stage (12 hpf) and retained at later stages (Supplementary Figure S8E−H’). The Tg(twist1a:EOS) embryos at 28 hpf and adults carried 3.23% and 0.04% of green-EOS+ hematopoietic cells, respectively (Supplementary Figure S8I and J). Photoconversion analysis in Tg(twist1a:EOS;fli1a:gfp) double-transgenic embryos also confirmed the hematogenic capacity of sclerotomal cells (Supplementary Figure S8G). Besides, we obtained a Tg(pax1a:gfp) transgenic line that expresses GFP in migrating sclerotomal cells (Supplementary Figure S9B−D, F−H, and J−L). As revealed by flow cytometry, on average 7.21% of circulating hematopoietic cells in Tg(pax1a:gfp) embryos at 28 hpf were GFP+ (Supplementary Figure S9N), and these cells could be seen in the heart at 36 hpf (Supplementary Figure S9M). GFP+ cells were found even in the blood and kidney marrow of adult Tg(pax1a:gfp) fish (Supplementary Figure S9O).
Taking these data together, we believe that hematogenic somitic cells include but are not limited to sclerotomal cells. As detailed below, these somite-derived hematopoietic precursors possess features of HSCs and thus are named somite-derived hematopoietic precursors and stem cells (sHPSCs).
Somite hematopoiesis is independent of LPM hematopoiesis and vasculature
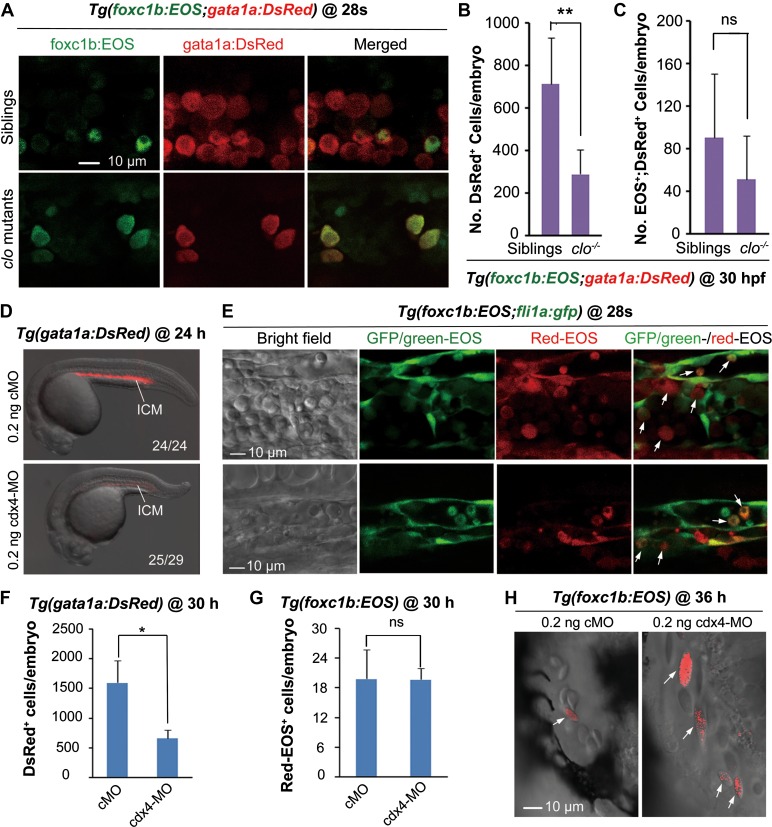
We were interested in potential relationship between somite hematopoiesis and classic LPM primitive hematopoiesis or AGM definitive hematopoiesis. Zebrafish cloche (clo) mutants are defective for early endothelial and hematopoietic lineages (Stainier et al., 1995). We found that the number of somite-derived (foxc1b:EOS+) gata1a+ blood cells in clo mutants was not significantly reduced (Figure 3A and C), contrasting to a significant reduction of total number of gata1a+ cells (Figure 3B). In zebrafish, cdx4 is an essential gene for primitive hematopoiesis (Davidson et al., 2003). Knockdown of cdx4 did not interfere in the generation of sHPSCs in Tg(foxc1b:EOS) embryos (Figure 3E, G, and H), although cdx4 knockdown led to a significant reduction in the number of gata1a:DsRed+ blood cells (Figure 3D and F). Besides, knockdown of trim33/tif1γ/mon, which is essential for primitive erythropoiesis (Ransom et al., 2004), had little effect on somite hematopoiesis either (Supplementary Figure S10). These results suggest that somite hematopoiesis and LPM primitive hematopoiesis are distinct hematopoietic processes.
Figure 3.
Somite hematopoiesis is unaffected in embryos lacking primitive hematopoietic cells. (A−C) Somite-derived blood cells existed in clo mutants. (A) Confocal microscopic observation of somite-derived blood cells in a segment of the ICM in Tg(foxc1b:EOS;gata1a:DsRed);clo mutant and sibling. (B and C) Flow cytometry revealed that the number of DsRed+ cells in clo mutants was significantly reduced (B) whereas the number of EOS+;DsRed+ blood cells was not changed (C). The cell number per embryo for each type was averaged from 10 embryos. **P < 0.01; ns, non-significance (P > 0.05). (D and F) The number of gata1a+ primitive hematopoietic cells was significantly reduced in Tg(gata1a:DsRed) cdx4 morphants as indicated by confocal observation (D) and flow cytometry (F). (D) The ratio of embryos with the representative pattern was indicated. (F) Circulating blood cells taken from a pool of five embryos at 30 hpf were sorted by flow cytometry and the number of DsRed+ blood cells per embryo was calculated. *P < 0.05. The average was based on three independent experiments. (E, G, and H) The generation of somite-derived Red-EOS+ blood cells was unaffected in cdx4 morphants. Tg(foxc1b:EOS;fli1a:gfp) (B), or Tg(foxc1b:EOS) (D and E) embryos were injected with cMO or cdx4-MO at the one-cell stage. Five pairs of nascent somites of the injected embryos were photoactivated at the 20s stage, followed by confocal observation of Red-EOS+ cells (indicated by arrows) in the ICM at the 28s stage (E) and in the heart at 36 hpf (H) and by sorting of Red-EOS+ blood cells (F and G). ns, statistically non-significance (P > 0.05).
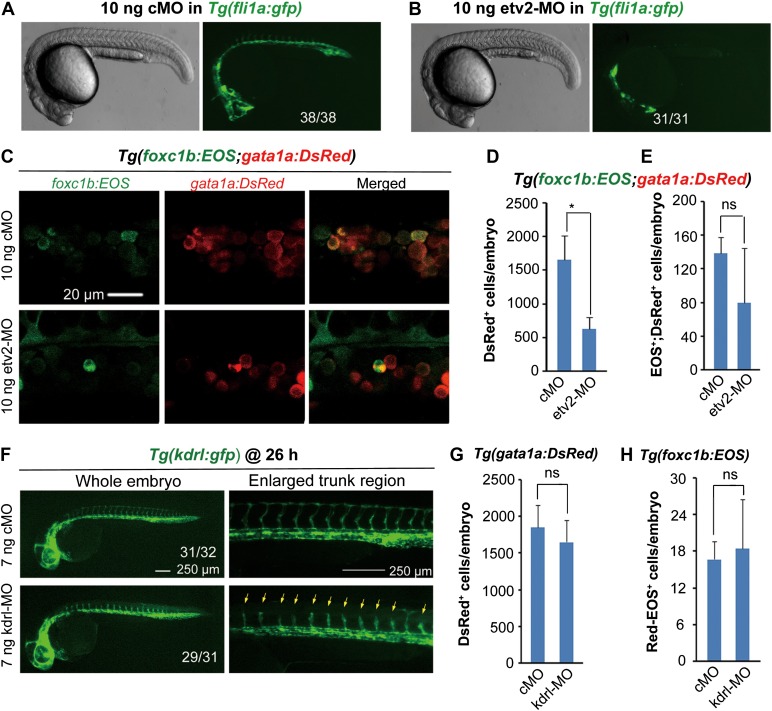
We next asked whether the embryonic vasculature, which can generate dHSCs from the dorsal aorta, plays a role in somite hematopoiesis. Zebrafish embryos depleted of etv2/etsrp fail to form vasculature, leading to a complete loss of circulation (Sumanas and Lin, 2006). We found that Tg(foxc1b:EOS) embryos still gave rise to sHPSCs in the absence of the dorsal aorta and cardinal vein when etv2 was knocked down (Figure 4A–E). Knockdown of kdrl, the VEGF receptor gene required for vascular formation (Covassin et al., 2006, 2009), did not affect sHPSCs (Figure 4F–H). These results together indicate that initial somite hematopoiesis is independent of LPM and aorta hematopoiesis.
Figure 4.
Vasculature imperfection has no impact on hematopoiesis. (A and B) Effect of etv2 knockdown on vasculature formation. Note that etv2 morphants had normal morphology but lacked both dorsal aorta and cardinal vein in the trunk region. The ratio of embryos with the representative pattern was indicated. cMO, control morpholino. (C) foxc1b:EOS;gata1a:DsRed double-positive cells in the ICM of etv2 morphant or control at the 28s stage. (D and E) Sorting results of gata1a:DsRed+ (D) and fox1b:EOS+;gata1a:DsRed+ blood cells (E) in etv2 and control morphants at 30 hpf. The fluorescent cell number per embryo was averaged from three independent experiments (10 embryos each). *P < 0.05; ns, non-significance (P > 0.05). (F) kdrl knockdown impaired growth of intersegmental vessels (indicated by arrows). The ratio of affected embryos was indicated. (G and H) Effect of kdrl knockdown on LPM hematopoiesis (G) and somite hematopoiesis (H). For the injected Tg(foxc1b:EOS) embryos, five pairs of new born somites were photoactivated at the 20s stage. All embryonic cells were collected from a group of five embryos at 30 hpf and were sorted by flow cytometry for DsRed+ (G) or red-EOS+ cells (H). The number of cells with expected fluorescence per embryo was averaged from three independent experiments. ns, non-significance (P > 0.05).
The somite-derived hematopoietic cells preserve expression of many muscle/skeletal genes
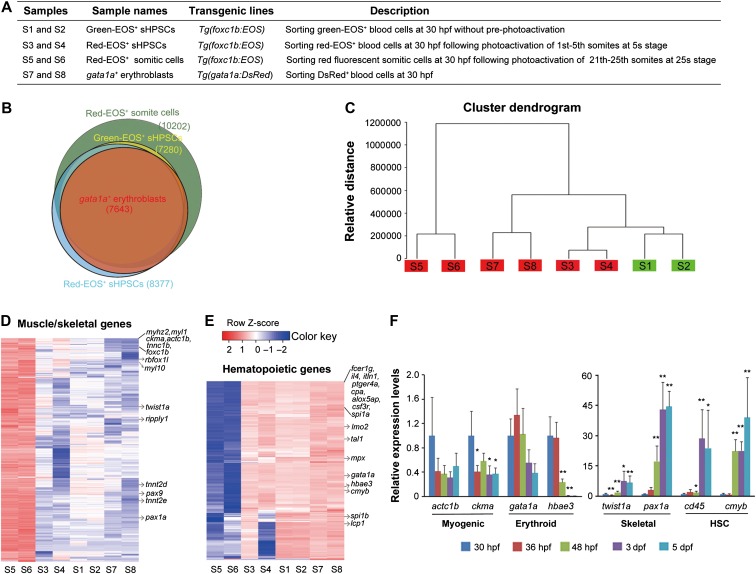
To investigate molecular features of embryonic sHPSCs, we performed RNA-seq analysis of four types of cells, with two independent samples for each, including non-photoactivated green-EOS+ sHPSCs (S1 and S2) from Tg(foxc1b:EOS) embryos, photoactivated red-EOS+ sHPSCs (S3 and S4) from Tg(foxc1b:EOS) embryos, red-EOS+ somitic cells (S5 and S6) from Tg(foxc1b:EOS) embryos, and gata1a+ (DsRed) blood progenitors (S7 and S8) from Tg(gata1a:DsRed) embryos, all of which were sorted by FACS at 30 hpf (Figure 5A). The number of genes detected in somitic cells was higher (10202 genes with fragments per kilobase of transcript per million fragments mapped (FPKM) ≥ 1 in both duplicates) than that (7280−8377 genes with FPKM ≥ 1 in both duplicates) in any of the other three types of hematopoietic cells (Figure 5B and Supplementary Table S1). Unsupervised hierarchical clustering of transcriptomes revealed that duplicates of the same cell type were most similar to each other (Figure 5C), indicative of high RNA-seq quality. The transcriptome of green-EOS+ sHPSCs clustered soon with red-EOS+ sHPSCs, suggesting that green-EOS+ blood cells in Tg(foxc1b:EOS) embryos are derived from somites rather than from LPM-derived blood cells. The gene expression patterns of foxc1b+ sHPSCs resembled those of gata1a+ cells, suggesting that the two types share common hematopoietic features (Figure 5C).
Figure 5.
Transcriptome profiling of nascent sHPSCs. (A) Description of cell samples used for RNA-seq analysis. (B) Overlapping of genes detected among four types of cell samples. Each circle reflected the genes in one type of cell samples, which were detected in both duplicates. (C) Unsupervised clustering of the transcriptomes of samples. Genes detected with FPKM > 0.1 in at least one sample were used for clustering. (D and E) Heatmaps showing expression patterns of 3056 muscle/skeletal genes (D) or 187 hematopoietic genes (E) differentially expressed between somite and gata1a+ cell types. Some well-known genes were indicated. (F) qRT-PCR analysis of a set of marker genes for their expression levels in green-sHPSCs hematopoietic cells sorted from the heart of Tg(foxc1b:EOS) embryos at indicated stages. Data were shown as the mean of triple samples with the error bar. Statistical significance levels (compared to the 30-hpf sample): *P < 0.05; **P < 0.01.
For comparison, we picked up 187 hematopoietic genes and 3056 muscle/skeletal-related genes that showed significantly different expression levels (fold change > 3, false discovery rate (FDR) < 0.05) between somite cell type and gata1a+ cell type. Generally, the expression levels of these genes in sHPSCs were closer to those in gata1a+ cell type (Figure 5D and E). Compared to the gata1a+ cell type, both green-EOS+ and red-EOS+ sHPSCs had higher expression levels of the muscle/skeletal-related genes (e.g. fmyhc2.1/myhz2, myl1, ckma, acta1b, tnnc1b, myl10, twist1a, pax9, and pax1a) (Figure 5D). However, both sHPSC types exhibited higher expression levels of the hematopoietic genes (e.g. fcer1g, spi1a, lmo2, tal1, mpx, gata1a, hbae3, cmyb, spi1b, and lcp1) compared to the somite cell type (Figure 5B). Thus, the sHPSCs have acquired hematopoietic characteristics and retained some muscle/skeletal markers. We note that the sHPSC types showed extremely low expression levels of the endothelial genes such as kdrl, flt1, flt4, and cdh5, which excludes the possibility of the endothelial origin. Compared to the red-EOS+ sHPSCs, the green-EOS+ sHPSCs displayed expression patterns of hematopoietic genes more like gata1a+ cells, which was presumably caused by earlier hematopoietic transformation of some green-EOS+ somitic cells than the photoactivated somitic cells. We further examined, by qRT-PCR, the expression levels of a set of genes for muscle (actc1b and ckma), sclerotome (twist1a and pax1a), hematopoietic precursors (cd45 and cmyb), and erythrocytes (gata1a and hbae3) in green-EOS+ sHPSCs sorted from Tg(foxc1b:EOS) embryos or larvae from 30 hpf to 5 days postfertilization (dpf). As shown in Figure 5F, actc1b and ckma expression levels started to drop at 36 hpf, while twist1a and pax1a expression levels started to rise at 2 dpf. The expression of gata1a and hbae3 started to decrease at 3 dpf and 2 dpf, respectively, but cd45 and cmyb markedly increased at 3 dpf and 2 dpf, respectively. It appears that following emergence of sHPSCs, they acquire more molecular natures of HSCs with increased expression of skeletal markers but progressive weakening or loss of muscle signatures.
The somite-derived primitive hematopoietic cells may exist in adult fish
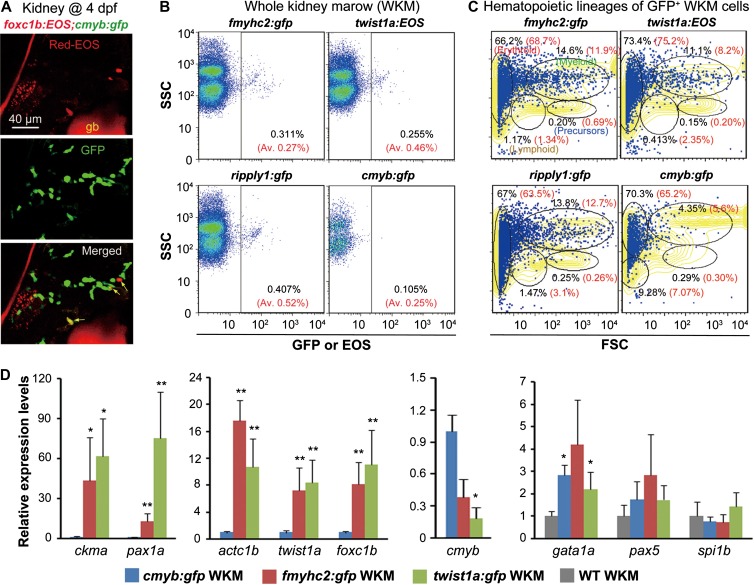
The sHPSCs are early hematopoietic cells. However, Tg(ripply1;gfp), Tg(fmyhc2:gfp), Tg(twist1a:EOS), and Tg(pax1a:gfp) adults contain circulating GFP+ blood cells, despite being low in number (Supplementary Figures S1G, S3F, S8J, and S9O). We wondered whether sHPSCs are truly HSCs with self-renewing potential and pluripotency and could last for life time. We found that photoconverted EOS-expressing somite-derived foxc1b+ cells existed in the kidney of Tg(foxc1b:EOS;cmyb:gfp) transgenic larvae at 4 dpf (Figure 6A), suggesting homing of sHPSCs in the kidney, a typical characteristic of fish dHSCs. Then, we checked the existence of fluorescent cells, putative sHPSCs descendants of sHPSCs, in the adult kidney of Tg(fmyhc2:gfp), Tg(twist1a:EOS), and Tg(rippy1:gfp) transgenic lines by flow cytometry using whole kidney marrow (WKM) of individual fish. We could not find any EOS+ cells in the circulation in Tg(foxc1b:EOS) adults and thus did not check this line. The EOS+ or GFP+ cells accounted for an average of 0.27%, 0.46%, and 0.52% (n = 3 adults) of WKM cells in Tg(fmyhc2:gfp), Tg(twist1a:EOS), and Tg(rippy1:gfp) transgenic lines, respectively, which were comparable to 0.25% of GFP+ WKM cells in Tg(cmyb:gfp) fish (Figure 6B). Further flow cytometry analysis of sorted fluorescent WKM cells indicated the presence of HSCs/precursors as well as erythroid, lymphoid, and myeloid lineages (Figure 6C). Compared to the GFP+ WKM cells sorted from Tg(cymb:gfp), the EOS+ or GFP+ WKM cells sorted from Tg(twist1a:EOS) or Tg(fmyhc2:gfp) showed significantly higher levels of actc1b, ckma, twist1a, pax1a, and foxc1b expression, lower level of cmyb expression, and comparable levels of gata1a (erythrocytes), pax5 (lymphocytes), and spi1b (myelocytes/lymphocytes) expression (Figure 6D). These data imply that embryonic hematopoietic cells may become HSCs but with distinct characteristics. However, we could not exclude the possibility that HSCs de novo express GFP or EOS in adult kidney in these lines.
Figure 6.
Analysis of sHPSCs in larvae and adults. (A) Co-colonization of foxc1+ sHPSCs (indicated by arrows) with cmyb+ HSCs in the kidney of a Tg(foxc1b:EOS;cmyb:gfp) transgenic larva, which was photoactivated in five pairs of posterior somites at the 25s stage. gb, gall bladder with red autofluorescence. (B) Detection of sPHSCs by flow cytometry in adult (3–6 months old) kidney marrow of various transgenic lines. The representative result was shown for one fish, and the average ratio (Av.) of EOS+ or GFP+ sHPSCs from three fish was indicated in parenthesis. (C) EOS+ or GFP+ sHPSCs sorted from the adult kidney marrow of a representative fish in various transgenic lines were re-analyzed by forward and side scatter. Different scatter fractions were circled with indicated percentage of cells. The yellowed area represented the sorting result of the WKM. The average percentage from three analyzed fish was written in red. (D) Expression levels of a set of muscle/skeletal or hematopoietic marker genes in twist1a+ or fmyhc2+ sHPSCs were analyzed by qRT-PCR compared to cmyb+ hematopoietic cells. GFP+ cells were sorted from the kidney marrow of corresponding transgenic adult fish. Data were shown as means from three experiments with the error bar indicated. *P < 0.05; **P < 0.01.
The somite-derived embryonic hematopoietic cells can become dHSCs
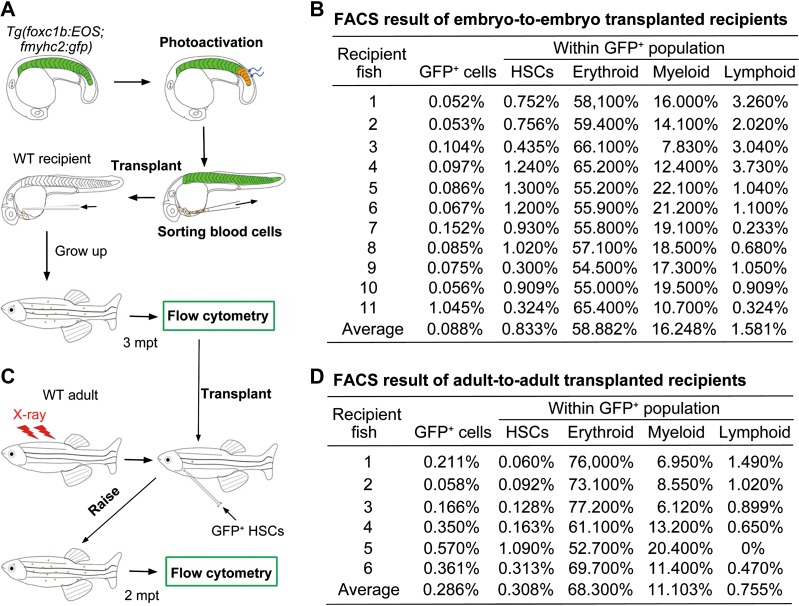
The next important question is whether sHPSCs in adults are descendants of embryonic sHPSCs via self-renewal. To address this issue, we performed transplantation of sHPSCs (Figure 7A) along with the transplantation of gata1a+ embryonic erythroblasts as controls. When gata1a+ erythroblasts sorted from 28-hpf Tg(gata1a:DsRed) transgenic embryos were transplanted into 36-hpf wild-type recipient embryos (> 100 donor cells per recipient), the donor gata1a+cells could be detected by flow cytometry in recipients at 3 dpf but were no longer detected (n = 15) by 7–9 dpf, suggesting their incapability of survival and self-renewal. For sHPSC transplantation, an average of 1.9 red-EOS+;GFP+ sHPSCs sorted from photoactivated Tg(foxc1b:EOS;fmyhc2:gfp) embryos at 28−30 hpf were injected into the circulation of a recipient embryo at 36 hpf. Among 78 recipients, 40 survived to adulthood (3 months old). We sacrificed 28 recipient adults for flow cytometry analysis of the kidney marrow and found that 11 of them harbored 0.052%–1.045% of GFP+ cells (Figure 7B), suggesting that embryonic sHPSCs survive for life time. The isolated GFP+ cells were subjected to a second-round flow cytometry and were found to contain all hematopoietic lineages with a stem cell proportion of 0.3%−1.3% (Figure 7B). Therefore, the sHPSCs of embryonic origin have hematopoietic pluripotency. To exclude the possibility that embryonic sHPSC donor cells may contain other uncharacterized types of stem cells, we performed a second-round transplantation using the GFP+ stem cells isolated from the kidney of the recipient adults. Three to five GFP+ stem cells along with ~30000 untreated helper WKM cells were transplanted into one X-ray-irradiated wild-type adults (Figure 7C). Two months post-transplantation, 6 of 18 survived adults (out of 40 recipients in total) were found to have 0.058%–0.57% of GFP+ cells in the kidney marrow and these GFP+ cells included all hematopoietic lineages (Figure 7D). Taking these results together, we conclude that embryonic sHPSCs can become everlasting HSCs.
Figure 7.
Embryonic sHPSCs become definitive HSCs in adult fish. (A and C) Illustration of between-embryo (A) or between-adult (C) transplantation of sHPSCs. (A) Five nascent somites of Tg(foxc1b:EOS;fmyhc2:gfp) embryos at the 20s stage were photoactivated, and red-EOS+/GFP+ circulating blood cells were sorted at 28−30 hpf and transplanted into 36-hpf wild-type (WT) recipient embryos (1.9 cells per recipient). (C) The GFP+ HSCs sorted from the transplanted recipient adults were re-transplanted into X-ray irradiated WT adults (3−5 GFP+ cells per recipient). (B and D) The kidney marrow of individual recipient adults was analyzed by flow cytometry for GFP+ cells and hematopoietic lineages. The between-embryo (B) and between-adult (D) transplanted recipients were analyzed 3 months and 2 months post-transplantation, respectively.
Discussion
Our data indicate that somitic cells in zebrafish embryos are able to transdifferentiate into hematopoietic cells, highlighting the somite as a novel embryonic hematopoietic site. Importantly, these early embryonic hematopoietic cells become definitive HSCs that provide all hematopoietic lineages throughout the lifespan. This finding challenges the long-existing view that early embryonic hematopoiesis only generates transitory hematopoietic cell populations for embryonic development (Dieterlen-Lievre, 1975; Orkin and Zon, 2008).
We used transgenic embryos expressing photoconvertible EOS in somites to investigate the hematogenic activity of somites. It is unlikely that the sHPSCs we identified are of the LPM origin. First, lateral irradiation of the middle region of somites at the 25s stage (Figure 1I) or the dorsal region of somites at the 20s stage (Figure 2A–D) in Tg(foxc1b:EOS) embryos would not flash the LPM-derived hematopoietic cells, because the latter are located ventral to the notochord level. The downward irradiation of single somites at 20s would also avoid flashing the ICM beneath the notochord. The red-EOS+ hematopoietic cells following these irradiation should arise from somites. Second, knockdown of the hematopoietic transcription factor cdx4 has little effect on the number of sHPSCs (Figure 3E, G, and H) but leads to a sharp reduction in the number of the LPM-derived gata1a+ primitive blood cells (Figure 3D and F), suggesting that sHPSCs and primitive blood cells are regulated differently. Third, the sorted sHPSCs express high levels of many muscle/skeletal markers, which are different from the LPM-derived gata1a+ hematopoietic cells (Figure 5C−F, and Supplementary Table S1). Recently, Feng Liu's group sorted transforming endothelial cells (to be HSCs) of the dorsa aorta from zebrafish Tg(kdrl:mCherry;runx1:gfp) transgenic embryos at 28 hpf and performed RNA-seq analysis (Zhang et al., 2015). Their data showed that the muscle/skeletal genes myl10, tpm2, tnnt2e, tnnt2d, tnnc1b, smyhc3, smyhc1, smyhc2, ckma, actc1b, pax1, and twist1 have low levels of expression in kdrl;runx1 double-positive cells, while the endothelia/hematopoietic genes kdrl, flt1, efnb2b, cmyb, ptprc/cd45, and gata1a express at high levels. Therefore, somite-derived hematopoietic cells have characteristics different from endothelia-derived kdrl;runx1-positive cells. Finally, the LPM-derived primitive hematopoietic precursors are believed to die during larva stages (Orkin and Zon, 2008; Tavian et al., 2010; Jagannathan-Bogdan and Zon, 2013). However, the embryonic sHPSCs could survive to the adulthood after being transplanted (Figure 7), differing from embryonic gata1a+ cells.
We have shown that sHPSCs in the adult can differentiate into multiple hematopoietic lineages. However, it remains unclear whether these cells have preference in differentiation in response to specific stimulations or stress. We observed that 5 out of 40 surviving adult fish that received transplanted sHPSCs from Tg(foxc1b:EOS; fmyhc2:gfp) embryos have GFP+ cells with unknown identities in the anal fin or tail fin (data not shown), while none of the adult fish receiving either cmyb:GFP+ HSCs (n = 28) or runx1:GFP+ HSCs (n = 35) at embryonic stages exhibits similar phenomena. We also observed that following tail fin cut of Tg(pax1a:gfp) transgenic adult fish, the GFP+ blood cells were rapidly expanded in the peripheral blood and subsequently regenerated fin bones were GFP+ (data not shown). The potential bias during differentiation of adult sHPSCs needs to be investigated in depth in the future.
Currently, it remains elusive whether embryonic hematopoiesis takes place in mammalian somites. It has been well demonstrated that the AGM region in mouse embryos around Day 10 possesses the highest hematopoietic activity (Medvinsky et al., 1993; Muller et al., 1994; Medvinsky and Dzierzak, 1996). It is possible that some somite-derived, hematopoietic fate-committed cells have already migrated into the AGM region at or before Day 10 and these cells transform into dHSCs. This possibility may be explored by carefully tracing the migrating routes of genetically labeled somitic cells in mouse embryos. According to the GEO database of NCBI (http://www.ncbi.nlm.nih.gov/geoprofiles/), human and murine adult bone marrow-derived HSCs express certain levels of the muscle/skeletal genes CKM, ACTC1, MYH4, TNNC1, TNNC2, RIPPLY, PAX1, TWIST1, TWIST2, NKX3.2, and so on (Scandura et al., 2004; Bystrykh et al., 2005; Rossi et al., 2005; Pellagatti et al., 2010; Pang et al., 2011). Human HSCs with high expression levels of these genes may represent a distinct subtype of HSCs and could have important clinical applications.
Materials and methods
Zebrafish strains and transgenic lines
The Tubingen wild-type strain was used. The transgenic lines Tg(fli1a:gfp) (Lawson and Weinstein, 2002), Tg(gata1a:DsRed) (Traver et al., 2003), and Tg(kdrl:gfp) (Jin et al., 2005) are described previously. The clochem39 was kindly provided by Dr Jing-Wei Xiong (Xiong et al., 2008). The transgenic line Tg(ripply1:gfp) was screened and established from the founders, which were injected with a Tol2-based construct consisting of 1.2-kb ripply1 promoter and gfp-coding sequence. The Tg(rbfox1l:gfp) line with an original code Bgs054 was established during a Tol2 transposon-mediated gene trap screen in our laboratory (Zhao et al., 2008), in which the gfp-coding sequence is inserted into the first intron of the rbfox1l locus. The Tg(fmyhc2:gfp) line was established during a Tol2 transposon-mediated enhancer trap screen, which was carried out by Dr Bo Zhang's laboratory at Peking University (Xue et al., 2010), and originally coded as mp523. We found that the Tol2 element in Tg(fmyhc2:gfp) line was located 2 kb downstream of 3′UTR of fmyhc2.1 and 2 kb upstream of 5′UTR of the fmyhc2.2. The 3.6-kb foxc1b promoter, 4.9-kb twist1a promoter, and 2.4-kb pax1a promoter upstream of the translation start site were used to generate Tg(foxc1b:EOS), Tg(twist1a:EOS), and Tg(pax1a:gfp) transgenic lines, respectively. When needed, fish from different transgenic lines and clo mutant line were crossed to obtain double-transgenic embryos or transgenic embryos in the mutant background. Embryos were raised in Holtfreter's solution at 28.5°C. Ethical approval was obtained from the Animal Care and Use Committee of Tsinghua University.
Photoconversion of EOS in live transgenic embryos
The target area of live Tg(foxc1b:EOS) and Tg(twist1a:EOS) embryos was flashed by 405 nm laser for 10 sec using an Olympus FV1200 Fluoview scanning confocal microscope. Unless otherwise stated, five nascent somites of an embryo, which was laterally orientated, were irradiated at a desired stage and then analyzed at later stages.
Microscopic observation and time-lapse imaging of live transgenic embryos
Embryos were dechorionated with pronase at desired stages, anaesthetized with 0.02% tricaine (Sigma) and mounted in desired orientation in 1% low melting point agarose. The embryos were then observed at 28.5°C under an Olympus FV1200 Fluoview scanning confocal microscope with 30× silicone oil objective. For time-lapse imaging, Z-stacks were acquired with a step size of 2 μm in an interval of 4−6 min over several hours. Images were three-dimensionally rendered and analyzed using Imaris software (Bitplane) and FV10-ASW 4.1 Viewer (Olympus). The Z-projections of the confocal stacks, cell migration tracing, and cross-section (segmentation) were performed with Imaris as well. The fluorescence of live embryos immobilized in 5% methylcellulose was also observed under a Zeiss SteREO Discovery V20 microscope.
Flow cytometry
For collecting embryonic blood, dechorionated, anaesthetized live embryos at 28−30 hpf were placed into blood buffer (0.9× PBS) containing 5% fetal calf serum and 3 U/ml heparin). Then, the heart chamber of individual embryos was torn using a syringe, allowing blood cells to flow into the buffer. Adult blood was obtained by cardiac puncture with a 10-μl micropipette tip coated with heparin and immediately sent into ice-cold blood buffer. For collecting kidney cells, the kidney of individual adult fish was taken out and placed into ice-cold blood buffer. The kidney was then disintegrated by gently absorbing and pushing out through a 1-ml syringe needle. Single-cell suspension was obtained by passing through a 40-μm nylon mesh filter (BD). For collecting red-EOS+ somitic cells, five nascent pairs of somites of Tg(foxc1b:EOS) transgenic embryos at the 25s stage were photo-switched from green to red fluorescence. When they developed to 30 hpf, their yolk was removed by pipetting through a 200-μl tip in Holtfreter's solution. The embryonic bodies were then dissociated in PBS containing 0.25% trypsin and 1 mM EDTA (pH 8.0) with gentle pipetting. For sorting gata1a+ blood cells from embryos without the intact vascular system (clo mutants and etv2 morphants), individual embryos were similarly processed to dissociate cells. The other steps were the same for kidney marrow cell preparation.
For fluorescence-activated cell sorting (FACS), 1 g/ml of DAPI was added to the cell suspension to exclude dead cells and debris. Flow cytometric acquisitions (FSC and SSC) were performed on a MoFlo XDP machine (BeckMan), and cell sorting was performed on a FACS ARIAIII (BD) cell sorter. The data were analyzed by using FlowJo software (Treestar).
Blood cell transplantation
Blood cells transplantation was performed essentially as described by Tamplin et al. (2015). For embryo-to-embryo transplantation, donor embryonic blood cells were taken from Tg(foxc1b:EOS;fmyhc2:gfp) double-transgenic embryos. Five pairs of new born somites of the transgenic embryos at about the 20s stage were illuminated by laser. When the photoactivated embryos developed to 28−30 hpf, blood cells were collected from the heart and subjected to sorting using the FACS ARIAIII cell sorter (BD). The collected red-EOS/GFP double-positive blood cells were resuspended in PBS at an estimated concentration of 1900 cells/μl. The cell suspension was injected into the sinus venosus of 36-hpf wild-type embryos using typical MPPI-2 quantitative injection equipment (Applied Scientific Instrumentation Co.) at a dose of 1 nl per embryo (1.9 cells). Of note, 40 out of 78 recipients survived to adulthood and 28 of surviving adults were subjected to cell sorting. When blood cells from Tg(gata1a:DsRed) embryos were used as donors, each recipient embryo received >100 DsRed+ blood cells. However, recipients of DsRed+ cells did not contain any donor cells as analyzed by FACS after ~1 week, and thus they were not analyzed later.
For adult-to-adult transplantation, GFP+ hematopoietic stem/precursor cells were collected following FACS sorting of the kidney marrow of embryo-to-embryo transplantation recipients at 3 months post-transplantation and used as donor cells. Wild-type adult fish to be used as recipients were first irradiated to ablate the blood forming system. A group of eight anaesthetized fish in a 50-ml centrifuge tube was placed in an RS 2000 X-ray irradiator (Rad Source Technologies, Inc.) and irradiated at a dose rate of 8 Gy/min. We determined that 30 Gy-irradiated fish had a survival rate <10% over 30 days, and thus 30 Gy was regarded as the minimum lethal dose (Traver et al., 2004). Each recipient fish was injected with 3−5 GFP+ HSCs together with ~30000 kidney marrow cells (as helper cells) derived from untreated wild-type adult fish. The kidney marrow cells of surviving recipients 2 months post-transplantation were collected for flow cytometry analysis.
RNA-seq library construction and next-generation sequencing
Total RNA was extracted from ~15000 blood cells or somitic cells sorted by FACS (~500 embryos) using RNeasy Micro Kit (QIAGEN), which gave ~150 ng RNA. Then, the RNA was used for library construction following the instructions of TruSeq RNA Sample Preparation Kit (Illumina), slightly modified to be suitable for low starting amount of total RNA. The libraries were sequenced on Illumina Hiseq2500 instrument and 100-bp pair-end reads passing the quality control were gained in fastq files.
RNA-seq data process and genes identification
Raw reads generated by Illumina sequencers were filtered with a QC pipeline in Perl to remove the low-quality reads with over 50% bases of quality value ≤5 and over 10% bases undetermined. The QC-passed reads were mapped to danRer7 downloaded from UCSC with TopHat2 (version 2.0.8b). Then, the gene expression levels were calculated using Cuffquant and Cuffnorm programs (Cufflinks 2.2.1).
By comparing the two groups of expression data, i.e. somitic cells and gata1a+ cells, with edgR in R-3.0.1, we defined genes with FPKM at least 8-fold higher in the somitic cell group (log2[FC(somitic+/gata1a+)] ≥3, P-value <0.05, FDR < 0.05) as somite genes. On the contrary, blood genes were those with FPKM at least 8-fold higher in the gata1a+ cell group (log2[FC(gata1a+/somitic+)] ≥3, P-value <0.05, FDR < 0.05).
Quantitative RT-PCR and whole-mount in situ hybridization
Total RNA was extracted from sorted cells using RNeasy Mini Kit (Qiagen) and treated by DNase I (Qiagen). cDNA synthesis and real-time PCR were performed as previously described. The relative mRNA expression level was normalized to the expression level of ef1a (Liu et al., 2013). The primer sequence information will be available upon request.
The whole-mount in situ hybridization was performed essentially following a standard protocol. High-resolution double fluorescent in situ hybridization was done as described by Brend and Holley (2009).
Morpholinos
To knock down gene expression, the standard control morpholinos (cMO) or gene-specific antisense MOs were purchased from Gene Tools. MOs were injected into one-cell stage embryos. The sequences of the morpholinos were as follows: cMO, 5′-CCTCTTACCTCAGTTACAATTTATA-3′; cdx4-MO, 5′-CGTACATGATTTGGAAGAAACCCCT-3′ (Davidson et al., 2003); trim33-MO, 5′-GCTCTCCGTACAATCTTGGCCTTTG-3′ (Monteiro et al., 2011); etv2-MO, 5′-CACTGAGTCCTTATTTCACTATATC-3′ (Sumanas and Lin, 2006); and kdrl-MO, 5′-CCGAATGATACTCCGTATGTCAC-3′ (Rottbauer et al., 2005).
Statistical analysis
All experiments were performed at least three times. The student's t-test (two-tailed, unequal variance) was used when two samples were compared. The standard error of the mean was calculated from the average of three samples in each case.
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Acknowledgements
The authors are grateful to Drs Zilong Wen (Hong Kong University of Science and Technology), Bo Zhang (Peking University), Shuo Lin (University of California, Los Angeles), Feng Liu (Institute of Zoology, Chinese Academy of Sciences), and Deli Shi (Shandong University) for providing transgenic zebrafish lines, and Dr Kenneth D. Poss (Duke University) for providing constructs. We thank the other members of the Meng Laboratory and staff at the Cell Facilities and Experimental Animals Center, Tsinghua University, for technical assistance.
Funding
This work was funded by the Major Science Programs of MOST of China (2012CB945101 and 2011CB943800), the National Natural Science Foundation of China (31221064), the China Postdoctoral Science Foundation (2013M540931), and the Tsinghua University Initiative Scientific Research Program (20141081299).
Conflict of interest: none declared.
References
- Bertrand J.Y., Chi N.C., Santoso B., et al. (2010). Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand J.Y., Kim A.D., Teng S., et al. (2008). CD41+ cmyb+ precursors colonize the zebrafish pronephros by a novel migration route to initiate adult hematopoiesis. Development 135, 1853–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brend T., and Holley S.A. (2009). Zebrafish whole mount high-resolution double fluorescent in situ hybridization. J. Vis. Exp. pii: 1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystrykh L., Weersing E., Dontje B., et al. (2005). Uncovering regulatory pathways that affect hematopoietic stem cell function using ‘genetical genomics’. Nat. Genet. 37, 225–232. [DOI] [PubMed] [Google Scholar]
- Cormier F., and Dieterlen-Lievre F. (1988). The wall of the chick embryo aorta harbours M-CFC, G-CFC, GM-CFC and BFU-E. Development 102, 279–285. [DOI] [PubMed] [Google Scholar]
- Covassin L.D., Siekmann A.F., Kacergis M.C., et al. (2009). A genetic screen for vascular mutants in zebrafish reveals dynamic roles for Vegf/Plcg1 signaling during artery development. Dev. Biol. 329, 212–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covassin L.D., Villefranc J.A., Kacergis M.C., et al. (2006). Distinct genetic interactions between multiple VEGF receptors are required for development of different blood vessel types in zebrafish. Proc. Natl Acad. Sci. USA 103, 6554–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A.J., Ernst P., Wang Y., et al. (2003). cdx4 mutants fail to specify blood progenitors and can be rescued by multiple hox genes. Nature 425, 300–306. [DOI] [PubMed] [Google Scholar]
- Detrich H.W. III, Kieran M.W., Chan F.Y., et al. (1995). Intraembryonic hematopoietic cell migration during vertebrate development. Proc. Natl Acad. Sci. USA 92, 10713–10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterlen-Lievre F. (1975). On the origin of haemopoietic stem cells in the avian embryo: an experimental approach. J. Embryol. Exp. Morphol. 33, 607–619. [PubMed] [Google Scholar]
- Gallagher T.L., Arribere J.A., Geurts P.A., et al. (2011). Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Dev. Biol. 359, 251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin I.E., Garcia-Porrero J.A., Coutinho A., et al. (1993). Para-aortic splanchnopleura from early mouse embryos contains B1a cell progenitors. Nature 364, 67–70. [DOI] [PubMed] [Google Scholar]
- Jagannathan-Bogdan M., and Zon L.I. (2013). Hematopoiesis. Development 140, 2463–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S.W., Beis D., Mitchell T., et al. (2005). Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 132, 5199–5209. [DOI] [PubMed] [Google Scholar]
- Jin H., Sood R., Xu J., et al. (2009). Definitive hematopoietic stem/progenitor cells manifest distinct differentiation output in the zebrafish VDA and PBI. Development 136, 647–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura A., Koshida S., Hijikata H., et al. (2005). Groucho-associated transcriptional repressor ripply1 is required for proper transition from the presomitic mesoderm to somites. Dev. Cell 9, 735–744. [DOI] [PubMed] [Google Scholar]
- Kissa K., and Herbomel P. (2010). Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464, 112–115. [DOI] [PubMed] [Google Scholar]
- Kissa K., Murayama E., Zapata A., et al. (2008). Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood 111, 1147–1156. [DOI] [PubMed] [Google Scholar]
- Lawson N.D., and Weinstein B.M. (2002). In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307–318. [DOI] [PubMed] [Google Scholar]
- Liu X., Xiong C., Jia S., et al. (2013). Araf kinase antagonizes Nodal-Smad2 activity in mesendoderm development by directly phosphorylating the Smad2 linker region. Nat. Commun. 4, 1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroto M., Bone R.A., and Dale J.K. (2012). Somitogenesis. Development 139, 2453–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maves L., Waskiewicz A.J., Paul B., et al. (2007). Pbx homeodomain proteins direct Myod activity to promote fast-muscle differentiation. Development 134, 3371–3382. [DOI] [PubMed] [Google Scholar]
- Medvinsky A., and Dzierzak E. (1996). Definitive hematopoiesis is autonomously initiated by the AGM region. Cell 86, 897–906. [DOI] [PubMed] [Google Scholar]
- Medvinsky A., Rybtsov S., and Taoudi S. (2011). Embryonic origin of the adult hematopoietic system: advances and questions. Development 138, 1017–1031. [DOI] [PubMed] [Google Scholar]
- Medvinsky A.L., Samoylina N.L., Muller A.M., et al. (1993). An early pre-liver intraembryonic source of CFU-S in the developing mouse. Nature 364, 64–67. [DOI] [PubMed] [Google Scholar]
- Monteiro R., Pouget C., and Patient R. (2011). The gata1/pu.1 lineage fate paradigm varies between blood populations and is modulated by tif1γ. EMBO J. 30, 1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin-Kensicki E.M., and Eisen J.S. (1997). Sclerotome development and peripheral nervous system segmentation in embryonic zebrafish. Development 124, 159–167. [DOI] [PubMed] [Google Scholar]
- Muller A.M., Medvinsky A., Strouboulis J., et al. (1994). Development of hematopoietic stem cell activity in the mouse embryo. Immunity 1, 291–301. [DOI] [PubMed] [Google Scholar]
- Murayama E., Kissa K., Zapata A., et al. (2006). Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity 25, 963–975. [DOI] [PubMed] [Google Scholar]
- Orkin S.H., and Zon L.I. (2008). Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132, 631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang W.W., Price E.A., Sahoo D., et al. (2011). Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc. Natl Acad. Sci. USA 108, 20012–20017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellagatti A., Cazzola M., Giagounidis A., et al. (2010). Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells. Leukemia 24, 756–764. [DOI] [PubMed] [Google Scholar]
- Pouget C., Gautier R., Teillet M.A., et al. (2006). Somite-derived cells replace ventral aortic hemangioblasts and provide aortic smooth muscle cells of the trunk. Development 133, 1013–1022. [DOI] [PubMed] [Google Scholar]
- Pouget C., Pottin K., and Jaffredo T. (2008). Sclerotomal origin of vascular smooth muscle cells and pericytes in the embryo. Dev. Biol. 315, 437–447. [DOI] [PubMed] [Google Scholar]
- Ransom D.G., Bahary N., Niss K., et al. (2004). The zebrafish moonshine gene encodes transcriptional intermediary factor 1γ, an essential regulator of hematopoiesis. PLoS Biol. 2, E237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D.J., Bryder D., Zahn J.M., et al. (2005). Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl Acad. Sci. USA 102, 9194–9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottbauer W., Just S., Wessels G., et al. (2005). VEGF-PLCγ1 pathway controls cardiac contractility in the embryonic heart. Genes Dev. 19, 1624–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandura J.M., Boccuni P., Massague J., et al. (2004). Transforming growth factor β-induced cell cycle arrest of human hematopoietic cells requires p57KIP2 up-regulation. Proc. Natl Acad. Sci. USA 101, 15231–15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver L., and Palis J. (1997). Initiation of murine embryonic erythropoiesis: a spatial analysis. Blood 89, 1154–1164. [PubMed] [Google Scholar]
- Stainier D.Y., Weinstein B.M., Detrich H.W. III, et al. (1995). Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development 121, 3141–3150. [DOI] [PubMed] [Google Scholar]
- Sumanas S., and Lin S. (2006). Ets1-related protein is a key regulator of vasculogenesis in zebrafish. PLoS Biol. 4, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamplin O.J., Durand E.M., Carr L.A., et al. (2015). Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell 160, 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoudi S., and Medvinsky A. (2007). Functional identification of the hematopoietic stem cell niche in the ventral domain of the embryonic dorsal aorta. Proc. Natl Acad. Sci. USA 104, 9399–9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavian M., Biasch K., Sinka L., et al. (2010). Embryonic origin of human hematopoiesis. Int. J. Dev. Biol. 54, 1061–1065. [DOI] [PubMed] [Google Scholar]
- Tavian M., Coulombel L., Luton D., et al. (1996). Aorta-associated CD34+ hematopoietic cells in the early human embryo. Blood 87, 67–72. [PubMed] [Google Scholar]
- Traver D., Paw B.H., Poss K.D., et al. (2003). Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat. Immunol. 4, 1238–1246. [DOI] [PubMed] [Google Scholar]
- Traver D., Winzeler A., Stern H.M., et al. (2004). Effects of lethal irradiation in zebrafish and rescue by hematopoietic cell transplantation. Blood 104, 1298–1305. [DOI] [PubMed] [Google Scholar]
- Warga R.M., Kane D.A., and Ho R.K. (2009). Fate mapping embryonic blood in zebrafish: multi- and unipotential lineages are segregated at gastrulation. Dev. Cell 16, 744–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenmann J., Ivanchenko S., Oswald F., et al. (2004). EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc. Natl Acad. Sci. USA 101, 15905–15910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J.W., Yu Q., Zhang J., et al. (2008). An acyltransferase controls the generation of hematopoietic and endothelial lineages in zebrafish. Circ. Res. 102, 1057–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y.L., Xiao A., Wen L., et al. (2010). Generation and characterization of blood vessel specific EGFP transgenic zebrafish via Tol2 transposon mediated enhancer trap screen. Prog. Biochem. Biophys. 37, 720–727. [Google Scholar]
- Yeo G.H., Cheah F.S., Jabs E.W., et al. (2007). Zebrafish twist1 is expressed in craniofacial, vertebral, and renal precursors. Dev. Genes Evol. 217, 783–789. [DOI] [PubMed] [Google Scholar]
- Zhang P., He Q., Chen D., et al. (2015). G protein-coupled receptor 183 facilitates endothelial-to-hematopoietic transition via Notch1 inhibition. Cell Res. 25, 1093–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Zhao X., Tian T., et al. (2008). Heart-specific isoform of tropomyosin4 is essential for heartbeat in zebrafish embryos. Cardiovasc. Res. 80, 200–208. [DOI] [PubMed] [Google Scholar]
- Zovein A.C., Hofmann J.J., Lynch M., et al. (2008). Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell 3, 625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.