Abstract
Apollon, an unusually large member of the inhibitors of apoptosis protein family, may be important for oncogenesis development. The aim of the present study was to assess the association between esophageal squamous cell carcinoma (ESCC) and Apollon expression levels, and to highlight the association between Apollon and the occurrence, development and prognosis of ESCC. Apollon expression was detected by immunohistochemical staining and reverse transcription-quantitative polymerase chain reaction in ESCC tissues, adjacent non-cancerous tissues and paired normal tissues respectively, in order to analyze the association between Apollon expression and the clinicopathological features of ESCC. Survival analysis was used to assess the prognostic significance of Apollon expression. It was determined that the mRNA and protein expression levels of Apollon were significantly higher in the carcinoma tissues compared with the adjacent non-cancerous tissues and normal control tissues (P<0.001). There was a significant difference in lymph node involvement and the tumor, nodes, and metastases stage in patients categorized according to different Apollon expression levels. The prognostic significance of Apollon was also determined using the log-rank method. The overexpression of Apollon was associated with shorter overall survival and disease-free survival rates. The present study indicates that Apollon expression is associated with the biological characteristics of ESCC, and may be a valuable prognostic factor and a novel chemotherapeutic target for ESCC treatment.
Keywords: esophageal squamous cell carcinoma, Apollon, apoptosis, inhibitors of apoptosis protein, prognosis
Introduction
The incidence of esophageal cancer ranks seventh in malignant diseases worldwide; the mortality rate ranks sixth (1). Esophageal squamous cell carcinoma (ESCC) is the dominant pathological type. Early ESCC clinical symptoms and signs are atypical and are frequently overlooked, thus early diagnosis is difficult. Additionally, it is easy for ESCC to progress to the late stage, owing to transfer to or an invasion of the surrounding organs, namely the trachea and aorta, which are adjacent to the esophagus; this process is facilitated by the esophagus not having a serous layer. Clinically, for more than half of patients with ESCC, the opportunity to have the tumor removed at diagnosis is missed, which is a cause of concern for the clinical doctors. The current treatment of ESCC is the combined strategy of surgical excision, radiotherapy and chemotherapy; however, it is largely ineffective, since the 5-year survival rate subsequent to surgery is 20–40% (2,3).
Apoptosis is a highly regulated process, necessary for homeostasis, as it removes excessively damaged and aging mutant cells (4). Dysfunction in the apoptotic process contributes to the pathogenesis of a variety of malignant tumors, and it is considered to be the key reason for their occurrence and development, being closely associated with tumor drug resistance (5). In the event that apoptosis is hindered, the mutant cells accumulate and increase in an uncontrolled manner (6). The members of the inhibitors of apoptosis protein (IAP) family are important negative regulators of apoptosis (7,8). The IAP family comprises eight members, which are the key regulators of cytokinesis, apoptosis and signal transduction and have been demonstrated to be tightly associated with oncogenesis. Apollon, also termed baculoviral IAP repeat containing 6 (BIRC6), a membrane-associated protein that is localized on the Golgi compartment and the vesicular system, is the largest member of the IAP family (9), which was identified by Chen et al (10) in 1999. Its gene is located on chromosome 2p21–22, encoding a large protein (530 kDa) that contains a single baculovirus inhibitor of apoptosis protein repeat domain and a ubiquitin-conjugating enzyme domain. Apollon is expressed in the brain, placenta, testes, lymphatic cells and secretory organs (10). A previous study indicated that Apollon was a dual regulator of cell proliferation and cell death, with multiple functions, due to its different functional domains and diverse binding patterns (9). It inhibits apoptosis by directly binding to cysteine/aspartate-specific proteases (the caspase family). Its antiapoptotic function is more marked compared with the B-cell chronic lymphocytic leukemia/lymphoma 2 family, primarily via a combination of diablo IAP-binding mitochondrial protein (Smac), HtrA serine peptidase 2 (HtrA2) and the caspases (11,12). Apollon has been identified to be important for apoptosis resistance in various types of cancer (10,11,13–16). Previous studies have determined that the upregulation of Apollon may be important for tumor generation (17–21). However, the association between ESCC and Apollon requires further elucidation.
The present study detected Apollon expression using immunohistochemistry and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) in ESCC tissues, adjacent non-cancerous tissues (ANCTs) and normal tissues. Apollon expression and clinicopathological features of ESCC were analyzed to highlight the association between Apollon and the occurrence, development and prognosis of ESCC.
Materials and methods
Patients and specimens
Patients were enrolled in the present study from Thoracic Surgery Xiangya Hospital, Central South University between May 2010 and May 2012, who were diagnosed with ESCC by more than two pathologists. Specimens were obtained, including 80 cases of ESCC tissues, 80 cases of paired ANCTs (~3–7 cm from the tumor margin) and 50 cases of normal tissues, from intraoperative radical esophagectomy specimens (>7 cm from the tumor margin). All specimens were immediately snap-frozen in liquid nitrogen and stored at −80°C until RNA and total protein extraction was performed. The patients were 69 males and 11 females, younger than 75 years of age and first-diagnosed cases, and they had not been exposed to chemotherapy, radiotherapy or other treatments prior to being sampled. The clinical and pathological data were complete and reliable. Informed consent was acquired from all patients prior to surgery. This study was approved by the Xiangya Hospital, Central South University Ethics Committee, and the usage of the information and specimens collected has been handled and anonymized according to the ethical and legal standards.
Reagents and instruments
Rabbit polyclonal anti-Apollon antibodies were purchased from Abcam (cat. nos. ab84429 and ab19609; Cambridge, UK); the PV-6001 two-step immunohistochemistry kit (cat. no. PV-6001) was from OriGene Technologies, Inc. (Beijing, China); Invitrogen® TRIzol reagent (cat. no. 15596026) was from Thermo Fisher Scientific, Inc. (Waltham, MA, USA); the GoScript Reverse Transcription kit (cat. no. A5001) was from Promega Corporation (Madison, WI, USA); the GoTaq RT-qPCR kit (cat. no. A6001) was from Promega Corporation; paraffin-embedded machines, the paraffin slicing machine and the automatic upright microscope system (DM5000 B) were from Leica Microsystems (GmbH, Wetzlar, Germany); the 400W UV imaging system was from Kodak (Kodak, Tokyo, Japan); and the ABI PRISM 7500 PCR applications were purchased from Thermo Fisher Scientific, Inc.
Immunohistochemistry
The rapid PV two-step staining method was performed with the following specifications: The paraffin slice thickness was 5 µm, and slices were grilled at 65°C for 60 min, followed by dewaxing, hydration and high-temperature antigen retrieval in a microwave in 0.1 M citrate solution (pH 6.0) for 10 min. Subsequently, the slices were incubated with 3% H2O2 at room temperature for 20 min, followed by an incubation with goat serum at room temperature for 20 min, and a subsequent incubation with the Apollon antibody (1:400) at 4°C overnight. The samples were then re-warmed and incubated with the anti-rabbit secondary antibodies in the PV-6001 two-step immunohistochemistry kit (cat. no. PV-6001; ZSGB-Bio Co., Ltd., Beijing, China) at room temperature for 20 min. The slices were stained with 3,3′-diaminobenzidine and hematoxylin and mounted, prior to microscopic examination. Immunohistochemical staining was scored independently by two pathologists without knowledge of the patients' characteristics. Any discrepancy was resolved by taking the consensus view. The score of immunoreactivity was performed by calculating the extent and intensity of staining positivity of the cells in a semi-quantitative manner. Interpretation of the scores was performed as previously described (22). The standards for evaluation included the following: Positive stain intensity (0, negative; 1, weak positive; 2, moderate positive; 3, strong positive) and proportion of positive areas (≤10%=1, 10–50%=2, ≥50%=3). The staining score was the multiplication of the positive stain intensity and the proportion of positive areas. Five high-power fields in each specimen were selected at random, and the final score was the average of the five scores.
RT-qPCR
Total RNA was extracted from 1 mg esophagus tissue using TRIzol, according to the manufacturers' protocol. Subsequently, samples were treated with a DNA-free kit (Ambion; Thermo Fisher Scientific, Inc.) to remove the chromosomal DNA. The complementary DNA (cDNA) was synthesized using the GoScript Reverse Transcription kit and stored at −20°C prior to use. The mRNA expression levels of Apollon and β-actin were determined by RT-qPCR using the ABI PRISM 7500 sequence detector system. The primer sequences were: Apollon, forward (F): 5′-TGACAGGGCATACATCACAG-3′, reverse (R): 5′-GCAACAATCTCCCACTGAAG-3′; and β-actin, F: 5′-GCACCACACCTTCTACAATGAG-3′, R: 5′-GATAGCACAGCCTGGATAGCA-3′. The mRNA expression levels of the target genes were normalized to the β-actin signal as a housekeeping gene. All the reactions were performed in triplicate using 20 µl samples containing 50 ng cDNA. The reaction protocol involved heating for 10 min at 95°C, followed by 40 cycles of amplification (15 sec at 95°C and 1 min at 60°C). The data were analyzed using the ABI PRISM 7500 Sequence Detection software v2.0.1. The levels of Apollon expression in unknown samples were calculated as a ratio of Apollon to β-actin. Data were analyzed using the 2−ΔΔCq method (23) and expressed as the log2 fold-change [−ΔΔCq (ESCC / ANCT)].
Statistical analysis
The patients were categorized into two groups according to the level of Apollon expression (high and low). The differences in Apollon expression with respect to the clinical factors at diagnosis, including age, gender, drinking history (>10 years), tumor size, differentiation grade, tumor (T)-classification, node (N)-classification and tumor, nodes, and metastases (TNM) stage, were analyzed. The expression levels are presented as median values. Counted data were compared using a one-way analysis of variance or an unpaired t-test. Qualitative data were compared using Pearson's χ2 test or Fisher's exact test. The variables were evaluated in a stepwise multivariate Cox analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Classification of the patients and their characteristics
A total of 80 patients were enrolled in the study. The mean age of the patients was 59.5 years (range 42–74 years). According to the 7th edition of the International Union Against Cancer's esophageal cancer staging (2009 edition) (24), the enrolled patients were classified as follows: 20 cases of stage I, 25 cases of stage II and 35 cases of stage III.
Apollon expression is upregulated in ESCC tissues
In the present study, Apollon protein was detected by immunohistochemical staining. Apollon protein was expressed in ESCC tissues, ANCTs and normal esophageal tissues, primarily in the cytoplasm of carcinoma and squamous cells, or in the normal and adjacent tissues in the underlying basal cells. In contrast with the staining of Apollon in normal esophagus tissues, which was mostly negative (Fig. 1A), the positive reactants of Apollon were yellow or brown substances that mainly existed in the cytoplasm (Fig. 1B and C). According to the standards used for evaluating the samples (as detailed in the Materials and methods section), the high expression level of Apollon protein was 60% (48/80) in the tumor tissues, whereas it was 2.5% (2/80) in the ANCTs and 0% (0/30) in the normal esophageal mucosa epithelial tissue. The difference between the different tissue groups was statistically significant (P<0.0001, Fig. 1D). No significant difference was identified in the Apollon expression levels in the ANCT group compared with the normal tissue group (P=0.132, Fig. 1D).
Figure 1.
Apollon protein and mRNA levels are frequently upregulated in ESCC. (A) Apollon was mostly negative in normal esophagus tissues. Magnification, ×400. The different expression levels of Apollon in various ESCC tissues are shown: (B) well differentiated ESCC and (C) poorly differentiated ESCC. A trend was observed that the lower the level of differentiation, the higher is the expression level. Magnification, ×400. (D) Compared with the ANCTs and the normal tissues, the ESCC tissues were revealed to express significantly higher levels of Apollon in the cytoplasm. ***P<0.001, Apollon immunohistochemical staining scores in the ESCC group vs. the ANCT group; *P=0.132, ANCT group vs. the normal group. (E) Expression of Apollon mRNA in 80 pairs of ESCC tissues and the ANCTs. Expression levels of Apollon were normalized to the corresponding levels of β-actin. (F) A positive correlation was observed between Apollon protein expression and the Apollon mRNA levels. ESCC, esophageal squamous cell carcinoma; ANCT, adjacent non-cancerous tissues.
The mRNA expression levels of Apollon in the 80 cases of ESCCs and the paired ANCTs were subsequently analyzed by RT-qPCR. A similar pattern of Apollon expression in terms of the mRNA levels was identified: Compared with the paired ANCTs, a 3-fold increase in Apollon expression was noted in the ESCC tissues (Fig. 1E). Comparative analysis of paired ESCCs with ANCTs revealed that increased Apollon expression (>2-fold [namely log2 (fold change)>1]) was observed in 75% of the cases (60/80). The immunohistochemical staining score was positively associated with the fold expression of Apollon mRNA (Fig. 1F); therefore, the overexpression of Apollon may be a frequent event in human ESCC.
Expression levels of Apollon are associated with the ESCC clinicopathological characteristics
The association between Apollon expression and the clinicopathological characteristics of ESCC was determined by the χ2 test. As summarized in Table I, the high expression of Apollon was significantly associated with the TNM stage (P<0.001) and lymph node (N classification) metastasis (P<0.001). However, no significant difference was identified between Apollon expression and age (P=1.000), gender (P=1.000), drinking history (P=0.587), tumor differentiation (P=0.119) or tumor size (P=0.301).
Table I.
Correlations between the Apollon expression level and clinicopathological characteristics of 80 cases of ESCC.
| Clinicopathological variable | n | Apollon expression
|
P-value | |
|---|---|---|---|---|
| Low | High | |||
| Age (years) | 1.000 | |||
| ≤60 | 39 | 10 | 29 | |
| >60 | 41 | 10 | 31 | |
| Gender | 1.000 | |||
| Female | 11 | 3 | 8 | |
| Male | 69 | 17 | 52 | |
| Drinking history (years) | 0.587 | |||
| ≤10 | 26 | 8 | 19 | |
| >10 | 54 | 12 | 41 | |
| Differentiation | 0.123 | |||
| Well | 21 | 6 | 15 | |
| Moderate | 42 | 13 | 29 | |
| Poor | 17 | 1 | 16 | |
| Tumor size (cm) | 0.301 | |||
| ≤4 | 45 | 9 | 36 | |
| >4 | 35 | 11 | 24 | |
| T classification | 0.464 | |||
| T1 | 28 | 10 | 18 | |
| T2 | 12 | 2 | 10 | |
| T3 | 30 | 6 | 24 | |
| T4 | 10 | 2 | 8 | |
| N classification | <0.001 | |||
| N0 | 40 | 18 | 22 | |
| N1 | 22 | 2 | 20 | |
| N2 | 18 | 0 | 18 | |
| TNM stage | <0.001 | |||
| I | 20 | 11 | 9 | |
| II | 25 | 6 | 19 | |
| III | 35 | 3 | 32 | |
Statistically significant P-values are shown in bold. Data were compared by using Pearson's χ2 or Fisher's exact test. T classification, tumor classification; N classification, node classification; TNM stage, tumor, nodes, and metastases stage; ESCC, esophageal squamous cell carcinoma.
Upregulation of Apollon expression is associated with the prognosis of ESCC
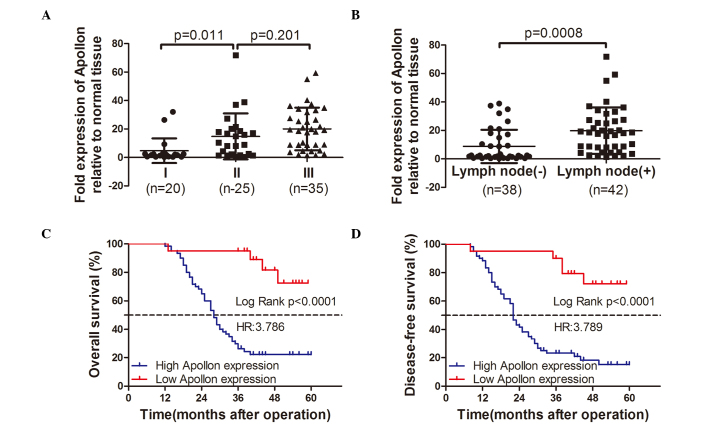
To assess the feasibility of using Apollon expression as an ESCC prognostic factor, the Cox proportional hazards regression model was introduced. Multivariate survival analysis on all parameters was performed. It was determined that overall survival time was significantly dependent on lymph node (N classification) metastasis (P=0.009), TNM stage (P=0.036) and Apollon expression levels (P=0.047; Table II), which indicated that patients with ESCC who had high levels of Apollon expression also had shorter overall survival times (median survival time: 28 vs. >60 months, P<0.001; Fig. 2) compared with the low Apollon expression group. Additionally, it was determined that lymph node metastasis (P= 0.044), TNM stage (P= 0.013) and Apollon expression levels (P=0.019; Table II) indicated significant differences for disease-free survival rates in the multivariate survival analysis. Patients with ESCC who had high Apollon expression also had shorter disease-free survival rates (median survival time: 22 vs. >60 months, P<0.001; Fig. 2) compared with the low Apollon expression group.
Table II.
Cox regression multivariate analysis of overall and disease-free survival in 80 patients with ESCC.
| Variables | n | Overall survival
|
Disease-free survival
|
||
|---|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | ||
| Age (years) | |||||
| <60 | 39 | 1 | 1 | ||
| ≥60 | 41 | 0.76 (0.39–1.48) | 0.415 | 0.64 (0.34–1.23) | 0.183 |
| Gender | |||||
| Female | 11 | 1 | 1 | ||
| Male | 69 | 1.00 (0.366–2.78) | 0.986 | 0.92 (0.32–2.61) | 0.874 |
| Drinking history (years) | |||||
| ≤10 | 26 | 1 | 1 | ||
| >10 | 54 | 0.91 (0.41–2.04) | 0.817 | 1.07 (0.49–2.35) | 0.86 |
| Differentiation | 0.864 | 0.59 | |||
| Well | 21 | 1 | 1 | ||
| Moderate | 42 | 0.99 (0.43–2.31) | 0.997 | 1.19 (0.54–2.68) | 0.659 |
| Poor | 17 | 1.23 (0.48–3.16) | 0.667 | 1.58 (0.64–3.89) | 0.318 |
| Tumor size (cm) | |||||
| ≤4 | 42 | 1 | 1 | ||
| >4 | 38 | 1.00 (0.49–2.03) | 0.987 | 1.29 (0.67–2.50) | 0.446 |
| T classification | 0.580 | 0.533 | |||
| T1 | 28 | 1 | 1 | ||
| T2 | 13 | 1.54 (0.54–4.44) | 0.419 | 1.39 (0.49–3.91) | 0.525 |
| T3 | 29 | 0.66 (0.23–1.93) | 0.451 | 0.55 (0.19–1.54) | 0.255 |
| T4 | 10 | 0.84 (0.13–5.55) | 0.856 | 0.49 (0.72–3.35) | 0.469 |
| N classification | 0.009 | 0.044 | |||
| N0 | 39 | 1 | 1 | ||
| N1 | 23 | 2.79 (0.80–9.69) | 0.107 | 1.98 (0.61–6.45) | 0.259 |
| N2 | 18 | 14.90 (2.3–96.39) | 0.005 | 7.70 (1.29–46.02) | 0.025 |
| TNM stage | 0.036 | 0.013 | |||
| I | 20 | 1 | 1 | ||
| II | 25 | 18.53 (1.82–188.6) | 0.014 | 12.25 (2.29–65.71) | 0.003 |
| III | 35 | 41.72 (2.37–733.9) | 0.011 | 27.66 (2.55–300.41) | 0.006 |
| Apollon expression | 0.019 | ||||
| Low | 20 | 1 | 1 | ||
| High | 60 | 3.63 (1.02–12.95) | 0.047 | 4.03 (1.25–12.98) | |
The statistically significant P-values are shown in bold. T classification, tumor clssification; N classification, node classification; TNM stage, tumor, nodes, and metastases stage, ESCC, esophageal squamous cell carcinoma; HR, hazard ratio.
Figure 2.
Apollon is a promising prognostic marker for ESCC. According to the reverse transcription-quantitative polymerase chain reaction data, when the expression of Apollon was classified into low expression (n=20) and high expression (n=60), the level of Apollon expression was significantly different between the (A) tumor, nodes, and metastases stage and (B) lymph nodal metastasis. Kaplan-Meier survival analysis of Apollon expression in ESCCs. A higher Apollon expression indicates a lower (C) disease-free survival (P<0.001) and (D) overall survival (P<0.001). ESCC, esophageal squamous cell carcinoma.
Discussion
Apollon is the largest member of the IAP family (10), and it has been associated with various types of cancer (10,11,13–16), including breast, colon, lung, cervical and prostate cancer, gliomas, fibrosarcomas and osteosarcomas. However, the importance of Apollon in ESCC has not been fully elucidated. The current study investigated the protein and mRNA expression levels of Apollon in a series of 80 paired ESCC specimens with intact follow-up data.
Immunohistochemical staining results revealed that Apollon protein expression was higher in ESCC tissues compared with ANCTs and normal esophageal tissues, as demonstrated by western blot analysis. RT-qPCR analysis revealed high Apollon mRNA expression in the majority of cases of ESCC (Fig. 1), which is in line with a previous study on ESCC (25) and various other cancer studies (13–16), indicating that Apollon may be important for ESCC progression. Apollon contributes to oncogenesis through its interference in the apoptotic balance. Apollon has a prosurvival role due to its antiapoptotic function, which is completed by its binding to the activated caspases (caspase-3, caspase-8, caspase-9) to block the apoptotic pathway directly. Additionally, it can ubiquitinate the IAPs antagonists, Smac and HtrA2, and induce their proteolytic and degradative functions, which indirectly enhances the antiapoptotic activity of IAPs (11,12). Apollon may be able to switch on ESCC via a different molecular mechanism; however, further studies are required to clarify this.
From the clinical data, the expression level of Apollon in the high and low groups was compared in terms of the various clinicopathological factors of 80 patients with ESCC. It was determined that the expression of Apollon was not associated with the age, gender, drinking history, tumor size or tumor differentiation of the patients; however, it was significantly associated with the lymph node metastasis and TNM stage. Therefore, it is possible that the higher expression of Apollon leads to malignant biological characteristics, despite the tumor differentiation, which is inconsistent with previous studies (26–30): More Apollon expression indicated poorer differentiation. Similar results have been published by Low et al (16) in terms of prostate cancer and Apollon, which indicate that increased expression of Apollon was a late event in prostate cancer and did not correlate with differentiation.
Previous research has revealed that a high expression of Apollon was well correlated with lymph node metastasis (21) and poor survival (26,27). Lymph node metastasis may be used as a prognostic factor for ESCC; therefore, a multivariate survival analysis was performed on Apollon expression and the lymph node metastasis status. The results revealed that patients with high Apollon expression/more lymph node metastasis (N2) had shorter overall and disease-free survival times compared with patients with neither. Higher lymph node metastasis or Apollon overexpression worsens the prognosis. Therefore, the evaluation of Apollon expression in conjunction with the lymph node status may provide novel information for the prognosis of patients, and provide an opportunity for improved planning of appropriate treatment strategies and management subsequent to surgery. Apollon is an indicator for metastasis and prognosis for ESCC, which is consistent with previous studies regarding Apollon (26–30). Previous studies have also determined that Apollon was an independent, unfavorable prognostic factor for overall survival and disease-free survival rates in human epithelial ovarian cancer (26). Notably, abundant previous research has demonstrated that a high level of Apollon expression is associated with poor chemotherapeutic outcomes in various carcinomas, including colon cancer, acute leukemia, prostate cancer and melanoma, whereas silencing Apollon may improve patient sensitivity to chemotherapy (19,28,31,32). Therefore, Apollon may be a novel tumor therapeutic target; however, further investigation is required in order to elucidate the association between ESCC and Apollon.
Previous studies revealed that a significant overexpression of IAPs in cancer tissue, including cIAP-1, cIAP-2, X-linked inhibitor of apoptosis and Survivin, was regarded as an early event in esophageal cancer and did not correlate with histological type or the stage of tumors (33–35). However, according to the data from the current study, Apollon is an antitype: High expression of Apollon is a late cancer event, indicative of a poor prognosis, and, as it is distributed in the cytoplasm and is not secreted into the extracellular space, its value in terms of early diagnosis is limited.
In conclusion, the present study indicates, to the best of our knowledge for the first time, that Apollon expression is associated with the prognosis of ESCC and may be a potentially valuable prognostic factor and novel chemotherapeutic target for ESCC. The mechanisms of Apollon in tumorigenesis and metastasis remain unclear, although the present study provides the first step for future researchers to perform further investigations in vitro and in vivo in order to clarify the regulatory mechanisms of Apollon expression in ESCC in association with the apoptotic signaling pathways.
Acknowledgments
This study was supported by grants from the Hunan Provincial Natural Science Foundation of China (grant no. 14JJ7091) and Hunan Provincial Tumor Hospital Fund (grant no. A2013-02).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Mariette C, Piessen G, Triboulet JP. Therapeutic strategies in oesophageal carcinoma: Role of surgery and other modalities. Lancet Oncol. 2007;8:545–553. doi: 10.1016/S1470-2045(07)70172-9. [DOI] [PubMed] [Google Scholar]
- 3.Lerut T, Coosemans W, Decker G, De Leyn P, Nafteux P, Van Raemdonck D. Cancer of the esophagus and gastroesophageal junction: Potentially curative therapies. Surg Oncol. 2001;10:113–122. doi: 10.1016/S0960-7404(01)00027-5. [DOI] [PubMed] [Google Scholar]
- 4.Giancotti FG, Tarone G. Positional control of cell fate through joint integrin/receptor protein kinase signaling. Annu Rev Cell Dev Biol. 2003;19:173–206. doi: 10.1146/annurev.cellbio.19.031103.133334. [DOI] [PubMed] [Google Scholar]
- 5.Naito M. Molecular mechanism of apoptosis inhibition by IAPs and its implication to cancer therapy. Seikagaku. 2006;78:525–528. In Japanese. [PubMed] [Google Scholar]
- 6.Evan G. Cancer - a matter of life and cell death. Int J Cancer. 1997;71:709–711. doi: 10.1002/(SICI)1097-0215(19970529)71:5<709::AID-IJC2>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 7.Srinivasula SM, Ashwell JD. IAPs: what's in a name? Mol Cell. 2008;30:123–135. doi: 10.1016/j.molcel.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong RS. Apoptosis in cancer: From pathogenesis to treatment. J Exp Clin Cancer Res. 2011;30:87. doi: 10.1186/1756-9966-30-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hauser HP, Bardroff M, Pyrowolakis G, Jentsch S. A giant ubiquitin-conjugating enzyme related to IAP apoptosis inhibitors. J Cell Biol. 1998;141:1415–1422. doi: 10.1083/jcb.141.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z, Naito M, Hori S, Mashima T, Yamori T, Tsuruo T. A human IAP-family gene, apollon, expressed in human brain cancer cells. Biochem Biophys Res Commun. 1999;264:847–854. doi: 10.1006/bbrc.1999.1585. [DOI] [PubMed] [Google Scholar]
- 11.Qiu XB, Goldberg AL. The membrane-associated inhibitor of apoptosis protein, BRUCE/Apollon, antagonizes both the precursor and mature forms of Smac and caspase-9. J Biol Chem. 2005;280:174–182. doi: 10.1074/jbc.M411430200. [DOI] [PubMed] [Google Scholar]
- 12.Qiu XB, Markant SL, Yuan J, Goldberg AL. Nrdp1-mediated degradation of the gigantic IAP, BRUCE, is a novel pathway for triggering apoptosis. EMBO J. 2004;23:800–810. doi: 10.1038/sj.emboj.7600075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao Y, Sekine K, Kawabata A, Nakamura H, Ishioka T, Ohata H, Katayama R, Hashimoto C, Zhang X, Noda T, et al. Apollon ubiquitinates SMAC and caspase-9, and has an essential cytoprotection function. Nat Cell Biol. 2004;6:849–860. doi: 10.1038/ncb1159. [DOI] [PubMed] [Google Scholar]
- 14.Lamers F, Schild L, Koster J, Speleman F, Øra I, Westerhout EM, van Sluis P, Versteeg R, Caron HN, Molenaar JJ. Identification of BIRC6 as a novel intervention target for neuroblastoma therapy. BMC Cancer. 2012;12:285. doi: 10.1186/1471-2407-12-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bianchini M, Levy E, Zucchini C, Pinski V, Macagno C, De Sanctis P, Valvassori L, Carinci P, Mordoh J. Comparative study of gene expression by cDNA microarray in human colorectal cancer tissues and normal mucosa. Int J Oncol. 2006;29:83–94. [PubMed] [Google Scholar]
- 16.Low CG, Luk IS, Lin D, Fazli L, Yang K, Xu Y, Gleave M, Gout PW, Wang Y. BIRC6 protein, an inhibitor of apoptosis: Role in survival of human prostate cancer cells. PloS One. 2013;8:e55837. doi: 10.1371/journal.pone.0055837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pohl C, Jentsch S. Final stages of cytokinesis and midbody ring formation are controlled by BRUCE. Cell. 2008;132:832–845. doi: 10.1016/j.cell.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 18.Ren J, Shi M, Liu R, Yang QH, Johnson T, Skarnes WC, Du C. The Birc6 (Bruce) gene regulates p53 and the mitochondrial pathway of apoptosis and is essential for mouse embryonic development. Proc Natl Acad Sci USA. 2005;102:565–570. doi: 10.1073/pnas.0408744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu L, Gu J, Sun L, Qian Q, Qian C, Liu X. Oncolytic adenovirus-mediated shRNA against Apollon inhibits tumor cell growth and enhances antitumor effect of 5-fluorouracil. Gene Ther. 2008;15:484–494. doi: 10.1038/gt.2008.6. [DOI] [PubMed] [Google Scholar]
- 20.Lopergolo A, Pennati M, Gandellini P, Orlotti NI, Poma P, Daidone MG, Folini M, Zaffaroni N. Apollon gene silencing induces apoptosis in breast cancer cells through p53 stabilisation and caspase-3 activation. Br J Cancer. 2009;100:739–746. doi: 10.1038/sj.bjc.6604927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Houdt WJ, Emmink BL, Pham TV, Piersma SR, Verheem A, Vries RG, Fratantoni SA, Pronk A, Clevers H, Borel Rinkes IH, et al. Comparative proteomics of colon cancer stem cells and differentiated tumor cells identifies BIRC6 as a potential therapeutic target. Mol Cell Proteomics. 2011;10:M111.011353. doi: 10.1074/mcp.M111.011353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Fu D, Xi J, Ji Z, Liu T, Ma Y, Zhao Y, Dong L, Wang Q, Shen X. Expression and clinical significance of UCH37 in human esophageal squamous cell carcinoma. Dig Dis Sci. 2012;57:2310–2317. doi: 10.1007/s10620-012-2181-9. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Sobin LH, Gospodarowicz MK, Wittekind C. TNM Classification of Malignant Tumors. 7th ed. Oxford: Wiley-Blackwell; 2010. [Google Scholar]
- 25.Zhang S, Tang W, Weng S, Liu X, Rao B, Gu J, Chen S, Wang Q, Shen X, Xue R, Dong L. Apollon modulates chemosensitivity in human esophageal squamous cell carcinoma. Oncotarget. 2014;5:7183–7197. doi: 10.18632/oncotarget.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Chen YJ, Hou J, Wang YY, Tang WQ, Shen XZ, Tu RQ. Expression and clinical significance of BIRC6 in human epithelial ovarian cancer. Tumour Biol. 2014;35:4891–4896. doi: 10.1007/s13277-014-1641-6. [DOI] [PubMed] [Google Scholar]
- 27.Sung KW, Choi J, Hwang YK, Lee SJ, Kim HJ, Lee SH, Yoo KH, Jung HL, Koo HH. Overexpression of Apollon, an anti-apoptotic protein, is associated with poor prognosis in childhood de novo acute myeloid leukemia. Clin Cancer Res. 2007;13:5109–5114. doi: 10.1158/1078-0432.CCR-07-0693. [DOI] [PubMed] [Google Scholar]
- 28.Ismail EA, Mahmoud HM, Tawfik LM, Habashy DM, Adly AA, El-Sherif NH, Abdelwahab MA. BIRC6/Apollon gene expression in childhood acute leukemia: Impact on therapeutic response and prognosis. Eur J Haematol. 2012;88:118–127. doi: 10.1111/j.1600-0609.2011.01734.x. [DOI] [PubMed] [Google Scholar]
- 29.Tang W, Xue R, Weng S, Wu J, Fang Y, Wang Y, Ji L, Hu T, Liu T, Huang X, et al. BIRC6 promotes hepatocellular carcinogenesis: Interaction of BIRC6 with p53 facilitating p53 degradation. Int J Cancer. 2015;136:E475–E487. doi: 10.1002/ijc.29194. [DOI] [PubMed] [Google Scholar]
- 30.Dong X, Lin D, Low C, Vucic EA, English JC, Yee J, Murray N, Lam WL, Ling V, Lam S, et al. Elevated expression of BIRC6 protein in non-small-cell lung cancers is associated with cancer recurrence and chemoresistance. J Thorac Oncol. 2013;8:161–170. doi: 10.1097/JTO.0b013e31827d5237. [DOI] [PubMed] [Google Scholar]
- 31.Abe S, Yamamoto K, Hasegawa M, Inoue M, Kurata M, Hirokawa K, Kitagawa M, Nakagawa Y, Suzuki K. Bone marrow cells of myelodysplastic syndromes exhibit significant expression of apollon, livin and ILP-2 with reduction after transformation to overt leukemia. Leuk Res. 2005;29:1095–1096. doi: 10.1016/j.leukres.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Tassi E, Zanon M, Vegetti C, Molla A, Bersani I, Perotti V, Pennati M, Zaffaroni N, Milella M, Ferrone S, et al. Role of Apollon in human melanoma resistance to antitumor agents that activate the intrinsic or the extrinsic apoptosis pathways. Clin Cancer Res. 2012;18:3316–3327. doi: 10.1158/1078-0432.CCR-11-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nemoto T, Kitagawa M, Hasegawa M, Ikeda S, Akashi T, Takizawa T, Hirokawa K, Koike M. Expression of IAP family proteins in esophageal cancer. Exp Mol Pathol. 2004;76:253–259. doi: 10.1016/j.yexmp.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Zhou S, Ye W, Shao Q, Qi Y, Zhang M, Liang J. Prognostic significance of XIAP and NF-κB expression in esophageal carcinoma with postoperative radiotherapy. World J Surg Oncol. 2013;11:288. doi: 10.1186/1477-7819-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaĭshlia NA, Zinov'eva MV, Sass AV, Kopantsev EP, Vinogradova TV, Sverdlov ED. Increase of BIRC5 gene expression in non-small cell lung cancer and esophageal squamous cell carcinoma does not correlate with expression of genes SMAC/DIABLO and PML encoding its inhibitors. Mol Biol (Mosk) 2008;42:652–661. [PubMed] [Google Scholar]