In most species that undergo sexual reproduction, early embryogenesis is dominated by maternal mRNAs and proteins. To ensure subsequent development, these macromolecules are degraded and zygotic transcription is activated during a highly conserved maternal-to-zygotic transition (MZT). Degradation of maternal mRNAs occurs before or during zygotic genome activation (ZGA) and is essential for normal embryonic development (Ramos et al., 2004; Semotok et al., 2005), but the underlying molecular mechanisms are poorly understood. Two recent studies (Liu et al., 2016; Yu et al., 2016) report that BTG4 (B-cell translocation gene 4) is required to degrade maternal mRNAs in mice, which significantly advance our limited understanding of this important process in mammals.
Mammalian BTG4 belongs to the TOB/BTG family of proteins. All BTG family members, except BTG4, interact with CNOT7 to regulate RNA turnover and with transcription factors to control transcription (Winkler, 2010). In both mouse and human, Btg4 transcripts are highly enriched in the ovary and testis. Yu et al. (2016) and Liu et al. (2016) independently report that mouse embryos lacking BTG4 arrest at the 1–2 cell stage and Btg4 knockout female mice are infertile. A major consequence of the absence of BTG4 is a global delay in maternal mRNA degradation during the MZT. Both papers document that BTG4 interacts with an RNA de-adenylation complex, CCR4-NOT, similar to other BTG family members. Furthermore, they demonstrate that the interaction between the BTG4 and the CCR4-NOT complex depends on the BTG domain of BTG4 and two CCR4-NOT components, CNOT7 and CNOT8. These interactions suggest that BTG4 fosters mRNA degradation by promoting RNA de-adenylation. Consistent with this formulation, ablation of Btg4 inhibited shortening of the polyA tails of several maternal mRNAs during oocyte maturation. The two groups also implicate additional RNA-binding proteins, including the translation initiation factor eIF4E (Yu et al., 2016) and the polyA-binding protein PABPC1L (Liu et al., 2016) in BTG4-dependent mRNA recognition and degradation. Yu et al. (2016) document that ZGA is impaired in embryos derived from Btg4-null female mice, which suggests that maternal mRNA clearance is a prerequisite for ZGA (Figure 1).
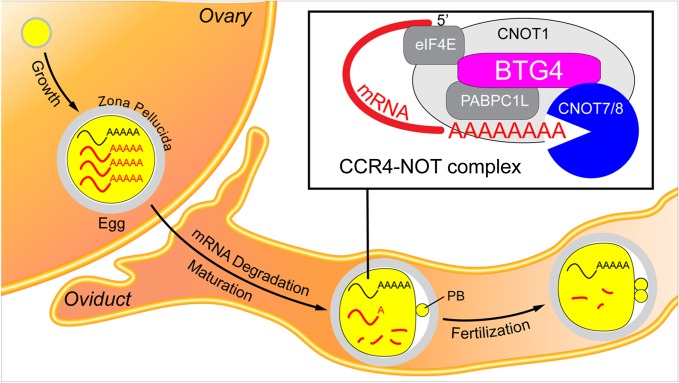
Figure 1.
BTG4-dependent maternal RNA clearance during oocyte maturation. Oocytes grow within the ovary and are ovulated into the oviduct where they are fertilized. Asymmetric meiotic divisions form polar bodies (PBs) present in the perivitelline space between the zona pellucida and the plasma membrane. During oocyte growth, maternal mRNAs accumulate, but are largely degraded during and after oocyte maturation. Rapid and efficient clearance of the majority of maternal mRNAs is required for normal embryonic development. Mouse BTG4 fosters maternal RNA degradation by recruiting the RNA de-adenylation complex CCR4-NOT onto target mRNAs and the de-adenylated mRNAs are subsequently degraded by RNA exonucleases.
Both Btg4 mRNA and protein are maternally produced. Yu et al. (2016) and Liu et al. (2016) report that Btg4 transcripts accumulate during oocyte growth in GV (germinal vesicle or nucleus)-intact oocytes, but are not translated until nuclear envelope breakdown (GVBD) when fully grown oocytes undergo meiotic maturation. Pharmacological blocking of oocyte maturation prevents the translation of Btg4 transcripts, which depends on three cytoplasmic polyadenylation elements in its 3′UTR. Yu et al. (2016) observed that pERK1/2 had a similar pattern of expression as the BTG4 protein in both mouse oocytes and embryos. They document that inhibiting pERK1/2 activity disrupted Btg4 mRNA polyadenylation and translation, suggesting that pERK1/2 functions upstream to activate BTG4 protein expression. Thus, accumulation of BTG4 is tightly regulated to ensure the expression in mature mouse oocytes and early embryos during the MZT. Together, both reports highlight a critical role of BTG4 in mouse maternal mRNA clearance and as a MZT-licensing factor.
These studies bring up several avenues for future investigation. First, the molecular basis for embryonic arrest in the absence of maternal BTG4 remains unclear. Both studies propose that delayed maternal mRNA clearance mediated by the BTG4 and the CCR4-NOT complex underlies the developmental arrest. Similar phenotypes are observed when the interactions between BTG4 and CNOT7 are disrupted. This model predicts that perturbation of the CCR4-NOT complex by removal of CNOT7 or CNOT8 will also result in embryonic arrest at the MZT. Among the long list of maternal mRNAs dependent on BTG4 for degradation, the persistence of a few key players could be responsible for the early developmental arrest. Therefore, overexpression of candidate maternal transcripts that are normally degraded could be assayed for their ability to disrupt development and phenocopy the Btg4-null mice. Second, the identification of interactions between the BTG4 and the translation initiation factor eIF4E suggests that temporal control of maternal mRNA degradation may be coupled with translation. This could be tested using 5′UTR-targeted morpholinos to block translation of maternal mRNA to determine the effect on their degradation. Third, the potential role of pERK1/2 in regulating oocyte maturation deserves further exploration. It would be of interest to determine whether ectopic expression of pERK1/2 in GV-intact oocytes leads to premature translation of Btg4 with concomitant acceleration of de-adenylation and degradation of maternal mRNAs.
Alternative strategies for the rapid clearance of maternal RNAs have been reported. In particular, small RNAs have generated considerable investigative interest for their role in shaping transcriptomes post-transcriptionally and in altering proteomic profiles during developmental transitions (He and Hannon, 2004). In zebrafish, the microRNA miR-430 is responsible for the rapid clearance of the majority of maternal RNA at the mid-blastula transition (comparable to the MZT, Giraldez et al., 2006). However, no counterpart has been identified in mice (Svoboda and Flemr, 2010) and the work by both Yu et al. (2016) and Liu et al. (2016) highlights the importance of RNA de-adenylation in the MZT. One of the key players of this machinery, BTG4, is specifically expressed in mouse oocytes and early embryos, which suggests that mice employ an MZT-specific RNA de-adenylation system for clearance of maternal mRNAs.
[This work was supported by the Intramural Research Program of NIDDK, NIH.]
References
- Giraldez A.J., Mishima Y., Rihel J., et al. (2006). Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–79. [DOI] [PubMed] [Google Scholar]
- He L., and Hannon G.J. (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531. [DOI] [PubMed] [Google Scholar]
- Liu Y., Lu X., Shi J., et al. (2016). BTG4 is a key regulator for maternal mRNA clearance during mouse early embryogenesis. J. Mol. Cell Biol. doi:10.1093/jmcb/mjw023. [DOI] [PubMed] [Google Scholar]
- Ramos S.B., Stumpo D.J., Kennington E.A., et al. (2004). The CCCH tandem zinc-finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development 131, 4883–4893. [DOI] [PubMed] [Google Scholar]
- Semotok J.L., Cooperstock R.L., Pinder B.D., et al. (2005). Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr. Biol. 15, 284–294. [DOI] [PubMed] [Google Scholar]
- Svoboda P., and Flemr M. (2010). The role of miRNAs and endogenous siRNAs in maternal-to-zygotic reprogramming and the establishment of pluripotency. EMBO Rep. 11, 590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler G.S. (2010). The mammalian anti-proliferative BTG/Tob protein family. J. Cell. Physiol. 222, 66–72. [DOI] [PubMed] [Google Scholar]
- Yu C., Ji S.Y., Sha Q.Q., et al. (2016). BTG4 is a meiotic cell cycle-coupled maternal-zygotic-transition licensing factor in oocytes. Nat. Struct. Mol. Biol. 23, 387–394. [DOI] [PubMed] [Google Scholar]