Abstract
Numerous clinical observations have confirmed that breakpoint cluster region-abelson fusion oncoprotein tyrosine kinase inhibitors used in leukemia treatment alter bone physiology in a complex manner. The aim of the present study was to analyze the whole transcriptome of cultured murine osteoblasts and determine the changes following treatment with imatinib and nilotinib using Sequencing by Oligonucleotide Ligation and Detection next generation RNA sequencing. This study also aimed to identify candidate signaling pathways and network regulators by multivariate Ingenuity Pathway Analysis. Based on the right-tailed Fisher's exact test, significantly altered pathways including upstream regulators were defined for each drug. The correlation between these pathways and bone metabolism was also examined. The preliminary results suggest the two drugs have different mechanisms of action on osteoblasts, and imatinib was shown to have a greater effect on gene expression. Data also indicated the potential role of a number of genes and signaling cascades that may contribute to identifying novel targets for the treatment of metabolic bone diseases.
Keywords: gene expression, MC3T3-E1 cells, pathway analysis, TKIs
Introduction
Tyrosine kinase inhibitors (TKI) have become a widely used class of drugs for the treatment of a range of solid tumors and hematological malignancies. In 2001, imatinib was the first TKI to be registered for the targeted therapy of chronic myelogenous leukemia (CML) (1). Imatinib is an ATP competitive inhibitor of the tyrosine kinase activity of abelson (ABL) kinase and the breakpoint cluster region-abelson fusion oncoprotein (BCR-ABL). Whereas ABL is found in the majority of cells, the fusion protein is only expressed in leukemic cells. Imatinib arrests proliferation and induces the apoptosis of CML cells. During long-term imatinib therapy, clinicians observed that a number of patients became resistant to the treatment. Therefore, a more selective and potent BCR-ABL inhibitor molecule, nilotinib, was developed, which was able to overcome resistance in numerous cases (2,3).
A number of TKIs have demonstrated marked effects on bone homeostasis and the remodeling balance. Imatinib directly inhibits osteoclastogenesis and the bone resorption activity of osteoclasts, and also reduces the survival of osteoclast precursors and mature osteoclasts (4–8). Based on in vitro studies, imatinib reduces osteoblast proliferation (3–7,9) and survival, but increases osteoblast cell differentiation (3,8). Similarly, nilotinib also effectively inhibits the proliferation rate of osteoblasts (3,10). However, nilotinib increases the secretion of osteoprotegerin (OPG) and decreases the expression of receptor activator of nuclear factor κ-B ligand (RANKL) (3). Other studies have shown increased osteoblast-specific gene expression, cell activity and mineralization induced by imatinib (3–9). It should be noted that the examined TKIs have differing effects on osteoblast function. The described differences may be dependent upon the concentration of the utilized TKI, the maturation stage of the osteoblasts and the distribution of various TKI-targeted receptors on cells (8,10,11). The direct influence of imatinib on osteoclasts and osteoblasts results from off-target effects on cell surface receptor tyrosine kinases [such as colony-stimulating factor 1 receptor, stem cell growth factor receptor (c-KIT), and platelet-derived growth factor receptor (PDGFR)] and carbonic anhydrase II (3,10). Nilotinib is a second-generation TKI with greater selectivity towards ABL/BCR-ABL over other tyrosine kinases (such as PDGFR, c-KIT and discoidin domain receptor kinases).
The clinical effects of TKI administration also show differences in bone metabolism. Changes in trabecular bone volume (TBV) were observed in patients with CML after imatinib therapy (7,10,12). TBV was measured in 17 patients with CML prior to treatment and 2- and 4-years after imatinib treatment. In 8 patients, there was a significant increase in TBV, although, serum phosphate and calcium levels of 9 participants were reduced (7). According to numerous clinical studies, hypophosphataemia (3,7,13–16), hypocalcemia (13–16) and hyperparathyroidism (13–16) have been documented during TKI administration. Vandyke et al (12) reported elevated bone mineral density (BMD) and bone volume:trabecular volume ratio at the femoral neck in imatinib-treated CML patients. During the 48-month observation period, trabecular bone area (TBA%) was decreased in 10 patients and increased in 24 patients (17). In other studies, diminished serum osteocalcin and N-telopeptide of type I collagen levels, as well as lower bone mineral content and impaired bone remodeling have also been reported (12–14,18).
Currently, there are numerous contradictory results regarding the effects of imatinib and nilotinib on bone metabolism, and there is no clear evidence to explain the results, either at the cellular level or in clinical observations. Furthermore, there is limited comprehensive transcription data available in relation to bone cell and/or tissue function and TKI treatment. Only targeted bone-specific gene expression [e.g. osteocalcin, alkaline phosphatase, OPG, RANKL and bone morphogenetic protein 2 (BMP2)] changes have been examined. Therefore, the aim of the present study was to analyze the whole transcriptome of cultured murine osteoblasts following imatinib and nilotinib treatment using Sequencing by Oligonucleotide Ligation and Detection (SOLiD) next generation RNA sequencing. This study aimed to identify candidate signaling pathways and network regulators by multivariate Ingenuity Pathway Analysis (IPA).
Materials and methods
In vitro cell culture
The MC3T3-E1 murine preosteoblast cell line was obtained from the American Type Culture Collection (Rockville, MD, USA). The cells were cultured in Minimum Essential Medium Eagle α-Modification (α-MEME, Sigma-Aldrich, St. Louis, MO, USA) supplemented with 0.292 g/l L-glutamin (Sigma-Aldrich), 5% fetal calf serum (FCS, Sigma-Aldrich) and 1% antibiotic solution (penicillin-streptomycin sulfate-amphotericin B) (Sigma-Aldrich). Cells were incubated at 37°C in a 5% CO2 atmosphere and 78% humidity. The cultured medium was changed twice a week. Cells were passaged when grown to 70% confluence using 0.25% Trypsin EDTA solution (Sigma-Aldrich). All experiments were conducted with MC3T3-E1 cells between passages 8 and 15. All used reagents were of analytical quality.
Effects of imatinib and nilotinib on cell viability
In the in vitro system, the following three sample groups were examined: Imatinib-treated, nilotinib-treated and untreated (control) osteoblast cell cultures.
The adequate incubation time and drug concentration were defined using a cell viability assay. Different imatinib (Glivec/Gleevec, STI571, CGP 57148B; Novartis, Basel Switzerland) and nilotinib (Tasigna; Novartis) concentrations (30 nM-20 µM) were administered to 40% confluent MC3T3-E1 cells for various incubation times (1–6 days) in 96-well plates. After removal of culture medium, cells were fixed with 100 µl/well trichloroacetic acid (Sigma-Aldrich) for 30 min. Then, cells were stained with 0.4% sulforhodamine-B (SRB, Sigma-Aldrich) protein dye solution for 30 min. After the excess dye solution removal, cell culture plates were rinsed with 1% acetic acid solution four times and dried at room temperature. The bound SRB was dissolved in 100 µl of 10 mM Trisma-Sol (Sigma-Aldrich) and the cell culture plates were shaken for 5 min. The measurements were performed by Multiskan Spectrum V1.2 1500-636 device (Thermo Fisher Scientific Inc, Waltham MA USA) at 520 nm. Three parallel measurements on 24-well cell culture plates were performed twice in all experiments.
RNA isolation
RNA isolation was performed using the High Pure Total RNA Isolation kit (Roche Diagnostics, Indianapolis, IN, USA) from the treated and untreated osteoblastic cells. The quality and quantity of isolated RNA were measured by Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and Qubit fluorometer (Thermo Fisher Scientific, Inc.). After pooling the parallel biological samples, the whole transcriptome analysis was performed by the Applied Biosystems SOLiD V4 device (Thermo Fisher Scientific, Inc.).
SOLiD next generation RNA sequencing
The whole transcriptome analysis of the purified, DNA-free total RNA molecules (>5 mg/sample, RIN >8.0, cc >400 ng/ml) were performed by SeqOmics Biotechnology Ltd. (SeqOmics Biotechnology Ltd., Szeged, Hungary; http://www.seqomics.hu/) with SOLiD next generation 50+20 bp reads paired-end technologies.
Statistical analysis
For the data analysis, Benjamini and Hochberg False Discovery Rate test was applied (19,20). P≤0.05 was considered to indicate a statistically significant difference. Genes showing significantly changed mRNA expression levels were evaluated by IPA 7.6 software (QIAGEN, Redwood City, CA, USA; www.ingenuity.com). Canonical pathway analysis utilizing the IPA library of canonical pathways identified the signaling routes that contained the differentially expressed genes in the input data set. The significance of the association between the data set and the canonical pathway was determined based on two parameters: i) A ratio of the number of genes from the data set that map to the pathway divided by the total number of genes that map to the canonical pathway and ii) a P-value calculated using Fischer's exact test determining the probability that the association between the genes in the data set and the canonical pathway is due to chance alone. The upstream regulator analysis identifies the cascade of upstream transcriptional regulators based on prior knowledge of expected effects between transcriptional regulators and their target genes stored in the Ingenuity Knowledge Base. The analysis examines the number of known targets of each transcription regulator present in the dataset, and also compares their direction of change (expression in the experimental sample relative to control) to what is expected from the literature in order to predict likely relevant transcriptional regulators. The aim of the overlap P-value is to identify transcriptional regulators that are able to explain observed gene expression changes. The overlap P-value measures whether there is a statistically significant overlap between the dataset genes and the genes that are regulated by a transcriptional regulator. It is calculated using Fisher's exact test, and significance is attributed to P<0.01. The most upregulated molecules were selected based on logarithmic fold change (logFC) values, which describe the rate of expression changes in the treated groups compared with the untreated control group.
Results
Differentially expressed genes in the treated groups
Based on the cell viability test, the 1 µM drug concentration and 6-day incubation period had the greatest effects on the expression profile of osteoblastic cells. Altogether, 16,383 (imatinib-treated group), 16,951 (nilotinib-treated group) and 17,290 (control group) annotated RNAs were found in the samples, respectively. Altered expression of 358 genes in the imatinib compared with the control and 21 genes in the nilotinib group compared with control were identified using the Benjamini and Hochberg statistical test. There were three common differentially expressed genes [C11orf87, chromosome 11 open reading frame 87 (AI593442), 3-hydroxyisobutyrate dehydrogenase pseudogene (Gm11225) and zinc finger protein 184 (Zfp184)] in the two treated groups compared with control. logFC values were as follows: logFC (AI593442)=2.95, logFC (Gm11225)=2.09 and logFC (Zfp184)=1.45 in the imatinib group, and logFC (AI593442)=2.82, logFC (Gm11225)=2.79 and logFC (Zfp184)=3.43 in the nilotinib group. These three genes had almost the same expression activity in the two groups. In the imatinib-treated group, all of the identified differentially expressed genes were upregulated. This was also observed in the nilotinib treatment, with one exception [logFC (RP23-390C13.1)=−2.95] based on logFC values.
Canonical pathway analysis
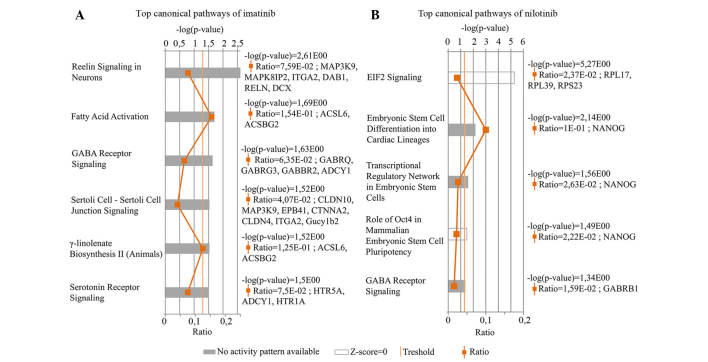
Genes showing significantly altered expression rates in response to the two drugs compared with control were evaluated by IPA canonical pathways analysis. Six most upregulated canonical pathways were identified in the imatinib-treated osteoblast cells (Fig. 1A). Reelin signaling in neurons pathway which participates in the formation of neuronal architectonic patterns had the lowest P-value. This pathway was represented by six genes, namely Mitogen-Activated Protein Kinase Kinase Kinase 9 MAP3K9, C-jun-amino-terminal kinase-interacting protein 2 (MAPK8IP2), Integrin subunit α2 (ITGA2), cytoplasmic adaptor disabled-1 (DAB1), Reelin gene (RELN) and doublecortin (DCX). The logFC values were: logFC (MAP3K9)=3.41, logFC (MAPK8IP2)=2.10, logFC (ITGA2)=2.35, logFC (DAB1)=2.24, logFC (RELN)=3.71 and logFC (DCX)=1,86. The fatty acid activation pathway supports adequate fatty acid and lipid biosynthesis. ACSL6 and ACSBG2 genes characterize this pathway. The logFC values of these genes were 3.39 and 2.42, respectively. Based on these two genes, IPA software indicated another signaling pathway, γ-linolenate biosynthesis II cascade, which is also responsible for lipid metabolism. The γ-aminobutyric acid (GABA) receptor signaling pathway primarily regulates signal transduction in the nervous system. In these results, two ion-channel (GABRQ and GABRG3), one G protein-coupled receptor B (GABBR2) and the adenylate cyclase type 1 genes (ADCY1) were shown to be associated with this pathway. The logFC values were as follows: 2.69, 1.95, 2.74 and 3.16, respectively. Sertolisertoli cell junction signaling pathway is involved in testicular cell growth, proliferation and development. This pathway was represented by seven differentially expressed genes. The logFC values were as follows: logFC (CLDN10)=1.94, logFC (MAP3K9)=3.41, logFC (EPB4.1)=2.51, logFC (CTNNA2)=2.76, logFC (CLDN4)=2.21, logFC (ITGA2)=2.35 and logFC (Gucy1b2)=2.86. Finally, the Serotonin receptor signaling pathway, which had been demonstrated to show a strong positive effect on bone mass was also identified. The logFC values of the three connected genes were: logFC (HTR5A)=2.06, logFC (ADCY1)=1.45 and logFC (HTR1A)=2.88. These pathways were shown to be significantly altered following treatment (P<0.05); however, the software could not predict the activity pattern of the most upregulated pathways. The most upregulated pathway is a relevant (significant) signaling pathway, where genes of the uploaded dataset were significantly enriched. The evaluation criteria of most upregulated pathways are the ratio and the P-value.
Figure 1.
Most upregulated canonical pathways of (A) imatinib and (B) nilotinib as determined by IPA analysis. These figures show the results of IPA canonical pathway analysis. The -log (P-value) of each pathway was calculated using right-tailed Fisher's exact test. In this method, the P-value for a given process annotation is calculated by considering the number of focus genes that participate in that process and the total number of genes that are known to be associated with that process in the selected reference set. The statistical threshold is indicated by an orange line at P=0.05. Gray and white bars indicate pathways where predictions are not currently possible, because either no activity pattern was available or the z-score was ~0.
Five most upregulated canonical pathways were identified in the transcriptome of the nilotinib-treated group (Fig. 1B). The Eukaryotic Initiation Factor 2 (EIF2) signaling pathway had the highest P-value, although the activation z-score was almost 0. EIF2 signaling has an important role in the initiation of translation. The logFC values of the three differentially expressed genes involved in this pathway were: logFC (RPL17)=2.41, logFC (RPL39)=1.99 and logFC (ribosomal protein S23; RPS23)=3.45. Embryonic stem cell differentiation into cardiac lineages, Transcriptional regulatory network in embryonic stem cells and Role of Oct4 in mammalian embryonic stem cell pluripotency pathways were denoted by the NANOG gene, which encodes a critical transcription factor for cell self-renewal. The GABA receptor signaling pathway is characterized with GABRB1 (γ-aminobutyric acid receptor A, subunit β1) gene, logFC=2.83. The GABA receptor signaling network was determined to be one of the most upregulated canonical pathway associated with the differentially expressed genes following treatment with imatinib and nilotinib compared with control.
Upstream regulator analysis
The IPA software defines the most upregulated upstream regulators on the basis of quality of expression changes. Five upstream regulators were identified in the imatinib group, namely: GLDN, FREM2, NRCAM, GRIP1 and very low density lipoprotein receptor (VLDLR). A summary of the identified upstream regulators, including their biological function associated with bone metabolism is shown in Table I (21–23).
Table I.
Most upregulated upstream regulators of imatinib using the ingenuity pathway analysis system.
| Gene symbol | Gene name | P-value of overlap | Regulates | General functions | Function related to bone biology |
|---|---|---|---|---|---|
| GLDN | Gliomedin | 7,54E-04 | Voltage-gated sodium channel, SPTBN4, ANK3, NFASC | Involved in the formation of the nodes of Ranvier along myelinated axons. | N/A |
| FREM2 | FRAS1 related extracellular matrix protein 2 | 7,54E-04 | FREM1, FRAS1, FREM2 | Extracellular matrix protein required for maintenance of the integrity of skin and renal epithelium. Also required for epidermal adhesion. | N/A |
| NRCAM | Neuronal cell adhesion molecule | 7,54E-04 | Voltage-gated sodium channel, NFASC, SPTBN4, ANK3, NRCAM, GLDN, neurotrophin | Ankyrin-binding protein involved in neuron-neuron cell adhesion. Role in the molecular assembly of the nodes of Ranvier. | N/A |
| GRIP1 | Glutamate receptor interacting protein 1 | 7,54E-04 | FRAS1, GRIA2, NR1I3, GRIP1, FREM2, FREM1, ESRRA, TFF1, HNF4A, ESR1, MEF2C, kinesin, ampa receptor | May be a localized scaffold for the assembly of a multiprotein signaling complex and as mediator of the trafficking of its binding partners at specific subcellular location in neurons. | N/A |
| VLDLR | Very low density lipoprotein receptor | 7,54E-04 | Cholesterol, triacylglycerol, DAB1, Ptk, DNA endogenous promoter, phospholipid, fatty acid, RAC1, LPL, SERPINE1, glycoprotein, RNA polymerase II, very low density lipoprotein | Binds VLDL and transport it into the cells by receptor-mediated endocytosis. In order to be internalized, the receptor-ligand complexes must first cluster into clathrin-coated pits. Important in VLDL-triglyceride metabolism and the reelin signaling pathway. | Support osteoblast differentiation and proliferation (21,22). Indirectly regulates bone mineral density and remodeling through cholesterol level (23). |
Data on the function of the most upregulated upstream regulators in bone biology were collected from the literature. Gene symbols, names and primary functions were listed to standard Genecards (http://www.genecards.org). P-values of overlap were calculated using Fisher's exact test and P<0.01 was considered to indicate a statistically significant difference.
In the nilotinib-treated osteoblast culture, five most upregulated upstream regulators were also found: FAAH, MARCH7, ACVR1B, RAD23B and ACVR1C. Details of these upstream regulators are listed in Table II (24).
Table II.
Most upregulated upstream regulators of nilotinib using the IPA system.
| Gene symbol | Gene name | P-value of overlap | Regulates | General functions | Function related to bone biology |
|---|---|---|---|---|---|
| FAAH | Fatty acid amide | 1.30E-04 | Anandamide, fatty acid, MAEL, FNDC3A, SEPT4, 2-arachidonoylglycerol, endocannabinoid, Prl3d1, oleoylethanolamide, MAP6D1, RPL7L1, RPL5, RPS18, acylethanolamide | Integral membrane protein that hydrolyzes a number of bioactive primary and secondary fatty acid amides, including the neuromodulatory compounds anandamide and oleamide to free fatty acid and ethanolamine. | Affects osteoblasts and osteoclasts, stimulates bone formation and inhibits bone resorption. |
| MARCH7 | Membrane- associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase | 6.61E-04 | NANOG | Adds ubiquitin to target lysines in substrate proteins, thereby signaling for their vesicular transport between membrane compartments. E3 ubiquitin ligases accept ubiquitin from an E2 ubiquitin-conjugating enzyme in the form of a thioester and then directly transfer the ubiquitin to targeted substrates. | N/A |
| ACVR1B | Activin receptor type-1B | 6.61E-04 | SMAD2, SMAD3, Xnr1, NODAL, L-threonine, DNA endogenous promoter, HP, activin, MAPK, Akt, LEFTY1, CYP19A1, LEFTY2, FSHR | Transmembrane serine/threonine kinase activin receptor type-1 forming receptor complex with receptor type-2. Activin signal regulates numerous physiological and pathological processes including neuronal differentiation and survival, hair follicle development and cycling, FSH production, wound healing, extracellular matrix production, immunosuppression and carcinogenesis. | Involved is bone homeostasis, bone cell proliferation and differentiation, skeletal development, bone turnover, bone mass and fracture risk (24). |
| RAD23B | RAD23 homolog B (S. cerevisiae) | 1.32E-03 | TDG, Lys48-linked polyubiquitin, 26s proteasome, PSMC1, TP53, NGLY1, XPC, MPG, DNA promoter | Multiubiquitin chain receptor involved in modulation of proteasomal degradation. Binds to polyubiquitin chains. Proposed to be capable to bind simultaneously to the 26S proteasome and to polyubiquitinated substrates and to deliver ubiquitinated proteins to the proteasome. | N/A |
| ACVR1C | Activin receptor type-1C | 1.32E-03 | SMAD2, SMAD3, FOS MAPK8, NODAL, Erk, LEFTY2, SERPINE1, Jnk, caspase, CASP9, JINK1/2, P38 MAPK, CASP3, LEFTY1 | Receptor for activin AB, activin B and NODAL. Involved in cell differentiation, growth arrest and apoptosis. | N/A |
Data on the function of the most upregulated upstream regulators in bone biology were collected from literature. Gene symbols, names and primary functions were listed to standard Genecards (http://www.genecards.org). P-values of overlap were calculated using Fisher's exact test and significance is attributed to P-values <0.01. The molecules were reported in IPA program. IPA, ingenuity pathway analysis.
Upregulated molecules
The most sensitive genes in response to imatinib and nilotinib treatment were determined based on fold changes using IPA core analysis. In the two groups, based on the degree of expression changes, the lists of the most sensitive genes with the most marked expression differences are shown in Tables III and IV (25–30). Following imatinib administration, the syntaxin binding protein 5-like (STXBP5L) gene showed the strongest upregulation (21-fold), while the RPS23 gene was most strongly upregulated following nilotinib treatment (11-fold) compared with control.
Table III.
Most upregulated molecules of imatinib treated cells in IPA analysis.
| Gene symbol | Gene name | LogFC | Regulates | General function | Function related to bone biology |
|---|---|---|---|---|---|
| STXBP5L | Syntaxin binding protein 5-like | 4.40 | Insulin, D-glucose | Involved in vesicle transport and exocytosis, as well as the regulation of protein secretion and protein transport. | Takes part in glucose homeostasis and negative regulation of insulin secretion. |
| GDF10 | Growth differentiation factor 10 | 4.33 | SMAD2, SERPINE1 | GDF10 (also known as bone morphogenetic protein 3B) is a member of transforming growth factor β superfamily. Involved in skeletal morphogenesis, apoptosis, cell development and fat cell differentiation. | Regulates osteoblast differentiation and ossification. Respond to transforming growth factor beta stimulus, supports skeletal system development. |
| HYDIN | HYDIN, axonemal central pair apparatus protein | 3.97 | N/A | Involved in cilia motility, brain development and axoneme assembly. | N/A |
| NYAP2 | Neuronal tyrosine- phosphorylated phosphoinositide- 3-kinase adaptor2 | 3.94 | N/A | Activates PI3K and concomitantly recruits the WAVE1 complex to the close vicinity of PI3K and regulates neuronal morphogenesis. Participates in protein binding and neuron projection morphogenesis. | N/A |
| AQP9 | Aquaporin 9 | 3.93 | Glycerol, water, D-glucose, urea, triacylglycerol, Gyk, Ins1, proinsulin, arsenite, PRKCB, MYC, BAX, CASP3 | Aquaporins are a family of water-selective membrane channels that allow the passage of a wide variety of non-charged solutes, including carbamides, polyols, purines and pyrimidines in a phloretin- and mercury-sensitive manner. Whereas amino acids, cyclic sugars, Na+, K+, Cl−, and deprotonated monocarboxylates are excluded. Also permeable to urea and glycerol. | Regulates bone resorption (25), osteoclastogenesis, (26) osteoclast differentiation(25-27) and bone mineral density (25). |
| CSMD1 | CUB and Sushi multiple domains 1 | 3.79 | D-glucose, complement protein | Takes part in glucose homeostasis and startle response. | Takes part in bone formation (28). |
| MYO3B | Myosin IIIB | 3.74 | L-threonin, peptide | Encodes one of the class III myosins. Myosins are activated by actins that move along actin filaments in the cell. Alternative splicing results in multiple transcript variants encoding different isoforms. | N/A |
| RELN | Reelin | 3.71 | DAB1, RELN, GSK3B, GRIN2B, NTRK2, AKT1, VLDLR, APP, SRC, FN1, NOTCH1, MAPT, L-tyrosine, CRK, FABP7 | Encodes a large secreted extracellular matrix protein that controls cell-cell interactions, and is critical for cell positioning and neuronal migration during brain development. Regulates microtubule function in neurons. Binds very-low-density lipoprotein particle receptor. | Thyroid hormone metabolic process, which regulates the bone remodeling. |
| EDA | Ectodysplasin-A | 3.69 | NFkB, WNT10B, WNT10A, PTHLH, AREG, EPGN, FGF20 | Transmembrane protein of the tumor necrosis factor (TNF) family which is important in the development of ectodermal tissues. Involved in epithelial-mesenchymal signaling during morphogenesis of ectodermal organs. Takes part in immune response. | Positively regulates canonical Wingless receptor signaling pathway. Wnt signaling pathway takes part in regulation of bone homeostasis. Positively regulates I-κB kinase/nuclear factor-κB cascade as well as NF-κB activity and import into nucleus. NF-κB has well known crucial role in osteoblast and osteoclast activities. |
| SLITRK5 | SLIT and NTRK- like family, member 5 | 3.69 | N/A | SLITRKs are expressed predominantly in neural tissues and have neurite-modulating activity. Suppresses neurite outgrowth. Involved in axonogenesis, dendrite morphogenesis, skin development and response to xenobiotic stimulus. | N/A |
Gene symbols, names were listed to standard GeneCards (http://www.genecards.org). Functions were listed to GeneCards (http://www.genecards.org), NCBI (http://www.ncbi.nlm.nih.gov/gene) and IPA (https://reports.ingenuity.com). The molecules were reported in IPA program. Data on the function of the most upregulated molecules in bone biology were collected from literature. IPA, ingenuity pathway analysis.
Table IV.
Most upregulated molecules of nilotinib treated cells in ingenuity pathway analysis.
| Gene symbol | Gene name | logFC | Regulates | General function | Function related to bone biology |
|---|---|---|---|---|---|
| RPS23 | Ribosomal protein S23 | 3.45 | rnr | Encodes a ribosomal protein which is a component of the 40S subunit. | N/A |
| ZFP184 | Zinc finger protein 184 | 3.43 | N/A | Takes part in sequence-specific DNA binding and in the regulation of transcription. | N/A |
| EDNRB | Endothelin receptor type B | 3.04 | Cyclooxygenase, SLC9A3, nitric oxide, VEGFA, EDN1, Nos, HIF1A, TYR, DNS promoter, Adcy, Ca2+, GNAI1, GNAO1, RNA polymerase II | Non-specific, G protein-coupled receptor for endothelin 1, 2, and 3. Located primarily in the vascular endothelial cells. Involved in vasoconstriction, vasodilation, bronchoconstriction and cell proliferation. | N/A |
| DKK4 | Dickkopf homolog 4 | 2.99 | N/A | A member of the dickkopf family. The secreted protein involved in embryonic development through interactions with the Wingless (WNT) signaling pathway. DKKs are important in vertebrate development. They locally inhibit Wnt regulated processes, such as antero-posterior axial patterning, limb development, somitogenesis and eye formation. DKKs are implicated in bone formation and bone disease. | Positively regulates osteoblast apoptosis, and negatively regulates osteoblast proliferation, differentiation and the Wnt/β catenin signaling pathway (29). |
| GABRB1 | γ-aminobutyric acid (GABA) A receptor, subunit β1 | 2.83 | Ion, chloride | A multisubunit chloride channel that mediates inhibitory synaptic transmission in the central nervous system. | N/A |
| RPL17 | Ribosomal protein L17 | 2.41 | N/A | Encodes a ribosomal protein that is a component of the 60S subunit. There are multiple processed pseudogenes of this gene dispersed through the genome, which is typical for genes encoding ribosomal proteins. Alternative splicing results in multiple transcript variants. | N/A |
| KLHL41 | Kelch-like 41 | 2.37 | N/A | Member of the kelch-like family. Thought to function in skeletal muscle development and differentiation. Regulates proliferation and differentiation of myoblasts and is involved in myofibril assembly. Required for pseudopod elongation in transformed cells. | N/A |
| NANOG | Nanog homeobox | 2.06 | GATA4, FAK, NANOG, FOXD3, POU5F1, PDCD4, CDK6, AHR, GATA6, ZFP42, DNA promoter, DNA endogenous promoter, RNA polymerase II, SEC22B, PCDHA2 | Transcription regulator involved in inner cell mass and ES cells proliferation and self-renewal. Prevents ES cell differentiation towards extraembryonic endoderm and trophectoderm lineages. Blocks bone morphogenetic protein-induced mesoderm differentiation of ES cells by physically interacting with SMAD1 and interfering with the recruitment of coactivators to the active SMAD transcriptional complexes. Acts as a transcriptional activator or repressor. | Involved in bone healing (30), negatively regulates bone morphogenetic cascade and Wingless signaling pathway. |
| RPL39 | Ribosomal protein L39 | 1.99 | N/A | Encodes a ribosomal protein that is a component of the 60S subunit. | N/A |
Gene symbols, names were listed to standard GeneCards (http://www.genecards.org). Functions were listed to GeneCards (http://www.genecards.org), NCBI (http://www.ncbi.nlm.nih.gov/gene) and IPA (https://reports.ingenuity.com). Data on the function of the most upregulated molecules in bone biology were collected from literature. ES, embryonic stem.
Discussion
In the present study, the effects of selective TKI imatinib and nilotinib on the global gene expression pattern of a murine osteoblastic cell line culture in vitro was observed. Previous studies have reported expression data in response to TKI only of strictly bone-related genes (RANKL, OPG, bone sialoprotein, osteocalcin, osterix, BMP2 and RUNX2) (2,3,7,8). To the best of our knowledge, this is the first study to observe the complete mRNA pattern of osteoblasts by whole transcriptome analysis. Thus, it was possible to demonstrate the most upregulated canonical pathways and upstream regulators that were affected in osteoblast cells by these compounds.
TKIs are widely used drugs for the treatment of certain oncohematological diseases, the treatment may continue for decades or the rest of the patient's lives. Therefore, there is a limitation in the present study as the results reflect the rapid primary drug effect on gene activities rather than the extended secondary expression changes. Thus future studies will aim to model the long-term effects of imatinib and nilotinib in an in vitro system.
Generally, imatinib and nilotinib inhibit the proliferation, growth and survival of BCR-ABL-positive cells. However, several studies have demonstrated contradicting drug effects on bone remodeling. These TKIs have been shown to attenuate osteoclastic bone resorption, reduce site-specific bone volume (7,12,13), change histomorphometric features of bone, support extracellular matrix mineralization (7) and cause concentration-dependent mixed effects on bone turnover markers and osteoblast activity (31). The results indicate different effects of imatinib and nilotinib on osteoblast function, which is reflected in altered gene expression patterns caused by the two drugs. This may be explained by the different chemical profiles, different target spectrums and the bound non-kinase molecules of the examined TKIs (32). Among the significantly altered genes in the two treatment groups, there were only three common genes. Zfp184, which participates in transcription regulation processes and may be important in osteoblast differentiation through chloride intracellular channel 1 interaction (https://reports.ingenuity.com, http://innatedb.com) (33). Biological roles of Gm11225 and AI593442 in bone metabolism have not yet been established.
GABA receptor signaling was found among the most upregulated canonical pathways in the imatinib and nilotinib groups. GABA receptors mediate signals of the neurotransmitter GABA. Osteoblasts constitutively express GABA (B) receptor subunits (34). The overexpression of several other GABA receptor family members (GABBR2, GABRQ, GABRG3 and GABRB1) was observed following after TKI treatment. Mentink et al (35) demonstrated that GABBR2 inhibits alkaline phosphatase activity and calcium accumulation in mesenchymal stromal cells. Signals via the GABA pathway can stimulate osteoblastogenesis, and the activation of GABA (B) receptors upregulates the key osteoblast differentiation markers: BMP2, secreted protein acidic and rich in cysteine/osteonectin (SPARC) and osteocalcin (36). During the bone remodeling process, this signal transduction network may be involved in bone cell proliferation, differentiation and development. Functional GABA (B) receptors appear to be predominantly expressed by osteoblasts rather than osteoclasts during remodeling and skeletogenesis (34,37).
The most significantly altered pathway in imatinib-treated osteoblasts was Reelin signaling in neurons, which is closely associated with nervous system function and development, as well as synaptogenesis and neurodegeneration. A genome-wide association study confirmed that genetic variants in the RELN gene have a crucial role in the development of otosclerosis, a complex bone remodeling disorder of the otic capsule (38). The extracellular reelin binds to VLDLR, which was shown to be one of the most upregulated upstream regulator molecules in the present study as well as apolipoprotein-E receptor 2 (ApoER2). This interaction indicates the involvement of tyrosine phosphorylation of DAB1 in the Reelin pathway (39). Reelin signals generated following binding to the lipoprotein receptors are also known to be involved in neuronal migration. The possible role of the Reelin signaling complex (Reelin/VLDLR/ApoER2/Dab1) in osteoblasts remains to be revealed.
The present study demonstrated that imatinib treatment elicited a significant change in the mRNA expression of genes involved in serotonin receptor signaling. Several types of serotonin (5-hydroxytryptamine, 5-HT) receptors can be found on major bone cells (osteoblasts, osteocytes and osteoclasts), including 5-HT1 and 5-HT2 (40–43). Expressional changes in another class of serotonin receptors (HTR5A, 5-HT5A receptor) was also identified. Serotonin may exhibit regulatory effects on bone and recent data suggested that gut-derived serotonin may mediate skeletal effects (44). Several in vitro studies have confirmed the functionality of serotonin signaling in bone cells, with mixed effects reported (44). Some suggest a direct stimulatory effect of serotonin on bone formation pathways, while others have found inhibitory effects. Serotonin produced in the periphery appears to act as a hormone and inhibits bone formation. Conversely, serotonin produced in the brain acts as a neurotransmitter and has a strong positive effect on bone mass by supporting bone formation and reducing bone resorption (45). The effect of serotonin signaling on bone resorption pathways also depends on the concentration present (46). However, the specific biochemical nature of serotonergic pathways influencing bone and their direct and/or indirect effects on bone metabolism remain unclear.
The EIF2 signaling pathway showed the greatest expression changes among pathways affected by nilotinib. Phosphorylated eIF2α and its downstream regulators are involved in the endoplasmic reticulum stress response of osteoblasts. Stress to the endoplasmic reticulum (e.g. protein misfolding, viral infection and nutritional deprivation) modifies transcriptional activation of bone remodeling and osteogenesis markers (47-49). In the present study, NANOG was significantly upregulated following nilotinib usage. This transcription factor is involved in three different pathways (Embryonic stem cell differentiation into cardiac lineages, Transcriptional regulatory network in embryonic stem cells and Role of Oct4 in mammalian embryonic stem cell pluripotency), which principally control cell pluripotency and self-renewal mechanisms. NANOG regulates the proliferation and differentiation of primary bone mesenchymal stem cells (MSCs), as well as skeletal tissue regeneration. NANOG in MSCs leads to increased proliferative capacity but expression is downregulated as MSCs undergo osteogenic differentiation (50). Another study revealed crosstalk between NANOG and the bone morphogenic protein cascade via direct binding to SMAD1 (51).
Using IPA analysis, the most upregulated upstream regulators in imatinib- and nilotinib-treated cells were identified, which are involved in a wide variety of biological processes. A number of these genes have been shown to influence bone metabolism. Nilotinib markedly increased the expression of Activin A receptors (ACVRs). ACVR1B and ACVR1C are transmembrane serine/threonine kinases that are members of the transforming growth factor β (TGF-β) pathway. The type-1 and type-2 receptors form a heteromeric complex. Within the complex, the type 1 receptors act as downstream transducers of activin signals and phosphory-late SMAD proteins. The activin signaling has well-known role in bone homeostasis (24). Activins affect proliferation, differentiation and activity of bone cells. Furthermore, they can influence bone turnover, bone mass and fracture risks (24). One of the most upregulated molecules following imatinib application was growth differentiation factor 10 (GDF10), also termed bone morphogenetic protein 3B, which is a member of the TGFβ superfamily. GDF10 is involved in skeletal morphogenesis, regulates osteoblast differentiation and supports skeletal system development (https://reports.ingenuity.com, http://www.genecards.org). Unlike the known bone-forming BMPs, GDF10 is a negative regulator of BMD (52). According to in vitro and in vivo observations, GDF10 inhibits osteogen-esis and bone formation (53).
STXBP5L and RPS23 genes were the most sensitive with the greatest fold changes following imatinib and nilotinib treatment. STXBP5L participates in vesicle transport and exocytosis (http://www.genecards.org). It is also involved in the regulation of protein secretion, protein transport, glucose homeostasis and regulation of insulin secretion (https://reports.ingenuity.com, http://www.ncbi.nlm.nih.gov/gene). RPS23 gene encodes a cytoplasm-located ribosomal protein that is a component of the 40S subunit (https://reports.ingenuity.com, http://www.genecards.org). However, the direct role of these two most upregulated molecules in bone metabolism is presently unknown.
The limitations of this preliminary study are that the experiments were only performed on MC3T3-E1 osteoblastic cells. Further investigation is required to confirm changes in a number of the reported genes by reverse transcription-quantitative polymerase chain reaction or functional tests, and parallel treatment of other cell lines in order to validate the biologically significant targets.
In conclusion, the specifically altered gene expression patterns in response to imatinib and nilotinib observed in the osteoblast cultures may explain the previously observed in vivo clinical changes in bone metabolism. Further studies to assess the biological significance of these observations are required.
Acknowledgments
The authors would like to thank Dr Paul Manley and Mrs. Andrea Khoshdel (Novartis, Budapest, Hungary) for providing imatinib and nilotinib compounds and Dr István Nagy, director of SeqOmics Biotechnology Ltd. (Szeged, Hungary) for high-throughput RNA sequencing. They would also like to acknowledge Dr Attila Patócs, Dr István Marczell and Dr Zsolt Nagy from the MTA-SE Molecular Genetic Research Group at the 2nd Department of Internal Medicine, Semmelweis University (Budapest, Hungary) for their professional assistance with Ingenuity Pathway Analyses.
Abbreviations
- TKI
tyrosine kinase inhibitors
- CML
chronic myelogenous leukemia
- ABL
abelson kinase
- BCR-ABL
breakpoint cluster region-abelson fusion oncoprotein
- OPG
osteoprotegerin
- RANKL
receptor activator of nuclear factor κ-B ligand
- c-KIT
stem cell growth factor receptor
- PDGFR
platelet-derived growth factor receptor
- TBV
trabecular bone volume
- BMD
bone mineral density
- SOLiD
Sequencing by Oligonucleotide Ligation and Detection
- IPA
ingenuity pathway analysis
- logFC
logarithmic fold change
References
- 1.Cohen MH, Moses ML, Pazdur R. Gleeve™ for the treatment of chronic myelogenous leukemia: U.S. food and drug administration regulatory mechanisms, accelerated approval, and orphan drug status. Oncologist. 2002;7:390–392. doi: 10.1634/theoncologist.7-5-390. [DOI] [PubMed] [Google Scholar]
- 2.Tibullo D, Barbagallo I, Giallongo C, La Cava P, Branca A, Conticello C, Stagno F, Chiarenza A, Palumbo GA, Di Raimondo F. Effects of second-generation tyrosine kinase inhibitors towards osteogenic differentiation of human mesenchymal cells of healthy donors. Hematol Oncol. 2012;30:27–33. doi: 10.1002/hon.988. [DOI] [PubMed] [Google Scholar]
- 3.O'Sullivan S, Lin JM, Watson M, Callon K, Tong PC, Naot D, Horne A, Aati O, Porteous F, Gamble G, et al. The skeletal effects of the tyrosine kinase inhibitor nilotinib. Bone. 2011;49:281–289. doi: 10.1016/j.bone.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 4.Wihlidal P, Karlic H, Pfeilstöcker M, Klaushofer K, Varga F. Imatinib mesylate (IM)-induced growth inhibition is associated with production of spliced osteocalcin-mRNA in cell lines. Leuk Res. 2008;32:437–443. doi: 10.1016/j.leukres.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 5.Tibullo D, Giallongo C, La Cava P, Berretta S, Stagno F, Chiarenza A, Conticello C, Palumbo GA, Di Raimondo F. Effects of imatinib mesylate in osteoblastogenesis. Exp Hematol. 2009;37:461–468. doi: 10.1016/j.exphem.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 6.O'Sullivan S, Naot D, Callon K, Porteous F, Horne A, Wattie D, Watson M, Cornish J, Browett P, Grey A. Imatinib promotes osteoblast differentiation by inhibiting PDGFR signaling and inhibits osteoclastogenesis by both direct and stromal cell-dependent mechanisms. J Bone Miner Res. 2007;22:1679–1689. doi: 10.1359/jbmr.070719. [DOI] [PubMed] [Google Scholar]
- 7.Fitter S, Dewar AL, Kostakis P, To LB, Hughes TP, Roberts MM, Lynch K, Vernon-Roberts B, Zannettino AC. Long-term imatinib therapy promotes bone formation in CML patients. Blood. 2008;111:2538–2547. doi: 10.1182/blood-2007-07-104281. [DOI] [PubMed] [Google Scholar]
- 8.Jönsson S, Hjorth-Hansen H, Olsson B, Wadenvik H, Sundan A, Standal T. Imatinib inhibits proliferation of human mesenchymal stem cells and promotes early but not late osteoblast differentiation in vitro. J Bone Miner Metab. 2012;30:119–123. doi: 10.1007/s00774-011-0323-3. [DOI] [PubMed] [Google Scholar]
- 9.Fierro F, Illmer T, Jing D, Schleyer E, Ehninger G, Boxberger S, Bornhauser M. Inhibition of platelet-derived growth factor receptorbeta by imatinib mesylate suppresses proliferation and alters differentiation of human mesenchymal stem cells in vitro. Cell Prolif. 2007;40:355–366. doi: 10.1111/j.1365-2184.2007.00438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vandyke K, Fitter S, Dewar AL, Hughes TP, Zannettino AC. Dysregulation of bone remodeling by imatinib mesylate. Blood. 2010;115:766–774. doi: 10.1182/blood-2009-08-237404. [DOI] [PubMed] [Google Scholar]
- 11.Benito R, Lumbreras E, Abáigar M, Gutiérrez NC, Delgado M, Robledo C, García JL, Rodríguez-Vicente AE, Cañizo MC, Rivas JM. Imatinib therapy of chronic myeloid leukemia restores the expression levels of key genes for DNA damage and cell-cycle progression. Pharmacogenet Genomics. 2012;22:381–388. doi: 10.1097/FPC.0b013e328351f3e9. [DOI] [PubMed] [Google Scholar]
- 12.Vandyke K, Fitter S, Drew J, Fukumoto S, Schultz CG, Sims NA, Yeung DT, Hughes TP, Zannettino AC. Prospective histomorphometric and DXA evaluation of bone remodeling in imatinib-treated CML patients: Evidence for site-specific skeletal effects. J Clin Endocrinol Metab. 2013;98:67–76. doi: 10.1210/jc.2012-2426. [DOI] [PubMed] [Google Scholar]
- 13.Berman E, Nicolaides M, Maki RG, Fleisher M, Chanel S, Scheu K, Wilson BA, Heller G, Sauter NP. Altered bone and mineral metabolism in patients receiving imatinib mesylate. N Engl J Med. 2006;354:2006–2013. doi: 10.1056/NEJMoa051140. [DOI] [PubMed] [Google Scholar]
- 14.Berman E, Girotra M, Cheng C, Chanel S, Maki R, Shelat M, Strauss HW, Fleisher M, Heller G, Farooki A. Effect of long term imatinib on bone in adults with chronic myelogenous leukemia and gastrointestinal stromal tumors. Leuk Res. 2013;37:790–794. doi: 10.1016/j.leukres.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Jonsson S, Olsson B, Ohlsson C, Lorentzon M, Mellström D, Wadenvik H. Increased cortical bone mineralization in imatinib treated patients with chronic myelogenous leukemia. Haematologica. 2008;93:1101–1103. doi: 10.3324/haematol.12373. [DOI] [PubMed] [Google Scholar]
- 16.O'Sullivan S, Horne A, Wattie D, Porteous F, Callon K, Gamble G, Ebeling P, Browett P, Grey A. Decreased bone turnover despite persistent secondary hyperparathyroidism during prolonged treatment with imatinib. J Clin Endocrinol Metab. 2009;94:1131–1136. doi: 10.1210/jc.2008-2324. [DOI] [PubMed] [Google Scholar]
- 17.Hoehn D, Medeiros LJ, Kantarjian HM, Cortes JE, Wang XM, Bueso-Ramos CE. Digital image analysis as a tool to assess the effects of imatinib on trabecular bone in patients with chronic myelogenous leukemia. Hum Pathol. 2012;43:2354–2359. doi: 10.1016/j.humpath.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 18.Lawrence L. Long-term treatment with imatinib affected bone mineral density. Cancer Network; 2013. [Google Scholar]
- 19.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 20.Yoav Benjamini YH. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 21.Onishi M, Fujita Y, Yoshikawa H, Yamashita T. Inhibition of Rac1 promotes BMP-2-induced osteoblastic differentiation. Cell Death Dis. 2013;4:e698. doi: 10.1038/cddis.2013.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma Z, Thomas KS, Webb DJ, Moravec R, Salicioni AM, Mars WM, Gonias SL. Regulation of Rac1 activation by the low density lipoprotein receptor-related protein. J Cell Biol. 2002;159:1061–1070. doi: 10.1083/jcb.200207070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S, Guo HB, Liu YS, Wu F, Zhang H, Zhang Z, Xie Z, Sheng Z, Liao E. Relationships of serum lipid profiles and bone mineral density in postmenopausal Chinese women. Clin Endocrinol (Oxf) 2015;82:53–58. doi: 10.1111/cen.12616. [DOI] [PubMed] [Google Scholar]
- 24.Lotinun S, Scott Pearsall R, Horne WC, Baron R. Activin receptor signaling: A potential therapeutic target for osteoporosis. Curr Mol Pharmacol. 2012;5:S195–S204. doi: 10.2174/1874467211205020195. [DOI] [PubMed] [Google Scholar]
- 25.Chanprasertyothin S, Saetung S, Rajatanavin R, Ongphiphadhanakul B. Genetic variant in the aquaporin 9 gene is associated with bone mineral density in postmenopausal women. Endocrine. 2010;38:83–86. doi: 10.1007/s12020-010-9353-1. [DOI] [PubMed] [Google Scholar]
- 26.Aharon R, Bar-Shavit Z. Involvement of aquaporin 9 in osteoclast differentiation. J Biol Chem. 2006;281:19305–19309. doi: 10.1074/jbc.M601728200. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Song L, Wang Y, Rojek A, Nielsen S, Agre P, Carbrey JM. Osteoclast differentiation and function in aquaglyceroporin AQP9-null mice. Biol Cell. 2009;101:133–140. doi: 10.1042/BC20080083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraus DM, Elliott GS, Chute H, Horan T, Pfenninger KH, Sanford SD, Foster S, Scully S, Welcher AA, Holers VM. CSMD1 is a novel multiple domain complement-regulatory protein highly expressed in the central nervous system and epithelial tissues. J Immunol. 2006;176:4419–4430. doi: 10.4049/jimmunol.176.7.4419. [DOI] [PubMed] [Google Scholar]
- 29.Hiramitsu S, Terauchi M, Kubota T. The effects of Dickkopf-4 on the proliferation, differentiation, and apoptosis of osteoblasts. Endocrinology. 2013;154:4618–4626. doi: 10.1210/en.2013-1387. [DOI] [PubMed] [Google Scholar]
- 30.Bais M, McLean J, Sebastiani P, Young M, Wigner N, Smith T, Kotton DN, Einhorn TA, Gerstenfeld LC. Transcriptional analysis of fracture healing and the induction of embryonic stem cell-related genes. PLoS One. 2009;4:e5393. doi: 10.1371/journal.pone.0005393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aleman JO, Farooki A, Girotra M. Effects of tyrosine kinase inhibition on bone metabolism: Untargeted consequences of targeted therapies. Endocr Relat Cancer. 2014;21:R247–R259. doi: 10.1530/ERC-12-0400. [DOI] [PubMed] [Google Scholar]
- 32.Rix U, Hantschel O, Düernberger G, Remsing Rix LL, Planyavsky M, Fernbach NV, Kaupe I, Bennett KL, Valent P, Colinge J, et al. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib and dasatinib, reveal novel kinase and nonkinase targets. Blood. 2007;110:4055–4063. doi: 10.1182/blood-2007-07-102061. [DOI] [PubMed] [Google Scholar]
- 33.Yang JY, Jung JY, Cho SW, Choi HJ, Kim SW, Kim SY, Kim HJ, Jang CH, Lee MG, Han J, Shin CS. Chloride intracellular channel 1 regulates osteoblast differentiation. Bone. 2009;45:1175–1185. doi: 10.1016/j.bone.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 34.Fujimori S, Hinoi E, Yoneda Y. Functional GABA (B) receptors expressed in cultured calvarial osteoblasts. Biochem Biophys Res Commun. 2002;293:1445–1452. doi: 10.1016/S0006-291X(02)00405-9. [DOI] [PubMed] [Google Scholar]
- 35.Mentink A, Hulsman M, Groen N, Licht R, Dechering KJ, van der Stok J, Alves HA, Dhert WJ, van Someren EP, Reinders MJ, et al. Predicting the therapeutic efficacy of MSC in bone tissue engineering using the molecular marker CADM1. Biomaterials. 2013;34:4592–4601. doi: 10.1016/j.biomaterials.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Muhammad SI, Maznah I, Mahmud R, Zuki AB, Imam MU. Upregulation of genes related to bone formation by γ-amino butyric acid and gamma-oryzanol in germinated brown rice is via the activation of GABA (B)-receptors and reduction of serum IL-6 in rats. Clin Interv Aging. 2013;8:1259–1271. doi: 10.2147/CIA.S45943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahata Y, Takarada T, Hinoi E, Nakamura Y, Fujita H, Yoneda Y. Osteoblastic γ-aminobutyric acid, type B receptors negatively regulate osteoblastogenesis toward disturbance of osteoclastogenesis mediated by receptor activator of nuclear factor κB ligand in mouse bone. J Biol Chem. 2011;286:32906–32917. doi: 10.1074/jbc.M111.253526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schrauwen I, Ealy M, Huentelman MJ, Thys M, Homer N, Vanderstraeten K, Fransen E, Corneveaux JJ, Craig DW, Claustres M, et al. A genome-wide analysis identifies genetic variants in the RELN gene associated with otosclerosis. Am J Hum Genet. 2009;84:328–338. doi: 10.1016/j.ajhg.2009.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.May P, Herz J, Bock HH. Molecular mechanisms of lipo-protein receptor signalling. Cell Mol Life Sci. 2005;62:2325–2338. doi: 10.1007/s00018-005-5231-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westbroek I, van der Plas A, de Rooij KE, Klein-Nulend J, Nijweide PJ. Expression of serotonin receptors in bone. J Biol Chem. 2001;276:28961–28968. doi: 10.1074/jbc.M101824200. [DOI] [PubMed] [Google Scholar]
- 41.Bliziotes MM, Eshleman AJ, Zhang XW, Wiren KM. Neurotransmitter action in osteoblasts: Expression of a functional system for serotonin receptor activation and reuptake. Bone. 2001;29:477–486. doi: 10.1016/S8756-3282(01)00593-2. [DOI] [PubMed] [Google Scholar]
- 42.Dai SQ, Yu LP, Shi X, Wu H, Shao P, Yin GY, Wei YZ. Serotonin regulates osteoblast proliferation and function in vitro. Braz J Med Biol Res. 2014;47:759–765. doi: 10.1590/1414-431X20143565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yadav VK, Karsenty G. Serotonin: A new player in the regulation of bone remodeling. Medicographia. 2010;32:357–363. [Google Scholar]
- 44.Bliziotes M. Update in serotonin and bone. J Clin Endocrinol Metab. 2010;95:4124–4132. doi: 10.1210/jc.2010-0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ducy P, Karsenty G. The two faces of serotonin in bone biology. J Cell Biol. 2010;191:7–13. doi: 10.1083/jcb.201006123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Battaglino R, Fu J, Späte U, Ersoy U, Joe M, Sedaghat L, Stashenko P. Serotonin regulates osteoclast differentiation through its transporter. J Bone Miner Res. 2004;19:1420–1431. doi: 10.1359/JBMR.040606. [DOI] [PubMed] [Google Scholar]
- 47.Saito A, Ochiai K, Kondo S, Tsumagari K, Murakami T, Cavener DR, Imaizumi K. Endoplasmic reticulum stress response mediated by the PERK-eIF2 alpha-ATF4 pathway is involved in osteoblast differentiation induced by BMP2. J Biol Chem. 2011;286:4809–4818. doi: 10.1074/jbc.M110.152900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamamura K, Yokota H. Stress to endoplasmic reticulum of mouse osteoblasts induces apoptosis and transcriptional activation for bone remodeling. FEBS Lett. 2007;581:1769–1774. doi: 10.1016/j.febslet.2007.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirasawa H, Jiang C, Zhang P, Yang FC, Yokota H. Mechanical stimulation suppresses phosphorylation of eIF2alpha and PERK-mediated responses to stress to the endoplasmic reticulum. FEBS Lett. 2010;584:745–752. doi: 10.1016/j.febslet.2009.12.028. [DOI] [PubMed] [Google Scholar]
- 50.Bais MV, Shabin ZM, Young M, Einhorn TA, Kotton DN, Gerstnefeld LC. Role of Nanog in the maintenance of marrow stromal stem cells during post natal bone regeneration. Biochem Biophys Res Commun. 2012;417:211–216. doi: 10.1016/j.bbrc.2011.11.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki A, Raya A, Kawakami Y, Morita M, Matsui T, Nakashima K, Gaget FH, Rodríguez-Esteban C, Izpisúa Belmonte JC. Nanog binds to Smad1 and blocks bone morphogenetic protein-induced differentiation of embryonic stem cells. Proc Natl Acad Sci USA. 2006;103:10294–10299. doi: 10.1073/pnas.0506945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daluiski A, Engstrand T, Bahamonde ME, Gamer LW, Agius E, Stevenson SL, Cox K, Rosen V, Lyons KM. Bone morphogenetic protein-3 is a negative regulator of bone density. Nat Genet. 2001;27:84–88. doi: 10.1038/83810. [DOI] [PubMed] [Google Scholar]
- 53.Bahamonde ME, Lyons KM. BMP3: To be or not to be a BMP. J Bone Joint Surg. 2001;83-A(Suppl 1):S56–S62. [PubMed] [Google Scholar]