Abstract
The next-generation sequencing studies of breast cancer have reported that the tumour suppressor P53 (TP53) gene is mutated in more than 40% of the tumours. We studied the levels of oxidative lesions, including 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG), along the coding strand of the exon 5 in breast cancer patients as well as in a reactive oxygen species (ROS)-attacked breast cancer cell line using the ligation-mediated polymerase chain reaction technique. We detected a significant ‘in vitro’ generation of 8-oxodG between the codons 163 and 175, corresponding to a TP53 region with high mutation prevalence, after treatment with xanthine plus xanthine oxidase, a ROS-generating system. Then, we evaluated the occurrence of oxidative lesions in the DNA-binding domain of the TP53 in the core needle biopsies of 113 of women undergoing breast investigation for diagnostic purpose. An increment of oxidative damage at the −G− residues into the codons 163 and 175 was found in the cancer cases as compared to the controls. We found significant associations with the pathological stage and the histological grade of tumours. As the major news of this study, this largest analysis of genomic footprinting of oxidative lesions at the TP53 sequence level to date provided a first roadmap describing the signatures of oxidative lesions in human breast cancer. Our results provide evidence that the generation of oxidative lesions at single nucleotide resolution is not an event highly stochastic, but causes a characteristic pattern of DNA lesions at the site of mutations in the TP53, suggesting causal relationship between oxidative DNA adducts and breast cancer.
Keywords: breast cancer, TP53, exon 5, oxidative DNA lesions, pT
1. Introduction
Breast cancer is the most common cancer occurring in women,1 affecting approximately one in eight women during their lifetime.2 So far, a number of risk factors of breast cancer have been discovered, including reproductive and hormonal factors, family history of breast cancer, alcohol consumption, obesity and overweight.3 Knowledge of how such risk factors induce cancer is not well defined, but it appears that an excessive generation of reactive oxygen species (ROS) is associated to increased breast cancer risk.4–6 ROS are highly reactive intermediates, which cause various kind of oxidative DNA lesions, including 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG) and 3-(2-deoxy-β-D-erythropentafuranosyl)pyrimido[1,2-α]purin-10(3H)-one–deoxyguanosine adducts,7 important hallmarks in carcinogenesis.8 The central question is whether oxidative DNA adducts, in the breast, truly cause cancer. If so, lowering them would reduce the incidence of breast cancer. Conversely, whether the oxidative lesions are caused by tumour progression, and thereby they are just a bystander, so reducing their frequency would not truly lower cancer risk.
One of the central pieces of the genome that is often damaged in cancer cells is the tumour suppressor P53 (TP53), involved in the cell cycle arrest, DNA repair, senescence and apoptosis.9 Inactivating mutations in the TP53 are the main genetic events in the sporadic cancers with the vast majority arising from a single point mutation in the segment encoding the DNA-binding domain of the TP53.10 Somatic TP53 mutations have been detected in almost every type of sporadic cancers at rates from 38% to 50%11 and the first next-generation sequencing studies of breast cancer have found that the TP53 is mutated in more than 40% of the tumours.12 Nevertheless, William Thilly has shown that mutation spectra can vary with dose and thereby such spectra are limited in the ability to define their origins. Also, people are always exposed to mixtures of environmental carcinogens.
Genotoxic carcinogens, including ROS, are known to produce characteristic patterns of somatic mutations in the DNA of malignant cells, which are generally preceded by the generation of characteristic patterns of DNA lesions at the site of mutations, as exemplified by the 8-oxodG, that, if unrepaired, may result in G:C to T:A transversions and, at lesser extent, in G:C to AT transitions.8,13 Somatic mutation theory may explain how DNA lesions in genomic DNA lead to the malignant transformation of cells. The ligation-mediated polymerase chain reaction (LMPCR) assay is a method of choice for the genomic footprinting of human genomic DNA,14 because it can be used to quantitatively investigate single-strand DNA breaks having phosphorylated 5′-ends within single-copy DNA sequences.
Our current aims were to investigate whether cancer related genes have altered frequencies of oxidative lesions in cancer susceptibility regions and to evaluate whether oxidative DNA adducts truly cause breast cancer. We examined the levels of oxidative DNA lesions, including of 8-oxodG, along the coding strand of the exon 5 of the TP53 in MDA-MB23 oestrogen receptor negative (ER−) breast cells after treatment with xanthine plus xanthine oxidase, a ROS-generating system. The genomic footprinting was performed using the LMPCR technique15 with some modifications accordingly to Davies and Murray.16 The enzymatic cleavage of DNA was obtained by the treatment with formamidopyrimidine DNA glycosylase (Fpg) and endonuclease III (EndoIII), two Escherichia coli repair enzymes that recognize various types of oxidative damage.17,18 The generation of oxidative DNA damage in the experimental cells was also analysed using the 32P-postlabelling technique.19 Next, we conducted a hospital-based study for evaluating the occurrence of oxidative DNA lesions at the TP53 sequence level in the core needle biopsies of 113 of women undergoing breast investigation for diagnostic purpose.
2. Materials and methods
2.1. Cell culture and DNA isolation
The MDA-MB23 ER− breast cells were grown under standard conditions in a 5% CO2 humidified incubator and treated at 30–40% confluence with 0.2 mM xanthine plus 1.0 or 5.0 mU xanthine oxidase for 24 h. DNA was isolated using a method requiring RNase and proteinase treatments and extraction with organic solvents.20 After DNA concentration and purity determination, coded samples were stored at −80 °C.
2.2. Study population
Breast cancer cases and controls were recruited among women undergoing breast investigation for diagnostic purpose at the Senology Unit. After being informed of the purpose of the study and signed an informed written consent, core needle biopsies were collected under radiographic guidance by interventional radiologists, snap-frozen and stored at −80°C until laboratory analysis. A questionnaire was completed by each participant after biological sampling collection. Histopathological diagnosis and laboratory test findings were obtained from the Pathological Anatomy Unit. Study procedures were performed in accordance with the guidelines of the General Hospital Institutional Committee that reviewed and approved the protocol.
2.3. Ligation-mediated polymerase chain assay
The levels of oxidative lesions have been analysed along the exon 5 using the LMPCR technique15 with some modifications.16 The melting temperatures (Tm) of the primers were calculated by the method of the nearest-neighbor,21 according to Allawi and SantaLucia.22
2.3.1. Enzymatic cleavage
DNA template (2 µg, 0.4 µg/µl) was incubated with a mix of reaction buffer (10 ×), 0.4 U Fpg (0.2 U/µl) and 200 U EndoIII (100 U/µl) at 37 °C for 60 min (final volume 10 µl) to create fragments with 5′-phosphate groups at the sites of DNA lesions.
2.3.2. Primer extension
The fragments containing ligatable breaks were amplified with 16.0 pmol of a primer with the following sequence 5′-GGCAACCAGCCCTGTCG and a calculated Tm of 56 °C, in a mix of 3 U Thermococcus litoralis exo- (Vent exo-) DNA polymerase (2 U/µl),23 0.6 mM dNTPs and 0.5 mM MgSO4. The following program was performed: denaturation at 95 °C for 2 min, annealing at 54 °C for 2 min and elongation at 72 °C for 2 min (1 cycle) followed by 20 cycles of 1 min at 95 °C, 2 min at 54 °C and 3 min at 72 °C.
2.3.3. Overnight ligation
The blunt ended fragments with 5′-phosphate termini were overnight incubated with a mix of ligase buffer (10×), 0.05 mg/ml bovine serum albumin (added as 10 mg/ml), 60 pmol of the asymmetric double-stranded unphosphorylated linker, and 90 U T4 ligase (400 U/µl) at 17 °C (final volume 30 μl) for generating a common sequence at all 5′-ends. After ligation, the linker-ligated fragments were evaporated to dryness under reduced pressure in a speed-vacuum concentrator and resuspended in 10 μl of ultrapure water.
2.3.4. PCR amplification and labelling
PCR amplification has been combined with direct labelling according to Davies and Murray.16 The amplification and labelling reactions were achieved with 12.0 pmol of a IRDye® 700/800 fluorescence-labelled primer with the following sequence 5′-TCTCTCCAGCCCCAGCTGCTCAC and a calculated Tm of 61 °C and 12.0 pmol of the LP25 universal linker primer in a mix of 1 U of Vent exo- (2 U/μl), 0.3 mM dNTPs and 2.5 mM MgSO4. The following PCR conditions were utilized: denaturation at 95 °C for 2 min (1 cycle), followed by 10 cycles of 45 s at 95 °C, 3 min at 59 °C and 3 min at 72 °C.
2.3.5. Detection and quantification
The IRDye® 700/800 fluorescence-labelled products (2 µl) were denaturated with formamide loading buffer (3 µl) at 95 °C for 5 min (final volume 5 µl), and, after 10 min on ice, denaturated labelled products (1 µl) were subjected to electrophoresis on a polyacrylamide sequencing gel, 80.0% acrylamide, 10% bis-acrylamide/urea and run in the LI-COR 4300 DNA analyzer at 65 °C for 7 h. DNA lesions were identified as the locations in which the presence of Fpg and EndoIII-sensitive sites stopped the Vent exo- from progressing and resulted in an intense dark band. The relative band intensity (RI), quantificated by ImageQuant, were calculated by following formula RI = Ij/Imax, where Ij is the intensity in pixels of each band after background subtraction and Imax is the mean intensity in pixels of the highest intensity bands.24 The RI values were corrected across experiments based on the recovery of the reference standard. The exact position of each base was determined by including labelled DNA fragments obtained by sequencing of the region of interest according to Ruano and Kidd25 and appropriate size IRDye® 700/800 sizing standards markers in the sequencing gel.
2.4. 32P-DNA postlabelling assay
The levels of 3-(2-deoxy-β-D-erythro-pentafuranosyl)pyrimido [1,2-α]purin-10(3H)-one deoxyguanosine (M1dG) adducts in the experimental cells were measured using a version of the 32P-DNA postlabelling assay,19 that has been developed for the specific detection of M1dG adducts.26 The levels of DNA damage were expressed as relative adduct labelling = pixels in adducted nucleotides/pixels in normal nucleotides. Adduct levels were corrected across experiments based in the recovery of the reference standard, which has been prepared as previously reported.27
2.5. Statistical analysis
Given the right-skewed distribution of the data, adduct levels were log-transformed to stabilize the variance and normalize the distribution. The Mann–Whitney U test was used to compare experimental groups in the ‘in vitro’ studies. The association of oxidative lesions with breast cancer was evaluated by the analysis of covariance, including terms for age (continuous), menopausal status and smoking history (non-smokers, current smokers), to estimate the effect of each variable on the outcome adjusting for the concomitant effect of the other variables included in the model. The analysis of covariance was also applied to examine the difference between the levels of oxidative DNA lesions according to the pathological diameter, the histologic cell grade and the frequency of hormonal receptors positive to oestrogen and progesterone. Post hoc Dunnett tests were performed for multiple comparisons among variable levels. All statistical tests were two-sided and P < 0.05 has been considered to be statistically significant. The data were analysed using SPSS 13.0 (IBM SPSS Statistics, New York, NY).
3. Results
3.1. Xanthine plus xanthine oxidase treatment
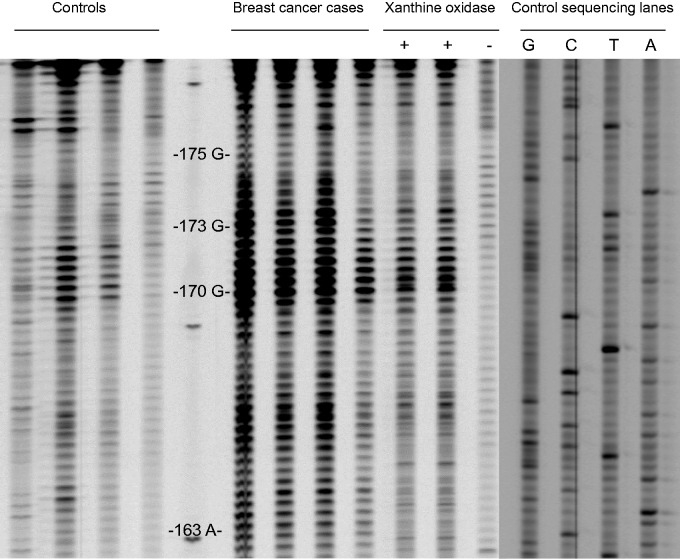
To look for the genomic footprints caused by electrophilic compounds in experimental cells treated with the xanthine plus xanthine oxidase system, we measured the levels of oxidative lesions along the coding strand of the exon 5 using the LMPCR assay. Figure 1 reports a representative map of the area with the cluster of oxidative DNA lesion products at the TP53 sequence level after treatment with 0.2 mM xanthine plus 5.0 mU xanthine oxidase system. The sequencing gel shows that various bases were hotspots for oxidative DNA damage between the codons 163 and 175. Other DNA damage sites were not observed. After showing the production of DNA damage at the DNA sequence level, we tried to quantify the levels of oxidative lesions at single nucleotide resolution following exposure of the same cell line to various doses of the ROS-generating system. The mean levels of oxidative lesions, expressed as RI,24 at selected positions of representative codons of the TP5311 were reported in Table 1. Significant dose-response relationships were found at the –T–and –G– residues into the codons 163, 170, 173 and 175, with the highest levels of DNA damage in the experimental cells treated with the highest dosage of the free radical-generating system, and with the lower levels in those exposed to the intermediate dosage. In cells without exposure to the xanthine plus xanthine oxidase system, a background level of oxidative DNA damage was found. The presence of oxidative damaged DNA in the experimental cells treated with increased concentration of the xanthine plus xanthine oxidase system was confirmed by the analysis of M1dG adducts, a biomarker of oxidative stress and lipid peroxidation,28–30 using the 32P-postlabelling technique. We observed that the frequency of M1dG was significantly increased within the cells treated with the ROS-generating system as compared to controls. The mean levels of M1dG adducts × 106 normal nucleotides were 2.87 ± 0.59 and 0.38 ± 0.10 of the treated cells and the controls, respectively, P = 0.007 (Table 1). A significant dose-response relationship was observed, P-value for trend < 0.001.
Figure 1.
Representative map of oxidative lesion along the coding strand of the exon 5 of the TP53 of women with breast cancer as compared to controls. MDA-MB23 ER− breast cells treated with 0.2 mM xanthine plus 5.0 mU xanthine oxidase system and untreated cells were on the left. Control sequencing lanes were on the right, whereas the numbers and letters to the right represent codon positions.
Table 1.
The mean levels of 3-(2-deoxy-β-D-erythro-pentafuranosyl)pyrimido[1,2-α]purin-10(3H)-one deoxyguanosine (M1dG) adducts, expressed as relative adduct labelling (RAL × 106), and the average amounts of oxidative lesions, expressed as relative intensity (RI), at the TP53 level in the MDA-MB23 oestrogen receptor-negative breast cells treated with increased concentrations of the xanthine plus xanthine oxidase system
| Genomic DNA |
Oxidative lesions along the coding strand of the exon 5 of the human TP53
gene |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| M1dG |
Codon 163 (-TAC-) |
Codon 170 (-ACG-) |
Codon 173 (-GTG-) |
Codon 175 (-CGC-) |
|||||||
| N | RAL ± s.e. | P-value | RI ± s.e. | P-value | RI ± s.e. | P-value | RI ± s.e. | P-value | RI ± s.e. | P-value | |
| Experimental dosages | |||||||||||
| Controlsa | 10 | 0.38 ± 0.10 | 0.08 ± 0.02 | 0.06 ± 0.01 | 0.10 ± 0.03 | 0.11 ± 0.03 | |||||
| 0.2 mM xanthine, 1.0 mU xanthine oxidase | 10 | 1.89 ± 0.10 | 0.016 | 0.10 ± 0.02 | 0.214 | 0.40 ± 0.07 | 0.016 | 0.18 ± 0.03 | 0.114 | 0.19 ± 0.03 | 0.164 |
| 0.2 mM xanthine, 5.0 mU xanthine oxidase | 10 | 2.87 ± 0.59 | 0.007 | 0.22 ± 0.03 | 0.004 | 0.55 ± 0.08 | 0.004 | 0.44 ± 0.07 | 0.005 | 0.28 ± 0.04 | 0.014 |
| P-value for trend | <0.001 | 0.001 | <0.001 | 0.001 | 0.012 | ||||||
aReference level.
3.2. Breast cancer case-control study
The core needle biopsies were obtained from 113 women, average aged 58.8 ± 15.5 yrs. Seventy-three women were confirmed as having breast cancer from histopatological analysis. Forty women with benign breast lesions were considered as controls. Nineteen women were classified as smokers. Breast cancer case and benign breast disease groups were comparable in terms of age, 60.6 yrs ± 15.0 (s.d.) vs. 55.7 yrs ± 16.1 (s.d.), respectively.
Having validated the use of the LMPCR assay and having showed that the ROS-generating system can produce highly localized damage in experimental cells, we tried to quantify the generation of oxidative lesions along the coding strand of the exon 5 of the TP53 in the study population. Our results show that a cluster of oxidative DNA lesions, including 8-oxodG, was present between the codons 170 and 173. A representative sequencing gel in Fig. 1 shows that specific bases were consistently hotspots for oxidative DNA damage in the breast cancer cases as compared to the controls. Other hotspots for oxidative DNA damage were not observed. The average levels of oxidative lesions, expressed as RI,24 at the levels of selected nucleotides of representative codons of the TP5311 were reported in Table 2. Our findings indicate that significant enhancements of the levels of 8-oxodG into the codons 170 and 173 were detected in the breast cancer cases as compared to the controls (Table 2). Specifically, the levels of oxidative lesions at the –T– position into the codon 163 of the cancer cases and the controls were 0.19 ± 0.02 and 0.15 ± 0.01, respectively, P = 0.493 after adjusting for age, menopausal status and smoking history; the values of 8-oxodG at the –G– residue into the codon 170 of the breast cancer cases and the controls were 0.51 ± 0.04 and 0.30 ± 0.02, respectively, P = 0.012 after correction for the confounding factors; the levels of 8-oxodG at the first –G – position into the codon 173 of the breast cancer patients and the controls were 0.44 ± 0.04 and 0.28 ± 0.02, respectively, P = 0.039 after adjusting for the confounding factors; and the levels of 8-oxodG at the –G – residue into the codon 175 of the cancer patients and the controls were 0.23 ± 0.02 and 0.18 ± 0.01, respectively, P = 0.270 after correction for the confounding factors.
Table 2.
The mean levels of oxidative lesions at the DNA sequence level, expressed as relative intensity (RI), at selected positions of representative codons along the coding strand of the exon 5 of the TP53 according to study variables
| Oxidative lesions along the coding strand of the exon 5 of the human TP53
gene | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Codon 163, (-TAC-) |
Codon 170 (-ACG-) |
Codon 173 (-GTG-) |
Codon 175 (-CGC-) |
||||||
| Na | RI ± s.e. | P-valueb | RI ± s.e. | P-valueb | RI ± s.e. | P-valueb | RI ± s.e. | P-valueb | |
| Menopausal status | |||||||||
| Premenopausalc | 43 | 0.19 ± 0.03 | 0.45 ± 0.05 | 0.41 ± 0.05 | 0.24 ± 0.02 | ||||
| Postmenopausal | 70 | 0.16 ± 0.01 | 0.897 | 0.43 ± 0.03 | 0.928 | 0.36 ± 0.03 | 0.949 | 0.20 ± 0.01 | 0.926 |
| Smoking habit | |||||||||
| Non-smokersc | 94 | 0.17 ± 0.01 | 0.45 ± 0.03 | 0.38 ± 0.03 | 0.22± 0.01 | ||||
| Smokers | 19 | 0.17 ± 0.03 | 0.937 | 0.39 ± 0.06 | 0.568 | 0.38 ± 0.07 | 0.738 | 0.22 ± 0.03 | 0.972 |
| Cancer status | |||||||||
| Controlsc | 40 | 0.15 ± 0.01 | 0.30 ± 0.02 | 0.28 ± 0.02 | 0.18 ± 0.01 | ||||
| Breast cancer cases | 73 | 0.19 ± 0.02 | 0.493 | 0.51 ± 0.04 | 0.012 | 0.44 ± 0.04 | 0.039 | 0.23 ± 0.02 | 0.270 |
| Pathologic stage of tumour | |||||||||
| pT1, tumour ≤20 mm in greatest dimension | 29 | 0.15 ± 0.02 | 0.997 | 0.45 ± 0.04 | 0.131 | 0.40 ± 0.05 | 0.599 | 0.21 ± 0.03 | 0.996 |
| pT2, tumour >20 mm but ≤50 mm in greatest dimension | 10 | 0.16 ± 0.05 | 0.998 | 0.51± 0.08 | 0.203 | 0.48 ± 0.08 | 0.182 | 0.23 ± 0.05 | 0.919 |
| pT3, tumour >50 mm in greatest dimension | 5 | 0.41 ± 0.11 | 0.062 | 0.76 ± 0.13 | 0.025 | 0.73 ± 0.19 | 0.037 | 0.46 ± 0.09 | 0.012 |
| P-value for trend | 0.120 | 0.001 | 0.003 | 0.036 | |||||
| Histologic cell grade | |||||||||
| G1, well differentiated | 9 | 0.09 ± 0.03 | 0.126 | 0.28 ± 0.07 | 0.756 | 0.25 ± 0.08 | 0.525 | 0.17 ± 0.06 | 0.533 |
| G2, moderately differentiated | 21 | 0.27 ± 0.05 | 0.094 | 0.64 ± 0.06 | <0.001 | 0.61 ± 0.07 | 0.001 | 0.33 ± 0.05 | 0.025 |
| G3, poorly differentiated | 15 | 0.19 ± 0.04 | 0.797 | 0.54 ± 0.06 | 0.013 | 0.48 ± 0.09 | 0.094 | 0.25 ± 0.05 | 0.612 |
| P-value for trend | 0.091 | <0.001 | 0.001 | 0.072 | |||||
| Oestrogen receptor | |||||||||
| Er − c | 8 | 0.15 ± 0.03 | 0.40 ± 0.09 | 0.40 ± 0.10 | 0.21 ± 0.04 | ||||
| Er + | 56 | 0.15 ± 0.15 | 0.655 | 0.45 ± 0.04 | 0.753 | 0.35 ± 0.03 | 0.417 | 0.19 ± 0.18 | 0.640 |
| Progesterone receptor | |||||||||
| Pr − c | 13 | 0.14 ± 0.02 | 0.43 ± 0.11 | 0.43 ± 0.09 | 0.19 ± 0.03 | ||||
| Pr + | 51 | 0.15 ± 0.01 | 0.841 | 0.43 ± 0.03 | 0.707 | 0.34 ± 0.03 | 0.483 | 0.20 ± 0.01 | 0.973 |
aSome figure do not add up to the total because of missing values.
bP-values after adjusting for the confounding factors.
cReference level.
When the pathologic stage of the breast tumours was examined, significant increments of oxidative lesions at the –G– positions into the codons 170, 173 and 175 with increased pathological stage were found (Table 2). Significant trends were also observed. In detail, the levels of oxidative lesions at single nucleotide resolution were 0.41 ± 0.11 and 0.15 ± 0.01 at the –T– residue into the codon 163 of the breast cancer cases with a pT3 stage and the controls, respectively, P = 0.062 after adjusting for the confounding factors; the values of 8-oxodG were 0.76 ± 0.13 and 0.30 ± 0.02 at the –G– position into the codon 170 of the breast cancer cases with a pT3 stage and the controls, respectively, P = 0.025 after correction for the confounding factors; the levels of 8-oxodG were 0.73 ± 0.19 and 0.28 ± 0.02 at the first –G– residue into the codon 173 of the breast cancer patients with a pT3 stage and the controls, respectively, P = 0.037 after adjusting for the confounding factors; and the amounts of 8-oxodG were 0.46 ± 0.09 and 0.18 ± 0.01 at the –G– position into the codon 175 of the cancer patients with a pT3 stage and in the controls, respectively, P = 0.012 after correction for the confounding factors. Statistically significant relationships were observed at the –G– residues into the codons 170, 173 and 175 with the highest levels of 8-oxodG in the patients with a pT3 stage, and intermediate values in those with early stages of cancer (Table 2).
On the other hand, when the histologic cell grade of tumours was analysed, a statistically significant enhancement of 8-oxodG levels into the codon 170 was observed with increasing grading (Table 2), but with the highest levels of DNA damage at the –G– residues into the codons 173 and 175 in patients with a G2 value. Significant trends of 8-oxodG generation into the codons 170 and 173 with increasing grade were found (Table 2). In detail, the levels of 8-oxodG were 0.54 ± 0.06, 0.64 ± 0.06 and 0.30 ± 0.02 at the –G– residue into the codon 170 in the breast cancer cases with G3 and G2 values and the controls, respectively, P < 0.013 and P < 0.001 after correction for the confounding factors.
4. Discussion
4.1. Single nucleotide TP53 lesions and free radicals
We evaluated the genomic footprints caused along the coding strand of the exon 5 of the TP53, a gene highly mutated in breast cancer,11 from the treatment with increased doses of the xanthine plus xanthine oxidase system using the LMPCR technique. The xanthine oxidase is an enzyme, which catalyses the conversion of hypoxanthine and xanthine into uric acid and uses oxygen as redox partner by producing various ROS, including superoxide anion radicals and hydrogen peroxide. As result, we observed a significant production of 8-oxodG between the codons 163 and 175 in the experimental cells (Fig. 1) corresponding to a region of the TP53 with high mutation rates.11 The presence of oxidative damage in the experimental cells treated with the ROS-generating system was confirmed using the 32P-postlabelling technique.19 After treatment with increased concentrations of the ROS-generating system, the production of oxidative lesions was increased of about 9-fold half level (Table 1), supporting that the damage at single nucleotide resolution results from ROS attack. Overall, our findings presented here indicate that the interaction of electrophilic species with genomic DNA causes the generation of oxidative DNA damage in a susceptible region along the coding sequence of the exon 5. Our results are consistent with the previous reports of Rodriguez et al.31 and Arakawa et al.,24 that analysed the frequency of oxidative lesions at the TP53 sequence level after hydrogen peroxide treatment in ‘in vitro’ studies. In particular, Arakawa et al. 24 reported that ROS treatment induced a similar pattern of fingerprints of oxidative DNA lesions within the exon 5.
4.2. Single nucleotide TP53 lesions and breast cancer
Having shown the production of base damage at the TP53 sequence level after ROS attack, we have focused on the largest analysis of genomic footprinting in humans to date. Our next result provides evidence that the generation of oxidative lesions at single nucleotide resolution is not an event highly stochastic with an overall low frequency, but causes site-specific DNA damage inducing a characteristic pattern of DNA lesions at the site of mutations in the TP53. This supports causal relationship between oxidative DNA lesions and breast cancer. Indeed, as expected, the mapping of the exon 5 indicates the presence of a cluster of oxidative damage between the codons 163 and 175 in the cancer patients as compared to the controls (Fig. 1). Although codon 175, known as one of the mutation hot spots in breast cancers,11 also exhibited the highest adduction rate in the breast cancer cases, no apparent difference is shown regarding the incidence of oxidative lesions between codons 163 and 175 (Table 2). Taken together, our results show the existence of a cluster of hotspots of DNA damage at the TP53 sequence level, that can be explained by a higher susceptibility of particular sites to be attacked from electrophilic compounds.8 Additionally, the fingerprint map of the breast cancer patients was comparable to that observed in the experimental cells treated with the ROS-generating system (Fig. 1). In particular, the amounts of oxidative damage at the –G– residues into the codons 170 and 173 were significantly increased in the breast cancer patients as compared to healthy tissues (Table 2). The fact that the exon 5 of the TP53 can be targeted to unique sites in the genome of breast cancer patients will be of considerable value. The increased levels of oxidative DNA damage along the DNA-binding domain can indeed be considered a cancer risk factor, since if unrepaired, they can lead to various mutations and subsequently trigger breast carcinogenesis. This depends on whether the oxidation causes cancer or the generation of oxidative DNA damage is influenced by early effects of cancer itself.
4.3. Single nucleotide TP53 lesions and prognostic factors
Enhanced levels of 8-oxodG at the TP53 sequence level were also significantly associated with conventional prognostic factors such as the pathological stage and the histological grade of breast tumours, indicating that high levels of 8-oxodG may exert detrimental effects during cancer development and tumour progression. Nevertheless, the possibility that ROS act such as a form of endogenous chemotherapy during tumour progression cannot be excluded. This association was even more evident at the –G– residue into the codon 170 of the patients with poorly differentiated tumours of >50 mm dimension (Table 2). Consistent with our results, a main role for oxidative stress in the pathogenesis of oestrogen-induced breast cancer has been suggested.32 Earlier studies have discovered augmented levels of biomarkers of oxidative stress in either urine and tumour tissue DNA.4,5,33–35 Recently, high 8-oxodG has been associated to subsequent risk of breast.5 Also, we have conducted a hospital-based study to analyse the amount of M1dG, a biomarker of oxidative stress,29,30,36 in breast cancer patients.4 In that study, we found a significant association between M1dG and breast cancer, as well as with increasing the degree of differentiation and severity of cancer. Various underlying mechanisms are involved in the generation of oxidative DNA lesions, such as the oestrogens,34 that are metabolically activated into catechol oestrogens from the cytochrome P450 enzymes.37 Metabolic redox cycling between catechol oestrogens and their corresponding quinones can generate oxidative stress and ROS.34 Chronic inflammation associated to adiposity3 as well activated inflammatory neutrophils38 can also induce ROS.
4.4. TP53 genomic fooprinting
As the major news of this study, this largest analysis of genomic footprinting of oxidative lesions at the TP53 sequence level to date provided a first roadmap describing the signatures of oxidative lesions in human breast cancer. Remarkably, our results indicate that the LMPCR technique15 a versatile DNA-lesion footprinting technique, which enables sensitive and specific detection of oxidative DNA damage at the level of sequence resolution, may be used for evaluating cancer risk in future clinical and epidemiological studies. In particular, the current LMPCR procedure involved the combination of PCR amplification with direct labelling developed by Davies and Murray.16 Furthermore, this system will allow detailed analysis of the effects of site-specific DNA lesions on cellular functions.
4.5. Single nucleotide TP53 lesions and mutation prevalence
Data on mutation prevalence in breast cancer may be easily accessed through the IARC TP53 database,11 therefore, we examined the levels of oxidative DNA lesions at single nucleotide resolution to the frequencies of mutations in the same positions along the DNA-binding domain11 in the attempt of evaluating the potential mechanisms underlying the pathogenesis of breast cancer. Table 3 shows that the generation of oxidative DNA damage along the DNA-binding domain of the TP53 was found in sites where the prevalence of transversion and transition mutations was ranging from 0.28% to 4.52%. The occurrence of special mutation patterns might suggest of the nature of the mutagens that have caused them. In this case, the mutation spectra reveal some of the characteristics expected from the effects of ROS exposure on DNA. It was apparent that the generation of 8-oxodG associated to the principal mutagenic events generated by this compound,13 may be in part responsible of the induction of these TP53 mutations. Theoretically, the colocalization of DNA lesions and mutational hotspots in the TP53 gene may be used for causality inference. This interpretation is further strengthened by previous findings reporting that G:C→T:A tranversions derived from 8-oxodG have been found in malignant tumours.8 Of course, other models have indicated that the deamination of 5-methylcytosine in methylated CpG sequences may be a mechanisms of CpG transitions.39 Interestingly, the process of 5-methylcytosine deamination may be enhanced by the exposure to ROS, especially in cancers arising from inflammatory precursors.40 These elements suggest that the present spectra of oxidative lesions may theoretically account for at least a part a percentage of the TP53 mutations present in breast tumours.
Table 3.
Relationships between oxidative damage at the TP53 level and the mutational spectrum of the TP53 in malignant breast tumours
| Codon number | Site of DNA lesione | RI ± s.e.a | Ratiob | Nomenclature | Tumour distribution (%) | Mutational event | Mutational rateb (%) |
|---|---|---|---|---|---|---|---|
| 163 | (-TAC-) | 0.19 ± 0.02 | 1.27 | c.487T>A | 0.28 | A:T→T:A | 6.57 |
| 170 | (-ACG-) | 0.51 ± 0.04 | 1.7 | c.511G>A | 0.03 | G:C→A:T | 20.68 |
| 173 | (-GTG-) | 0.44 ± 0.05 | 1.57 | c.517G>T | 0.31 | G:C→T:A | 10.58 |
| 175 | (-CGC-) | 0.23 ± 0.03 | 1.27 at CpG | c.524G>A | 4.52 | G:C→A:T at CpG | 18.37 |
aThe mean levels of oxidative lesions, expressed as relative intensity (RI), in breast cancer patients.
bRatio = levels of oxidative damage at the TP53 sequence levels in breast cancer cases/levels of oxidative DNA damage of controls.
cPrevalence of mutations at the site of DNA lesions at the TP53 sequence level.
dPrevalence of transversion and transition mutations in the TP53.
eUnderlined letters indicate the residues for which oxidative lesions are reported.
4.6. Conclusions
The integration of DNA adduct data with cancer mutations, as exemplified in our report, may provide an improved knowledge of the complex relationship between the TP53 gene and breast cancer development and progression. Considering that DNA-lesion footprinting in conjunction with mutation analysis are used to correlate DNA damage and mutagenesis and to demonstrate causality inference, we may argue that oxidative lesions occurring at the –G– residues into the codons 170 and 173 might serve as a necessary hit required to drive the process of breast carcinogenesis and tumour progression. Our study provides evidence that the production of oxidative lesions at the TP53 sequence level is not an event stochastic, but induce a characteristic spectrum of DNA adducts at the site of mutations, supporting causal relationship between oxidative DNA lesions and breast cancer. Also, the data suggest the possibility that the level of oxidative DNA lesions may be a prognostic factor in breast cancer. Analysing the generation of DNA damage at single nucleotide resolution will serve as a foundation for future work examining the mechanisms of oxidative DNA damage in breast carcinogenesis.
Acknowledgements
We are grateful to Cecilia M. Beccani, Cristina M. Chilleni, Verena Ludovici, Paola Frullini, Fiorenza Noferini, Claudia Simonetto and Francesca Tinti for technical assistance and to all the women participating to the study.
Conflict of interest
None declared.
Funding
This work has received financial support from the Istituto Toscano Tumori, Florence, Italy. Funding to pay the Open Access publication charges for this article was provided by Fondo per gli investimenti della ricerca scientifica (FIRB), [No. RBAP10MY35-002].
References
- 1.Vandeloo M.J., Bruckers L.M., Janssens J.P. 2007, Effects of lifestyle on the onset of puberty as determinant for breast cancer. Eur. J. Cancer Prev., 16, 17–25. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Soerjomataram I., Dikshit R., et al. 2015, Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer, 136, E359–386. [DOI] [PubMed] [Google Scholar]
- 3.Madeddu C., Gramignano G., Floris C., Murenu G., Sollai G., Maccio A. 2014, Role of inflammation and oxidative stress in post-menopausal oestrogen-dependent breast cancer. J. Cell. Mol. Med., 18, 2519–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peluso M., Munnia A., Risso G.G., et al. 2011, Breast fine-needle aspiration malondialdehyde deoxyguanosine adduct in breast cancer. Free Radic. Res., 45, 477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loft S., Olsen A., Moller P., Poulsen H.E., Tjonneland A. 2013, Association between 8-oxo-7,8-dihydro-2'-deoxyguanosine excretion and risk of postmenopausal breast cancer: nested case-control study. Cancer Epidemiol. Biomarkers Prev., 22, 1289–96. [DOI] [PubMed] [Google Scholar]
- 6.Huang X. 2008, Does iron have a role in breast cancer? Lancet Oncol., 9, 803–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marnett L.J. 2000, Oxyradicals and DNA damage. Carcinogenesis, 21, 361–70. [DOI] [PubMed] [Google Scholar]
- 8.Cooke M.S., Evans M.D., Dizdaroglu M., Lunec J. 2003, Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J., 17, 1195–214. [DOI] [PubMed] [Google Scholar]
- 9.Bargonetti J., Manfredi J.J. 2002, Multiple roles of the tumor suppressor p53. Curr. Opin. Oncol., 14, 86–91. [DOI] [PubMed] [Google Scholar]
- 10.Olivier M., Hussain S.P., Caron de Fromentel C., Hainaut P., Harris C.C. 2004, TP53 mutation spectra and load: a tool for generating hypotheses on the etiology of cancer. IARC Sci. Publ., 247–70. [PubMed] [Google Scholar]
- 11.Petitjean A., Mathe E., Kato S., et al. 2007, Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat., 28, 622–9. [DOI] [PubMed] [Google Scholar]
- 12.Ellis M.J., Ding L., Shen D., et al. 2012, Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature, 486, 353–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan X., Grollman A.P., Shibutani S. 1999, Comparison of the mutagenic properties of 8-oxo-7,8-dihydro-2'-deoxyadenosine and 8-oxo-7,8-dihydro-2'-deoxyguanosine DNA lesions in mammalian cells. Carcinogenesis, 20, 2287–92. [DOI] [PubMed] [Google Scholar]
- 14.Besaratinia A., Pfeifer G.P. 2006, Investigating human cancer etiology by DNA lesion footprinting and mutagenicity analysis. Carcinogenesis, 27, 1526–37. [DOI] [PubMed] [Google Scholar]
- 15.Besaratinia A., Pfeifer G.P. 2009, DNA-lesion mapping in mammalian cells. Methods, 48, 35–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies N.P., Murray V. 2000, Shortened protocol for terminal deoxynucleotidyl transferase-dependent PCR. BioTechniques, 29, 1168–70. [DOI] [PubMed] [Google Scholar]
- 17.Karakaya A., Jaruga P., Bohr V.A., Grollman A.P., Dizdaroglu M. 1997, Kinetics of excision of purine lesions from DNA by Escherichia coli Fpg protein. Nucleic Acids Res., 25, 474–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dizdaroglu M. 2005, Base-excision repair of oxidative DNA damage by DNA glycosylases. Mutat. Res., 591, 45–59. [DOI] [PubMed] [Google Scholar]
- 19.Munnia A., Bonassi S., Verna A., et al. 2006, Bronchial malondialdehyde DNA adducts, tobacco smoking, and lung cancer. Free Radic. Biol. Med., 41, 1499–505. [DOI] [PubMed] [Google Scholar]
- 20.Peluso M., Castegnaro M., Malaveille C., et al. 1990, 32P-postlabelling analysis of DNA adducted with urinary mutagens from smokers of black tobacco. Carcinogenesis, 11, 1307–11. [DOI] [PubMed] [Google Scholar]
- 21.SantaLucia J., Jr. 1998, A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA., 95, 1460–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allawi H.T., SantaLucia J., Jr. 1997, Thermodynamics and NMR of internal G.T mismatches in DNA. Biochemistry, 36, 10581–94. [DOI] [PubMed] [Google Scholar]
- 23.Vigneault F., Drouin R. 2005, Optimal conditions and specific characteristics of Vent exo- DNA polymerase in ligation-mediated polymerase chain reaction protocols. Biochem. Cell Biol., 83, 147–65. [DOI] [PubMed] [Google Scholar]
- 24.Arakawa H., Weng M.W., Chen W.C., Tang M.S. 2012, Chromium (VI) induces both bulky DNA adducts and oxidative DNA damage at adenines and guanines in the p53 gene of human lung cells. Carcinogenesis, 33, 1993–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruano G., Kidd K.K. 1991, Coupled amplification and sequencing of genomic DNA. Proc. Natl. Acad. Sci. USA., 88, 2815–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munnia A., Amasio M.E., Peluso M. 2004, Exocyclic malondialdehyde and aromatic DNA adducts in larynx tissues. Free Radic. Biol. Med., 37, 850–8. [DOI] [PubMed] [Google Scholar]
- 27.Bono R., Romanazzi V., Munnia A., et al. 2010, Malondialdehyde-deoxyguanosine adduct formation in workers of pathology wards: the role of air formaldehyde exposure. Chem. Res. Toxicol., 23, 1342–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peluso M.E., Munnia A., Giese R.W., Chellini E., Ceppi M., Capacci F. 2015, Oxidatively damaged DNA in the nasal epithelium of workers occupationally exposed to silica dust in Tuscany region, Italy. Mutagenesis, 30, 519–25. [DOI] [PubMed] [Google Scholar]
- 29.Peluso M., Srivatanakul P., Munnia A, et al. 2010, Malondialdehyde-deoxyguanosine adducts among workers of a Thai industrial estate and nearby residents. Environ. Health Perspect., 118, 55–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peluso M., Munnia A., Ceppi M., et al. 2013, Malondialdehyde-deoxyguanosine and bulky DNA adducts in schoolchildren resident in the proximity of the Sarroch industrial estate on Sardinia Island, Italy. Mutagenesis, 28, 315–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez H., Akman S.A., Holmquist G.P., Wilson G.L., Driggers W.J., LeDoux S.P. 2000, Mapping oxidative DNA damage using ligation-mediated polymerase chain reaction technology. Methods, 22, 148–56. [DOI] [PubMed] [Google Scholar]
- 32.Key T., Appleby P., Barnes I., Reeves G., Endogenous H. and Breast Cancer Collaborative, G. 2002, Endogenous sex hormones and breast cancer in postmenopausal women: reanalysis of nine prospective studies. J. Natl. Cancer Inst., 94, 606–16. [DOI] [PubMed] [Google Scholar]
- 33.Ray G., Batra S., Shukla N.K., et al. 2000, Lipid peroxidation, free radical production and antioxidant status in breast cancer. Breast Cancer Res. Treat., 59, 163–70. [DOI] [PubMed] [Google Scholar]
- 34.Cavalieri E.L., Rogan E.G. 2011, Unbalanced metabolism of endogenous estrogens in the etiology and prevention of human cancer. J. Steroid Biochem. Mol. Biol., 125, 169–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsui A., Ikeda T., Enomoto K., et al. 2000, Increased formation of oxidative DNA damage, 8-hydroxy-2'-deoxyguanosine, in human breast cancer tissue and its relationship to GSTP1 and COMT genotypes. Cancer Lett., 151, 87–95. [DOI] [PubMed] [Google Scholar]
- 36.Peluso M.E., Munnia A., Bollati V., et al. 2014, Aberrant methylation of hypermethylated-in-cancer-1 and exocyclic DNA adducts in tobacco smokers. Toxicol Sci., 137, 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolton J.L., Thatcher G.R. 2008, Potential mechanisms of estrogen quinone carcinogenesis. Chem. Res. Toxicol., 21, 93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gungor N., Haegens A., Knaapen A.M., et al. 2010, Lung inflammation is associated with reduced pulmonary nucleotide excision repair in vivo. Mutagenesis, 25, 77–82. [DOI] [PubMed] [Google Scholar]
- 39.Pfeifer G.P. 2000, p53 mutational spectra and the role of methylated CpG sequences. Mutat. Res., 450, 155–66. [DOI] [PubMed] [Google Scholar]
- 40.Vaninetti N.M., Geldenhuys L., Porter G.A., et al. 2008, Inducible nitric oxide synthase, nitrotyrosine and p53 mutations in the molecular pathogenesis of Barrett's esophagus and esophageal adenocarcinoma. Mol. Carcinog., 47, 275–85. [DOI] [PubMed] [Google Scholar]