Abstract
Arthrospira platensis is a multi-cellular and filamentous non-N2-fixing cyanobacterium that is capable of performing oxygenic photosynthesis. In this study, we determined the nearly complete genome sequence of A. platensis YZ. A. platensis YZ genome is a single, circular chromosome of 6.62 Mb in size. Phylogenetic and comparative genomic analyses revealed that A. platensis YZ was more closely related to A. platensis NIES-39 than Arthrospira sp. PCC 8005 and A. platensis C1. Broad gene gains were identified between A. platensis YZ and three other Arthrospira speices, some of which have been previously demonstrated that can be laterally transferred among different species, such as restriction-modification systems-coding genes. Moreover, unprecedented extensive chromosomal rearrangements among different strains were observed. The chromosomal rearrangements, particularly the chromosomal inversions, were analysed and estimated to be closely related to palindromes that involved long inverted repeat sequences and the extensively distributed type IIR restriction enzyme in the Arthrospira genome. In addition, species from genus Arthrospira unanimously contained the highest rate of repetitive sequence compared with the other species of order Oscillatoriales, suggested that sequence duplication significantly contributed to Arthrospira genome phylogeny. These results provided in-depth views into the genomic phylogeny and structural variation of A. platensis, as well as provide a valuable resource for functional genomics studies.
Keywords: evolution, lateral gene transfer, comparative genomics, chromosomal rearrangement
1. Introduction
Spirulina is a multi-cellular and filamentous blue-green microalga that has gained considerable popularity in the health food industry and increasingly as a protein and vitamin supplement. Spirulina taxonomically belongs to two separate genera, namely Spirulina and Arthrospira, and consists of about 15 species.1 Arthrospira is the most common and widely available spirulina that has been extensively investigated in various health-related studies. 1 It is composed of closely related, genetically and physiologically distinct lineages whose differences enable them to adapt to specific ecological niches. Arthrospira platensis from Lake Chenghai is a well-known representative in China, and many previous studies associated with stress tolerance and gene functional characterization were regarding to this cyanobacterial strain. 2–5 Arthrospira also contains high amount of proteins, polyunsaturated fatty acids, vitamins, minerals, and photosynthetic pigments. Despite its highly useful applications, very little is known about the phylogeny, physiological and genetic system in A. platensis. Unlike other bacteria, the genome of microorganisms belonging to phylum Cyanobacterium generally possesses a large amount of dispersed repetitive sequences, particularly Arthrospira, thereby causing difficulty in the complete assembly of its genome.6 In the past 6 years, several genomes from genus Arthrospira that belong to geographically distinct lineages have been nearly completely recovered by independent sequencing and assembly into single super-scaffolds, respectively, including A. platensis NIES-39, Arthrospira sp. PCC 8005 and A. platensis C1.6–8 Previous studies have generated on its genomic constitution, annotation, classification of respective isolates, as well as provided the first opportunity to reconstruct its phylogenetic relationship with other different strains. Nevertheless, there were no in-depth comparative genomics analyses revealing the molecular implications associated with genetic diversity affecting the evolutionary origin, and how these genetic differences generated among distinct lineages. In addition, considering the ubiquity of dispersed repeated sequences and its emerging role in genome evolution, we can hardly be said to comprehend Arthrospira genome without giving an account of what these repetitive sequences are, what they do and how they arise. Present study reports the nearly complete genome sequence of one cultivated strain, namely, A. platensis YZ. We then reconstructed the genomic evolution events involving this strain and compared the gene and genomic structure with those of closely related species to assess the phylogenetic relationship, genetic diversity and genomic structure variation.
2. Materials and methods
Arthrospira platensis YZ was initially isolated from Chenghai Lake in Yunnan Province, China. The cyanobacterial strain was cultured under 0.02 M NaCl Zarrouk medium at 30 °C, light intensity of 8 kilolux and 75% humidity. After total DNA was extracted, two genomic shotgun libraries with 1.5- and 4.5-kb insert sizes were constructed described as previously.9 A fosmid (pCC1FOS fosmid vector) library was constructed by using the CopyControl Fosmid library production kit (Epicentre), following the manufacturer’s recommendations. In addition, a 10-kb Illumina mate-pair library was also constructed as described elsewhere.10 All libraries were sequenced from both ends. Reads that were derived from the two small-insert sized libraries were initially mixed and assembled by using the Phrap software. Consed was used in the finishing process.11 Initially, the primary assembly generated a total of 2,294 contigs, many of which were determined to be mis-assembled. Then, the primary assembly was manually checked via assessment of the paired-end reads distances of the cloned libraries. The mis-assembled regions were disassembled, and local blocks were established for re-assembly. The Illumina and fosmid reads were mapped onto the assembly to scaffold the contigs. The contiguous relationships between all the contigs at non-repetitive regions were anchored by the Illumina mate-pair or fosmid paired-end reads. Gaps were closed by primer-walking on the PCR products from neighbour extremity of the linked contigs or corresponding fosmid clones. The Illumina reads were also employed to confirm and correct those with low-quality (but not repeated sequence) assembly regions by using the Genome Analysis Toolkit.12
We used the Glimmer software to predict protein-coding genes with potential open reading frames of >150 bp in size.13 RNAmmer and tRNAscan-SE were utilized to identify rRNA and tRNA genes, respectively.14 , 15 Gview was used to construct basic genomic features.16 BLASTX was used to annotate predicted protein-coding genes against the public protein database with an e-value threshold of 1e-5. Genome-wide identification of restriction-modification (RM) systems were conducted by using BLASTP searching against REbase with a >50% amino acid identity and >50% query coverage after all ORFs were theoretically translated.17 Comparative analysis of RM enzymes between Arthrospira strains was performed by using BLASTP with an e-value threshold of 1e-20. We selected 31 partial or complete genomes of determined species from Oscillatoriales for phylogenetic analysis. A species tree was generated by concatenation of 27 of 31 conserved proteins as previously reported,18 including dnaG, frr, nusA, pgk, pyrG, rplA, rplB, rplC, rplD, rplE, rplF, rplK, rplL, rplM, rplN, rplP, rplS, rpoB, rpsB, rpsC, rpsE, rpsI, rpsK, rpsM, rpsS, smpB, and tsf, because some species did not harbour all the genes due to various reasons. Gloeobacter violaceus PCC 7421 from order Gloeobacterales was used as outgroup for rooting the tree. Maximum-likelihood phylogenetic trees were generated by using PhyML 3.0 with 1,000 bootstrap replications.19
The protein-coding gene derived from four Arthrospira genomes was predicted using the same criteria as earlier described. Gview was used to construct general features that were subsequently employed in comparative analysis. In silico hybridization was performed using the BLASTP program, with an e-value threshold of 1e-5. Using A. platensis YZ as reference, gene order in the other three species was established accordingly, followed by the identification of potential laterally transferred genes (LTGs) by using a custom-derived script combined with a manual check. Insertion sequences were predicted by using IS finder.20 Assessment of global genomic co-linearity relationships among species was performed by using the mauve software.21 BLASTN was used to identify direct and inverted repeats (IRs) as well as palindromes. Given that all the theoretically predicted genes were protein-coding genes, genome-wide clustering of all genes was performed. Only those species from Oscillatoriales with relatively intact genome sequence (genome scaffolds of < 10) were selected for whole genome clustering analysis, including Lyngbya majuscula 3L, Geitlerinema sp. PCC 7105, Leptolyngbya sp. PCC 7376, Leptolyngbya sp. PCC 7375, Oscillatoria nigro-viridis PCC 7112, Pseudanabaena sp. PCC 6802, Oscillatoria sp. PCC 10802, Microcoleus sp. PCC 7113, Crinalium epipsammum PCC 9333, Oscillatoria acuminata PCC 6304, Leptolyngbya boryanar PCC 6306, Spirulina major PCC 6313, Oscillatoriales cyanobacterium JSC-12, Spirulina subsalsa PCC 9445, Pseudanabaena sp. PCC 7367, Geitlerinema sp. PCC 7407, and four strains from genus Arthrospira, because many genes were doomed to be undetermined in the incomplete genome assembly. All genome sequences were obtained from Integrated Microbial Genomes at DOE Joint Genome Institute or NCBI. Cluster Database at High Identity with Tolerance (CD-HIT) was used to perform gene clustering analysis with 50% alignment coverage for both sequences (-aL 0.5, -aS 0.5) at nine resolutions.22 C/W ratio was calculated as the number of total clusters divided by the number of total genes. R/T ratio represented the number of redundant gene clusters (the clusters which comprised at least two genes) divided by the total number of clusters. Smaller C/W ratios indicated a higher level of gene redundancy, whereas larger R/T ratios suggested broader gene redundancy spectra. Tandem repeats (TRs) were identified by using TR Finder with the following parameters (2 7 7 80 10 100 2000 -f -d -m).23 Clustering of repeat motifs was conducted by CD-HIT with both 100% identity and 100% query, and hit alignment coverage. Clustered regularly interspaced short palindromic repeat (CRISPR) Finder was used to identify CRISPR arrays.24 The complete genome sequence of A. platensis YZ has been deposited to DDBJ/EMBL/GenBank under accession CP013008.
3. Results and Discussion
3.1. General features of A. platensis YZ genome
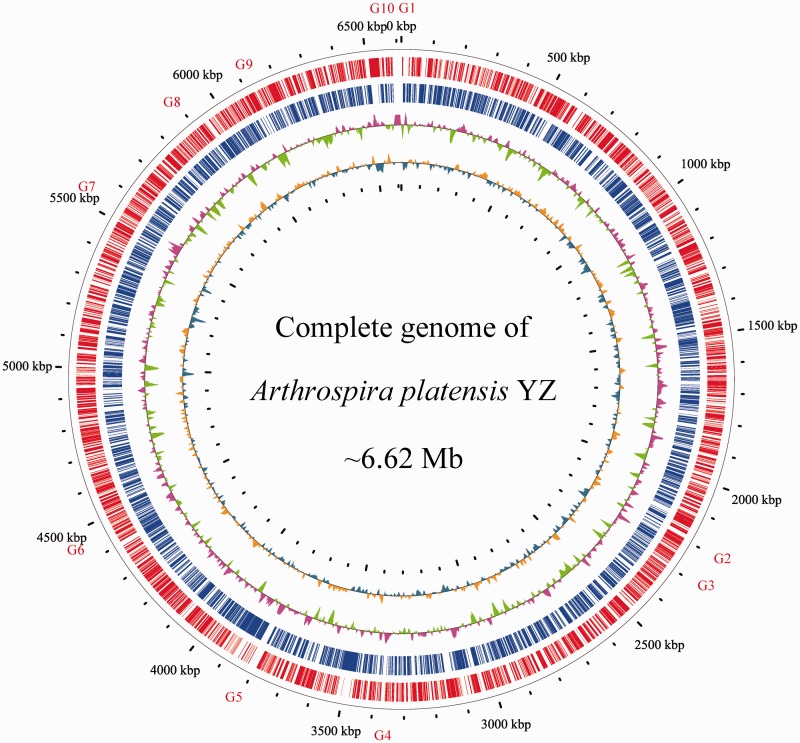
We selected one single cultivated cyanobacterial strain, which we designated as A. platensis YZ, for whole genome sequencing. Genomic DNA was sequenced using a whole-genome shotgun sequencing strategy, which was conducted by using a ‘hybrid’ approach that combined Sanger and Illumina sequencing technologies. Four different libraries, including 1.5-, 4.5-, 40-kb insert-sizes clone libraries, and an Illumina mate-pair library with a 10-kb insert-size were constructed and sequenced. Over 130,000 high-quality Sanger reads (∼8.9-fold genome coverage) and 10,434,858 Illumina reads (∼77.2-fold genome coverage) were obtained. We assembled the A. platensis genome into one super-scaffold with terminal overlaps. Taken together, A. platensis has a single, circular chromosome of 6.62 Mb in size, with an average GC content of 44.2% (Fig. 1). The current version of the A. platensis YZ genome contains 10 undetermined regions with a totally estimated gap size of 96 kb, and the remaining gaps apparently consist of TRs and Arthrospira-specific repeats. The genome size of A. platensis YZ is a little smaller than the previously sequenced A. platensis NIES-39 (6.79 Mb),8 but larger than Arthrospira sp. PCC 8005 (6.23 Mb)7 and A. platensis C1 (6.09 Mb).6 GC skew analysis to anchor potential origin and terminator for chromosome replication did not show obvious skew shift. This is different from those bacteria whose chromosomal replication origin and terminator could be clearly identified,9 , 25 but is similar to other cyanobacterial genomes, suggesting the occurrence of potential chromosomal rearrangements, sequence duplications and lateral gene transfer (LGT) events,26 because the bias of nucleotide compositions between leading and lagging strands were disrupted. In addition, mapping Illumina short reads back to the genome resulted in 99.6% of the total reads uniquely aligned to the assembly and 98.4% paired-end reads represented correct orientation and distance, which was indicative of the high credibility of the assembled genome.
Figure 1.
Circular representation of the A. platensis YZ genome. From outside to inside: circle 1, physical map scaled to 100 kb; Circles 2 and 3, coding sequences transcribed in the clockwise and counterclockwise directions; Circles 4 and 5 represent GC content and GC skew, respectively. Gaps were labelled with G1-G10.
The A. platensis YZ genome was predicted to harbour 6,784 protein-coding genes with average gene length of 795 bp. A total of 3,149 and 6,711 protein-coding genes can be annotated by known function or hypothetical protein in the UniProt/Swiss-Prot and non-redundant protein databases, respectively. Cluster of Orthologous Groups of proteins database annotation showed that a number of genes participated in its replication, recombination, and repair (Supplementary Figure S1). Moreover, the genome sequence was also predicted to contain two sets of rRNA genes and 39 tRNA genes that were predicted to translate 19 types of amino acids, except for lysine. The number of rRNA gene clusters in the A. platensis YZ genome is similar to that observed in A. platensis NIES-39, Arthrospira sp. PCC 8005, and A. platensis C1, whereas the number of tRNA genes differed from those of cynaobacteria (40, 42, and 39 tRNA, respectively).
3.2. Phylogenetic and comparative genomics analyses of Arthrospira
Because Arthrospira belongs to order Oscillatoriales, a species tree was constructed for phylogenomic analysis by concatenating 27 conserved proteins coexisted in the 31 Oscillatoriales species (Fig. 2). As expected, A. platensis YZ was clustered together with the other members of genus Arthrospira, which is highly congruent with the findings of previous study on whole cyanobacteria phylum phylogeny.27 Subclade D of the tree showed that A. platensis YZ was phylogenetically closer to A. platensis str. Paraca and A. platensis NIES-39 than Arthrospira sp. PCC 8005 and A. platensis C1, which was indicative of intimate intragenus relationship among A. platensis YZ, A. platensis str. Paraca, and A. platensis NIES-39.
Figure 2.
A species tree from Oscillatoriales. Phylogenetic reconstruction of members of Oscillatoriales using the maximum-likelihood method was performed in the present study. Taxa are colour-coded according to the same genus. Nodes supported with a bootstrap of > 70% are indicated by black dots. Phylogenetic subclades are grouped into seven major subclades (A–G).
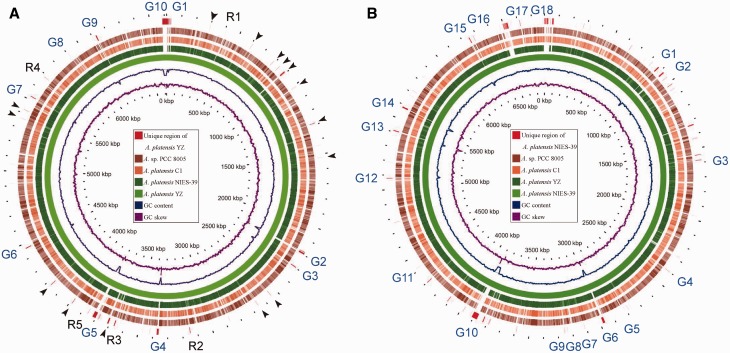
Gene gains and losses have been considered as one of the most significant contributors to functional changes.28 To further characterize the differences among several A. platensis genomes, we performed comparative genomics analysis of A. platensis YZ with three other species, including A. platensis NIES-39, Arthrospira sp. PCC 8005 and A. platensis C1, whereas the complete genome of A. platensis str. Paraca is currently unavailable.29 Extensive sequence gains were identified when using A. platensis YZ as backbone during its comparative analysis to the other three species. At least 20 apparently unique regions, excluding the undetermined sequence, were detected in the A. platensis YZ genome (Fig. 3A). More particular regions in the A. platensis NIES-39 genome were observed, which might be explained by its larger genome (Fig. 3B). However, the unique regions in Arthrospira sp. PCC 8005 and A. platensis C1 were markedly reduced relative to that in A. platensis YZ and A. platensis NIES-39. Furthermore, in silico hybridization identified 100 A. platensis YZ-specific genes, of which 82 could not be annotated by the known proteins.
Figure 3.
Comparative genomic analysis of four complete sequenced Arthrospira speices. A. platensis YZ (A) and A. platensis NIES-39 (B) were used as backbone (reference or hit) for the comparison to three query genomes, respectively. The genomes scaled to 100 kb. Counting from the outside toward the centre: red colour (slot 1) indicated that the nucleotide sequences were only detected in the reference genomes and undetermined gaps of the reference where query genomes were not mapped by ‘Ns’. Black arrow and R1-R5 in (A) indicate specific or unique regions in A. platensis YZ genome, and the regions R1-R5 were selected for analyses of gene gains in A. platensis YZ relative to other three genomes. G1-G10 in (A) and G1-G18 in (B) indicate 10 and 18 undetermined gaps in A. platensis YZ and A. platensis NIES-39 genome, respectively. Slots 2-5 showed that the corresponding query regions had higher sequence similarity (≥70%) with the reference sequence. Empty regions on the query slots indicated parts without similar hits between the backbone sequence and the query sequences, and those undetermined bases in the reference genomes. The last two circles respectively indicate the GC content and GC skew of the reference genomes. The Arthrospira sp. PCC 8005 was abbreviated as A. sp. PCC 8005 in this and the following figures.
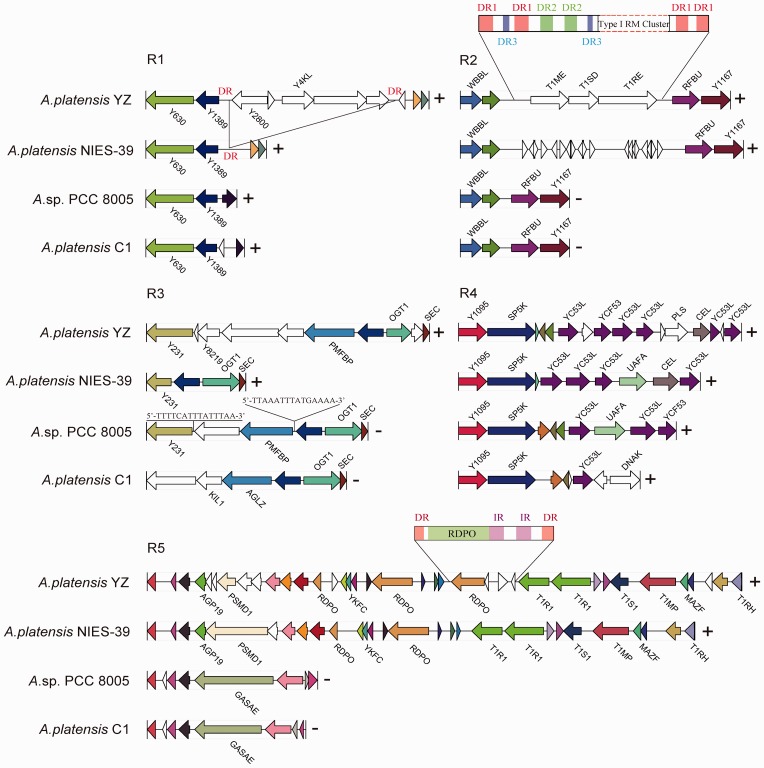
The most powerful approach for identification of gene gains and losses is by conducting a direct molecular genetic analysis of DNA sequences.30 We analysed several syntenic blocks with extensive gene gains in A. platensis YZ relative to the other three A. platensis genomes (Fig. 4). These blocks involved addition of unknown functions and well characterized genes, which indicated the occurrence of potential LGT events. Scanning the upstream and downstream sequences surrounding these additional genes in A. platensis YZ did not identify known transposons or insertion elements. Furthermore, comparative analysis showed an additional 5-kb sequence that encoded five genes that were flanked by a pair of 170-bp direct repeats (DRs) with high similar sequence identity (>91%) that were inserted into the A. platensis YZ genome relative to that of the A. platensis NIES-39, as indicated in R1 (Fig. 4). These potential LTGs included two ATPases that belonged to the AAA superfamily and three hypothetical proteins that were extensively distributed in the various Oscillatoriales species. Moreover, we did not identify typical IRs that flanked these genes, suggesting that the acquisition of these genes might have not been mediated by transposable elements, but was possibly mediated by recombinase through recombination where a pair of 170-bp DRs was presented. However, there was no known recombinase encoded by these genes, which in turn suggesting that this accessory region could have been lost from A. platensis NIES-39 by recombination between two DR sequences. Nucleotides composition analysis did not show obviously abnormal discrepancy of GC content between acquired and flanking regions. Potential LGT associated with acquisition of type I RM system was observed (R2). A complete RM system was acquired in A. platensis YZ relative to Arthrospira sp. PCC 8005 and A. platensis C1. More complexity of nested DRs’ composition (DR1, DR2, and DR3) surrounding the acquired gene cluster in A. platensis YZ was identified at this area, whereas only DR3 sequence appeared as a single event in the whole genomes of Arthrospira sp. PCC 8005 and A. platensis C1. Supplementary Material S1 listed the three DRs appeared in the A. platensis YZ. Moreover, these nested DRs were sporadically scattered across the counterpart region in A. platensis NIES-39 but not throughout the genome, and showed an increased frequency of emergence compared with A. platensis YZ, suggestive of the high instability of this region. Interestingly, we also identified a cluster of probably non-Arthrospira-derived genes including methyltransferase (Y8219) encoding gene that are present in A. platensis YZ genome (R3). The Y8219 and the immediate downstream two function-unknown genes in A. platensis YZ shared only 51.0, 51.2, and 24.2% amino acid identities with the orthologues from Microcoleus vaginatus, Arthrospira sp. PCC 8005 and Pseudoalteromonas agarivorans, respectively, suggesting that these genes were more likely derived from distantly related organisms accompanied by extensive mutations, because two of these three genes had no orthologues in recently sequenced Arthrospira genome at all. The genomic structure in this area among four strains is also largely different, which indicated high variability of this region. In addition, potential LGT between closely related organisms was also observed. Three genes from Arthrospira sp. PCC 8005 that were inserted into the A. platensis NIES-39 backbone resembled the local genomic structure of A. platensis YZ (R4, Fig. 4). Finally, LGT potentially mediated by mobile genetic element (MGE) was also identified in A. platensis YZ genome relative to that observed in A. platensis NIES-39 (R5, Fig. 4). A reverse transcriptase (RTase)-encoding gene was immediately followed by a pair of long terminal repeats (LTRs, >90% identity), which was further globally enwrapped by a pair of long DRs (LDRs) (>85% identity) that leads to the formation of a typical retrotransposon. The retrotransposons consist of two subclasses, LTR and non-LTR retrotransposons, based on the phylogeny of RTase which they encodes.31 Duplication of retrotransposons is a replicative event through transcribing themselves to RNA intermediate and then reversely transcribing back to DNA that has been demonstrated to be directed by RTase which is the key component of the retrotransposons.32 However, its integration into the A. platensis genome seems to be a recombination event, because it is unreasonable to generate such LDRs by using transposase.
Figure 4.
Five typical regions involved in potential LGT events in A. platensis YZ compared with three other Arthrospira species. R1-R5 corresponds to the R1-R5 in the Figure 3. Homologous genes are marked with same colour, whereas non-homologous genes are left blank. Gene-coding products were labelled with abbreviated names provided in the UniProt database, whereas unlabelled arrows indicate unannotated or unknown gene. (+) or (−) suggests the block is located on the non-complementary or complementary strand of the corresponding genome. R1: Five additional genes were gained in the A. platensis YZ genome. A pair of 170-bp sequence (DR) with >95% identity flanked the LTGs in A. platensis YZ, whereas only one copy was detected in A. platensis NIES-39, and entirely absent in Arthrospira sp. PCC 8005 and A. platensis C1; R2: DR1 is 87-bp in length. DR2 and DR3 are parts of DR1. The genome of A. platensis NIES-39 in this region is rich in three types of DRs; R3: A 15-bp imperfect IR flanked the LTGs in Arthrospira sp. PCC 8005 compared with that of A. platensis NIES-39. No DR or IR surrounding the LTG at this region was detected in the A. platensis YZ genome; R4: No DR flanking the LTG was detected in A. platensis YZ compared with that of A. platensis NIES-39; R5: IR and DR are 349- and 187-bp in size, respectively.
In some eubacteria and archaea, such as Acinetobacter baumannii, Escherichia coli, Enterococcus faecium, Lactococcus lactis, and Mycobacterium smegmatis, transposon-mediated LGT usually generates a pair of DRs varying from several to a dozen base pairs flanking the foreign gene after its integration into the host genome.33–36 Interestingly, the size of DRs in the A. platensis was significantly longer than those of other species. For instance, a 170-bp sequence was only observed in the A. platensis NIES-39 genome, where it duplicated only once and thus generated a DR that flanked the acquired genes in the A. platensis YZ (R1, Fig. 4). Likewise, distributions of the 87-bp DR and its’ nested copies were strictly restricted to the neighbouring region of the type I RM system gene cluster, but not ubiquitously in the genome (R2, Fig. 4). LGT involved in the RM systems has been widely observed, including insert element-,37 phage-38, and integrative element-mediated transfer.39 Previous bacterial pan-genome statistical analysis suggested that solitary RM genes were most likely to be transferred by large MGEs, whereas complete RM systems were more frequently transferred autonomously or in small MGEs.40 However, there were no MGE involving in or flanking these acquired genes in R1 and R2. Therefore, it could not exclude that the formation of respective genomic architecture in the three other genomes underwent gene losses at the counterpart regions. The locally distributed DRs may indicate that these DRs were closely related to the gene gains or losses in R1 and R2.
3.3. Chromosomal rearrangements in Arthrospira
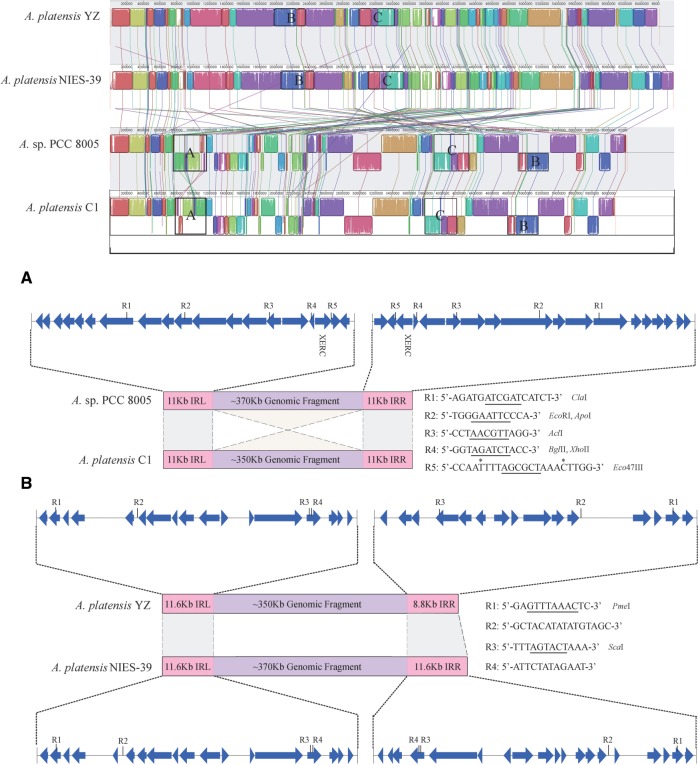
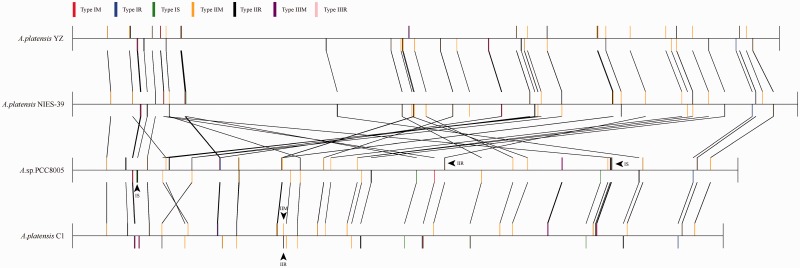
Comparative analysis of the four Arthrospira strains showed that orthologous genes within the same block often possessed distinct coding orientations, which in turn was suggestive of the presence of chromosomal rearrangements. Comparison of global genomic synteny revealed that A. platensis YZ and A. platensis NIES-39 had strong co-linearity, whereas Arthrospira sp. PCC 8005 was highly collinear with A. platensis C1 (Fig. 5), which was consistent with the evolutionary relationship among these species. The observed co-linearity divided the four Arthrospira genomes into two groups. There were few structural variations within groups, except for a 350-kb fragment that was detected in situ to be inverted in orientation between Arthrospira sp. PCC 8005 and A. platensis C1 (region A, Fig. 5). However, extensive chromosomal rearrangements were observed between the two groups, including inversion, translocation, deletion, as well as combinations of these events, suggesting that speciation between groups underwent multiple and extremely complicated structural variations that are very difficult to interpret. We analysed the sequences surrounding the inverted genomic fragment between Arthrospira sp. PCC 8005 and A. platensis C1. Result showed that a pair of 11-kb long inverted repeats (LIRs) with >97% global sequence identities flanked and further extended into the inverted fragment, which resulted in an obscure boundary for the chromosomal inversion. Within the LIR, we identified five pairs of palindromes that constituted a series of restriction enzyme recognition sites (RERS). These molecular evidences suggested that the chromosomal inversion was probably initiated from the symmetrically distributed RERS of the internal region of the LIR and mediated by the restriction enzyme as previously proposed.41 , 42 This LIR seems not to be occasionally emerged at the edge of this inverted fragment, but was closely related to this region to some extent, because it appeared only three times and twice throughout the genomes of Arthrospira sp. PCC 8005 and A. platensis C1, respectively. The 11-kb LIR encoded a tyrosine recombinase that belonged to the integrase family.43 , 44 After a sequence replication event, fusion of the 11-kb copy into adjacent loci of genome might have resulted from the integrase-mediated site-specific recombination, which well explains the duplication events. In addition, the predicted protein-coding genes that harboured RERS that probably interacted with the restriction enzyme within the LIR apparently remained intact after the chromosomal inversion, because the sequence identities between LIR flanking the inverted genomic fragment were extremely high. Chromosomal rearrangements that were potentially associated with sequence duplication were also identified between two groups, such as Figure 5 regions B and C. We analysed the sequences that flanked potentially invertible genomic fragment (region B) between the two groups, and two RERS were identified within the LIR in A. platensis YZ and A. platensis NIES-39, albeit chromosomal inversion at this region was not recurrent among species in the same group. Five and seven copies of the intact 11.6-kb sequence were respectively detected in the genomes of A. platensis NIES-39 and A. platensis YZ. However, all the coding genes derived from the 11.6-kb LIR could not be annotated with known functions; therefore, its potential involvement in particular molecular function remains elusive. Likewise, a pair of 2-kb LIR flanking the potentially invertible genomic fragment was also identified in the region C. Therefore, generation of LIR, but not LDR, in the neighbouring region of genome via sequence duplication would not only enhance the frequency of occurrence of RERS and paired RERS, but also would not destroy gene order after chromosomal inversion. Taken together, the sequence duplication followed by the formation of LIRs flanking the genomic fragments, might play an important role in the chromosomal inversions in A. platensis.
Figure 5.
Comparison of global genomic synteny of A. platensis YZ to three other species. Corresponding blocks from these four genomes are shown based on sequence similarities. The homologous sequences at corresponding blocks are lined together and marked with the same colour. Three genomic fragment rearrangements are indicated by frames and labelled A–C. L-LIR and R-LIR represent left and right LIRs, respectively. In these regions, the L-LIR and R-LIR within and between groups both shared >95% global sequence similarities. In the region B, R-LIR in A. platensis YZ is a truncated IR that shared > 95% sequence similarity with L-LIR. R1-R5 showed the palindromic sequences that were distributed in the L-LIR and R-LIR, except for those labelled with an asterisk. The palindromes involved in the restriction enzymes dissection are underlined.
The RM system, an important immune system for bacteria that prevents the uptake of exogenous DNA, is largely variable in terms of copy number and constitution.45 , 46 It was also proposed to possess the potential role in chromosomal rearrangements, because they can cut DNA.41 Genome-wide identification of RMs in the Oscillatoriales order revealed that species from Arthrospira harboured the most abundant RM enzymes (Table 1 and Fig. 6). In the Arthrospira, four strains all contained types I and II RM enzymes, with type IIR enzymes predomination. Besides types I and II RM, Arthrospira sp. PCC 8005 and A. platensis C1 both harboured a type III RM system, while it was absent in A. platensis NIES-39 and A. platensis YZ.
Table 1.
Distribution of RM systems in Oscillatoriales order
| Species | 1M | 1R | 1S | 2M | 2R | 3M | 3R | Total |
|---|---|---|---|---|---|---|---|---|
| Geitlerinema sp. PCC 7407 | 1 | 1 | 0 | 4 | 1 | 0 | 0 | 7 |
| Leptolyngbya sp. 2LT21S03 | 2 | 1 | 2 | 4 | 1 | 0 | 0 | 10 |
| Leptolyngbya sp. PCC 7376 | 1 | 1 | 0 | 8 | 1 | 0 | 0 | 11 |
| Leptolyngbya sp. PCC 7104 | 1 | 1 | 0 | 6 | 4 | 0 | 0 | 12 |
| Pseudanabaena sp. PCC 7367 | 1 | 1 | 0 | 7 | 3 | 0 | 0 | 12 |
| L. boryanar PCC 6306 | 2 | 2 | 0 | 8 | 1 | 0 | 0 | 13 |
| L. majuscula 3L | 0 | 0 | 0 | 10 | 3 | 0 | 0 | 13 |
| Leptolyngbya sp. PCC 7375 | 3 | 2 | 0 | 9 | 2 | 0 | 0 | 16 |
| Geitlerinema sp. PCC 7105 | 1 | 3 | 1 | 9 | 4 | 0 | 0 | 18 |
| Trichodesmium erythraeum 21-75 | 2 | 2 | 0 | 10 | 5 | 0 | 0 | 19 |
| O. cyanobacterium JSC-12 | 1 | 1 | 0 | 13 | 5 | 0 | 0 | 20 |
| Pseudanabaena sp. PCC 6802 | 1 | 2 | 0 | 13 | 4 | 0 | 0 | 20 |
| Pseudanabaena sp. PCC 7429 | 1 | 1 | 0 | 11 | 9 | 0 | 0 | 22 |
| M. vaginatus PCC 9802 | 1 | 0 | 0 | 14 | 8 | 0 | 0 | 23 |
| Oscillatoria formosa PCC 6407 | 1 | 2 | 1 | 16 | 6 | 1 | 1 | 28 |
| Lyngbya sp. CCY8106 | 1 | 1 | 0 | 13 | 14 | 0 | 0 | 29 |
| Oscillatoria sp. PCC 6506 | 1 | 2 | 1 | 16 | 7 | 1 | 1 | 29 |
| Microcoleus sp. PCC 7113 | 5 | 5 | 1 | 14 | 5 | 0 | 0 | 30 |
| S. major PCC 6313 | 3 | 3 | 1 | 11 | 13 | 0 | 0 | 31 |
| Leptolyngbya sp. PCC 6406 | 7 | 6 | 2 | 9 | 9 | 0 | 0 | 33 |
| Oscillatoria sp. PCC 10802 | 1 | 0 | 0 | 22 | 10 | 0 | 0 | 33 |
| O. acuminata PCC 6304 | 4 | 4 | 0 | 20 | 9 | 0 | 0 | 37 |
| S. subsalsa PCC 9445 | 3 | 3 | 1 | 23 | 7 | 0 | 0 | 37 |
| Coleofasciculus chthonoplastes PCC 7420 | 4 | 2 | 0 | 22 | 11 | 0 | 0 | 39 |
| O. nigro-viridis PCC 7112 | 4 | 1 | 0 | 21 | 14 | 1 | 1 | 42 |
| C. epipsammum PCC 9333 | 5 | 6 | 5 | 17 | 10 | 0 | 0 | 43 |
| A. platensis str. Paraca | 4 | 3 | 3 | 23 | 18 | 0 | 0 | 51 |
| A. platensis NIES-39 | 4 | 4 | 4 | 26 | 20 | 0 | 0 | 58 |
| Arthrospira sp. PCC 8005 | 4 | 4 | 7 | 23 | 19 | 1 | 1 | 59 |
| A. platensis C1 | 5 | 6 | 5 | 24 | 18 | 1 | 1 | 60 |
| A. platensis YZ | 5 | 5 | 3 | 26 | 21 | 0 | 0 | 60 |
Figure 6.
Overview of RM systems distributed in the four Arthrospira genomes. Colour bars above or below the central line from each strain represent genes with two distinct coding orientations in the genome. Black line is used to connect those best hit RM enzyme between different strains. Arrow indicates the unique enzyme compared with the other three strains.
Based on the whole genome co-linear relationship, we performed comparative analyses of RM systems intra- and inter-groups for four Arthrospira strains. As expected, the co-linearity of RM systems-coding genes within groups was consistent with the whole genomic synteny (Fig. 6). Within the groups, besides those common RM systems, there were three unique enzymes in the A. platensis YZ genome compared with A. platensis NIES-39, including one type IIR, one type IM and one type IR enzyme, whereas only a type IS was uniquely existence in the A. platensis NIES-39 compared with A. platensis YZ. Likewise, seven (one type IR, one IS, three IIR, two IIM) and six (three type IS and three IIR) enzymes were uniquely presented to each other in A. platensis C1 and Arthrospira sp. PCC 8005, respectively. For inter-groups comparison, two (one IIR and one IIM) and three (two type IS and one IIR) enzymes were uniquely appeared in the A. platensis C1 and Arthrospira sp. PCC 8005 response to the other strains, respectively, while no unique RM enzyme occurred in the A. platensis YZ and A. platensis NIES-39 genomes (Fig. 6). Generally, the more unique RM gene relative to the smaller genome size was observed in these closely related strains, which may indicate the importance of RM systems for genome plasticity in the Arthrospira. The FASTA sequences of unique enzymes presented in each strain were listed in the Supplementary Material S2.
Chromosomal inversions have been frequently identified in other closely related cyanobacterial strains. For instance, an inverted 1.3-Mb fragment was detected between Cyanothece sp. PCC 8801 and PCC 8802, and a 188-kb inversion was identified between Synechococcus elongates PCC 6301 and PCC 7942 chromosome.47 , 48 However, these authors did not provide possible suggestions underlying the generation of the chromosomal inversion. Indeed, the 1.3-Mb inverted fragment was flanked by a pair of 5.2-kb LIRs with extremely high identities (>99%) both in Cyanothece sp. PCC 8801 and PCC 8802,48 and this LIR sequences were only detected in the flanking regions of the inverted fragment from these two closely related strains. Likewise, the 188-kb inversion between S . elongates PCC 6301 and PCC 7942 was also flanked by a pair of 0.5-kb LIRs with >81% identities in both strains.47 The above evidences may suggest that LIRs were closely related to the chromosomal inversion. Previous study on phylum Proteobacteria demonstrated that extensively rearranged genomes contain a high number of repeats among closely related bacteria.49 The negative association between the number of repeats and genome stability has been observed and statistically confirmed in γ-proteobacteria, where the genomes contained more repeats undergoing accelerated rearrangement rates, while genomes lacking repeats were more stable.50 Based on these molecular evidences, previous explanations for the possible mechanisms underlying chromosomal inversion included illegitimate recombination,51–53 intra-chromosomally homologous recombination,54 as well as integrase-mediated site-specific recombination.55 , 56 When compared with these mechanisms, restriction enzyme-mediated chromosomal inversions might be oversimplified, yet could serve as a new mechanistic model for chromosomal rearrangements. The proposed model for restriction enzyme-mediated chromosomal inversion has been described in Supplementary Figure S2.
3.4. Repeat sequences in Arthrospira
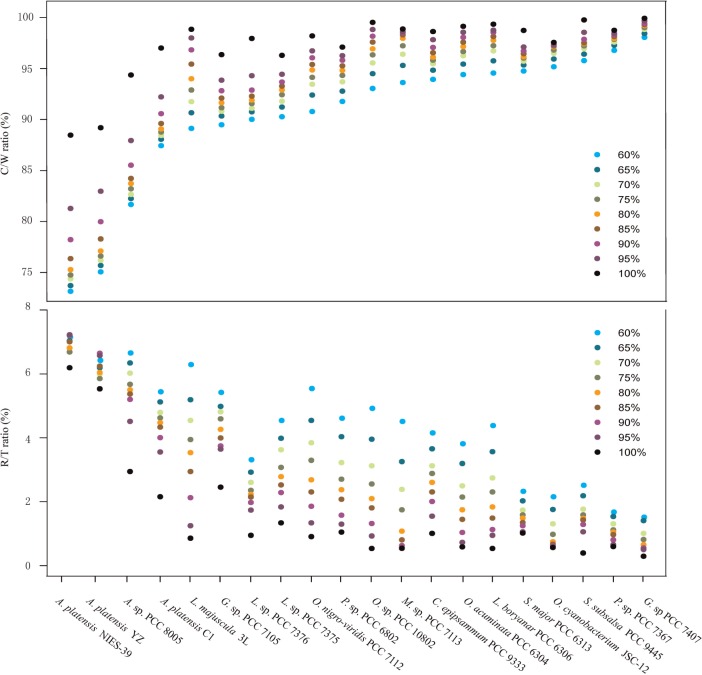
One of the most notable features of the cyanobacterial genome is the predominance of various repeated sequences. Genome-wide clustering analysis of all the genes of Oscillatoriales genomes, respectively, revealed that members of genus Arthrospira possessed the lowest C/W ratios at any level of homology, which in turn suggested that these underwent multiple duplication events during speciation (Fig. 7). C/W ratio represented the number of total clusters divided by the number of total genes. Moreover, members of Arthrospira, particularly A. platensis NIES-39 and A. platensis YZ, showed the highest R/T ratios, which was suggested that gene duplication in Arthrospira widely occurred. This ratio was calculated as the number of redundant gene clusters that comprised at least two genes divided by the total number of clusters. In A. platensis YZ, a total of 327 genes were determined to have been duplicated at least once, which in turn resulted in the generation of 2,020 redundant genes constituting approximately one-third of the total gene and one-sixth of the complete genome size (Supplementary Table S1). In general, in addition to a large number of uncharacterized genes, these duplicated genes are functionally involved in replication, recombination, energy production, transcription, and post-translational modification. Among these redundant sequences, the most frequently duplicated genes were RTase, transposase, and integrase (194, 159, and 12 copies, respectively), which were derived from 4, 23, and 1 clusters, respectively. We then investigated the probable origin of these enzymes. First, sequence alignment of 12 integrases showed >84% global amino acid similarity, and these integrases were further subdivided into three subtypes, which was suggestive that multiplication of integrase-encoding genes in A. platensis YZ genome was most probably generated by multiple gene duplication events that were accompanied by amino acid substitutions (Supplementary Figure S3). Second, we constructed the phylogenetic relationship of the two largest RTase clusters (clusters 527 and 4099, Supplementary Table S1), which contained 79 and 73 members, respectively. The RTases derived from the same cluster were located together, yet showed distinct boundaries between clusters (Supplementary Figure S4). This indicated that hundreds of RTase paralogs might have been derived from several common ancestors. Another significant characteristic of duplicated genes was that they were usually adjacently located or duplication-linked with a plenty of unknown functions, some of which can be annotated by one or two of the three enzymes (RTase, transposase, and integrase), and these duplicated fragments varied in size from several to dozens of kilobases.57–59 This was also partially identified by the observation of contiguous relationships of duplicated genes after sorted by their coordinates in the genome. Therefore, sequence duplication might significantly contribute to the genomic architecture of Arthrospira genomes, which resulted in the formation of various long repeats at both orientations. Previous studies have shown that a large proportion of the transposases and RTase constituted the long repeated sequences in some cyanobacteria.57–60 For example, 93% of 362 transposases of Microcystis aeruginosa PCC 7806 emerged in the long repeats.59 However, because most of the duplicated genes are functionally unknown, besides chromosomal rearrangements, the potential phylogenetic roles of multiple sequence duplication events in the Arthrospira are remaining elusive.
Figure 7.
Gene duplication in selected species of Oscillatoriales. The total genes from each species are clustered at 60, 65, 70, 75, 80, 85, 90, 95, and 100% amino acid sequence identities. For one certain species at one certain resolution, the C/W ratio represents the total number of clusters divided by the total number of genes. R/W ratio represents the number of redundant gene clusters (the clusters that comprised at least two genes) divided by the total number of clusters.
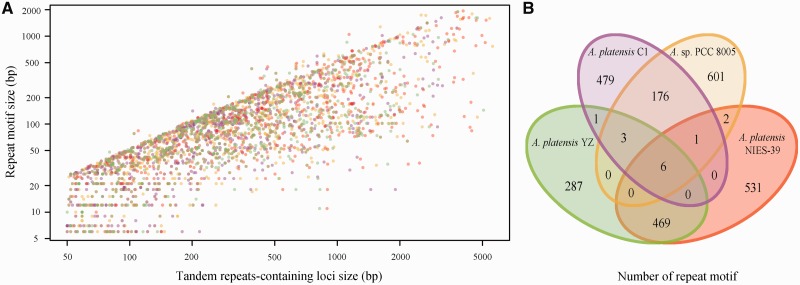
TRs are patterns of nucleotide sequence directly adjacent to each other. Distributions of TRs in the genomic loci are also recognized as the variable number of TR regions that have been widely used to determine an individual's inherited traits as well as for molecular typing both in prokaryotic and eukaryotic species.61 Based on the size of the repeat motif, TRs were classified into microsatellites, minisatellites, and macrosatellites with the motif sizes varying from 1 to 9, 10 to 100, and >100 bp, respectively.62 Examining the four Arthrospira genomes showed that 870, 736, 1,148, and 848 TRs-containing loci (TRL) were presented in Arthrospira sp. PCC 8005, A. platensis C1, A. platensis NIES-39, and A. platensis YZ, respectively (Fig. 8A), which represented 789, 666, 1,009, and 766 non-redundant repeat motifs (Fig. 8B). The size of these core repeat motifs varied from several base pairs to 1.9 kb that led to form the longest single TRL up to 8.1 kb. Comparative analyses showed that the number of shared repeat motifs among strains derived from the same groups was significantly more than those from different groups (Fig. 8B). Only six commonly shared repeat motifs were identified, suggesting that conservation of TRs among four strains was weak. Additionally, more overlapped repeats at both quantity and proportion level may indicate that the evolutionary relationship between A. platensis NIES-39 and A. platensis YZ was closer than that between Arthrospira sp. PCC 8005 and A. platensis C1.
Figure 8.
Distribution of TRs and their relationships among four Arthrospira genomes. (A) length distributions of 870, 736, 1,148, and 848 repeat motifs and their corresponding TRL in Arthrospira sp. PCC 8005, A. platensis C1, A. platensis NIES-39 and A. platensis YZ, respectively. Colour key for each strain is responded to the B panel. (B) 870, 736, 1,148, and 848 repeat motifs from each strain were independently clustered with 100% nucleotide identity, respectively. The non-redundant repeat motifs (789, 666, 1,009, and 766) from each strain were pooled and further clustered with 100% identity to show the common and unique features of these repeat motifs derived from different strains.
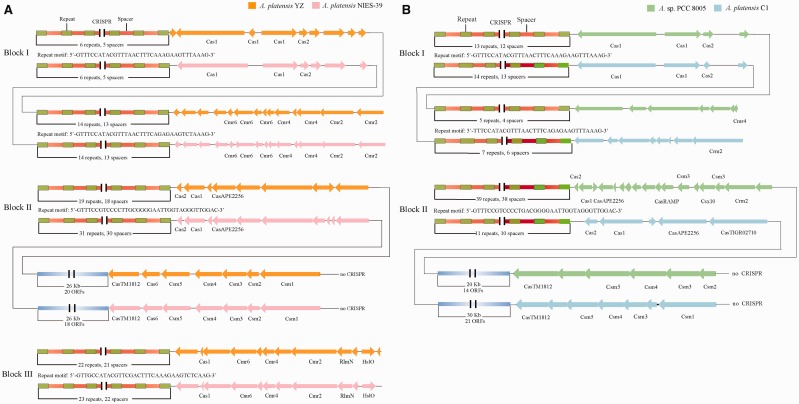
CRISPRs, a type of important repeat elements, are related to the adaptive immunity system that has been widely identified and characterized in many archaea and bacteria.63 , 64 We comparatively analysed the distribution of CRISPR and CRISPR-associated protein (Cas) systems among four Arthrospira genomes. On the whole, A. platensis YZ and A. platensis NIES-39 harboured three typical regions (Block I-III of Fig. 9A) that contained the CRISPR-Cas system, whereas only two regions were identified in A. platensis C1 and Arthrospira sp. PCC 8005 (Block I and II of Fig. 9B). It is interesting to find that two CRISPR arrays were adjacently located next to different cas gene organization in all strains as shown in Figure 9 Blocks I. The repeat motif of the corresponding CRISPR array was approximately identical to the ones from the members belonging to the same or different groups represented as global genomic synteny, except the slight diversity was presented in the latter array between members from the different groups. Strains derived from the same groups also showed the high similarity in cas gene organization. The main differences for the same CRISPR-Cas systems among strains involved copy number of repeats and the diversity of spacers (Supplementary Material S3). A. platensis NIES-39 and A. platensis C1 harboured more repeats and spacers than A. platensis YZ and Arthrospira sp. PCC 8005, respectively. Furthermore, we also found that one cas gene cluser in all the four strains (Csm1-Csm2-Csm3-Csm4-Csm5-Cas6-CasTM1812 in A. platensis YZ and A. platensis NIES-39, Csm1/Csm2-Csm3-Csm4-Csm5-CasTM1812 in A. platensis C1 and Arthrospira sp. PCC 8005) was not CRISPR-associated, which contradicted the general description of cas gene distribution, because we did not identify any CRISPR array that was proximal to the cas gene cluster. In addition to these typical CRISPR-Cas systems, several CRISPR arrays were also detected in the four genomes without a cas gene located within their proximal regions.
Figure 9.
The CRISPR-Cas system loci in the four Arthrospira genomes. Block I-III from left panel (A) showed the organization of CRISPR-Cas systems in A. platensis YZ and A. platensis NIES-39, and the Block I-II from right panel (B) showed the counterparts in Arthrospira sp. PCC 8005 and A. platensis C1. Repeat motifs amid the CRISPR arrays are shared by both strains at the corresponding loci. The unlabelled genes represent unannotated or functionally unknown genes.
In conclusion, A. platensis YZ was more closely related to A. platensis NIES-39 than Arthrospira sp. PCC 8005 and A. platensis C1 at various levels. Comparative genomics and clustering analyses suggested that the Arthrospira genome was highly fluid and exceptionally plastic, which was characterized by extensive chromosomal rearrangements, a large number of repetitive sequences as well as genes encoding transposases, RTases, recombinases, and restriction enzymes. Therefore, speciation of Arthrospira and the specialization for an ecological niche were accompanied by a large scale reorganization of its genomic structure. These findings will facilitate in better understanding the process of speciation and diversification in A . platensis.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Acknowledgements
We thank the members of ‘Spirulina Consortium’ at Beijing Genomics Institute, Shenzhen for whole genome DNA sequencing.
Accession number
Funding
Funding to pay the Open Access publication charges for this article was provided by National Natural Science Foundation of China (30571009), Natural Science Foundation of Zhejiang Province, China (Z307471) and Science and Technology Foundation of Zhejiang Province, China (2009C33040).
References
- 1. Habib M. A. B., Mashuda P., Huntington T. C., Hasan M. R. 2008, A Review on Culture, Production and Use of Spirulina as Food for Humans and Feeds for Domestic Animals and Fish. Food and Agriculture Organization of The United Nations: Rome, Italy. [Google Scholar]
- 2. Huili W., Xiaokai Z., Meili L., et al. 2013, Proteomic analysis and qRT-PCR verification of temperature response to Arthrospira (Spirulina) platensis. PLoS One, 8, e83485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang H., Yang Y., Chen W., et al. 2013, Identification of differentially expressed proteins of Arthrospira (Spirulina) plantensis-YZ under salt-stress conditions by proteomics and qRT-PCR analysis. Proteome Sci., 11, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pan R., Lu R., Zhang Y., et al. 2015, Spirulina phycocyanin induces differential protein expression and apoptosis in SKOV-3 cells. Int. J. Biol. Macromol., 81, 951–9. [DOI] [PubMed] [Google Scholar]
- 5. Zhao F., Zhang X., Liang C., Wu J., Bao Q., Qin S. 2006, Genome-wide analysis of restriction-modification system in unicellular and filamentous cyanobacteria. Physiol. Genomics, 24, 181–90. [DOI] [PubMed] [Google Scholar]
- 6. Cheevadhanarak S., Paithoonrangsarid K., Prommeenate P., et al. 2012, Draft genome sequence of Arthrospira platensis C1 (PCC9438). Stand. Genomic Sci., 6, 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Janssen P. J., Morin N., Mergeay M., et al. 2010, Genome sequence of the edible cyanobacterium Arthrospira sp. PCC 8005. J. Bacteriol., 192, 2465–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fujisawa T., Narikawa R., Okamoto S., et al. 2010, Genomic structure of an economically important cyanobacterium, Arthrospira (Spirulina) platensis NIES-39. DNA Res., 17, 85–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bao Q., Tian Y., Li W., et al. 2002, A complete sequence of the T. tengcongensis genome. Genome Res., 12, 689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dong Y., Xie M., Jiang Y., et al. 2013, Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat. Biotechnol., 31, 135–41. [DOI] [PubMed] [Google Scholar]
- 11. Gordon D., Abajian C., Green P. 1998, Consed: a graphical tool for sequence finishing. Genome Res., 8, 195–202. [DOI] [PubMed] [Google Scholar]
- 12. McKenna A., Hanna M., Banks E., et al. 2010, The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res., 20, 1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delcher A. L., Bratke K. A., Powers E. C., Salzberg S. L. 2007, Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics, 23, 673–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lagesen K., Hallin P., Rodland E. A., Staerfeldt H. H., Rognes T., Ussery D. W. 2007, RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res., 35, 3100–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lowe T. M., Eddy S. R. 1997, tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res., 25, 955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Petkau A., Stuart-Edwards M., Stothard P., Van Domselaar G. 2010, Interactive microbial genome visualization with GView. Bioinformatics, 26, 3125–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roberts R. J., Vincze T., Posfai J., Macelis D. 2010, REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res., 38, D234-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu M., Eisen J. A. 2008, A simple, fast, and accurate method of phylogenomic inference. Genome Biol., 9, R151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. 2010, New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol., 59, 307-321. [DOI] [PubMed] [Google Scholar]
- 20. Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. 2006, ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res., 34, D32–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Darling A. E., Mau B., Perna N. T. 2010, progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One, 5, e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li W., Godzik A. 2006, Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics, 22, 1658–9. [DOI] [PubMed] [Google Scholar]
- 23. Benson G. 1999, Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res., 27, 573–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grissa I., Vergnaud G., Pourcel C. 2007, CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res., 35, W52–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kawai M., Higashiura N., Hayasaki K., et al. 2015, Complete genome and gene expression analyses of Asaia bogorensis reveal unique responses to culture with mammalian cells as a potential opportunistic human pathogen. DNA Res., 22, 357–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Achaz G., Coissac E., Netter P., Rocha E. P. 2003, Associations between inverted repeats and the structural evolution of bacterial genomes. Genetics, 164, 1279–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shih P. M., Wu D., Latifi A., et al. 2013, Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. U S A, 110, 1053–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nei M., Rooney A. P. 2005, Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet., 39, 121–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lefort F., Calmin G., Crovadore J., et al. 2014, Whole-Genome Shotgun Sequence of Arthrospira platensis Strain Paraca, a Cultivated and Edible Cyanobacterium. Genome Announc., 2, 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ochman H., Lawrence J. G., Groisman E. A. 2000, Lateral gene transfer and the nature of bacterial innovation. Nature, 405, 299–304. [DOI] [PubMed] [Google Scholar]
- 31. Xiong Y., Eickbush T. H. 1990, Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J., 9, 3353–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fassbender S., Bruhl K. H., Ciriacy M., Kuck U. 1994, Reverse transcriptase activity of an intron encoded polypeptide. EMBO J., 13, 2075–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Johnsrud L., Calos M. P., Miller J. H. 1978, The transposon Tn9 generates a 9 bp repeated sequence during integration. Cell, 15, 1209–19. [DOI] [PubMed] [Google Scholar]
- 34. Garcia-Migura L., Hasman H., Svendsen C., Jensen L. B. 2008, Relevance of hot spots in the evolution and transmission of Tn1546 in glycopeptide-resistant Enterococcus faecium (GREF) from broiler origin. J. Antimicrob. Chemother., 62, 681-687. [DOI] [PubMed] [Google Scholar]
- 35. Machielsen R., Siezen R. J., van Hijum S. A., van Hylckama Vlieg J. E. 2011, Molecular description and industrial potential of Tn6098 conjugative transfer conferring alpha-galactoside metabolism in Lactococcus lactis. Appl. Environ. Microbiol., 77, 555-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roberts A. P., Davis I. J., Seville L., Villedieu A., Mullany P. 2006, Characterization of the ends and target site of a novel tetracycline resistance-encoding conjugative transposon from Enterococcus faecium 664.1H1. J. Bacteriol., 188, 4356–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Takahashi N., Ohashi S., Sadykov M. R., Mizutani-Ui Y., Kobayashi I. 2011, IS-linked movement of a restriction-modification system. PLoS One, 6, e16554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kita K., Kawakami H., Tanaka H. 2003, Evidence for horizontal transfer of the EcoT38I restriction-modification gene to chromosomal DNA by the P2 phage and diversity of defective P2 prophages in Escherichia coli TH38 strains. J. Bacteriol., 185, 2296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Burrus V., Bontemps C., Decaris B., Guedon G. 2001, Characterization of a novel type II restriction-modification system, Sth368I, encoded by the integrative element ICESt1 of Streptococcus thermophilus CNRZ368. Appl. Environ. Microbiol., 67, 1522–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Oliveira P. H., Touchon M., Rocha E. P. 2014, The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res., 42, 10618–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suyama M., Bork P. 2001, Evolution of prokaryotic gene order: genome rearrangements in closely related species. Trends Genet., 17, 10–3. [DOI] [PubMed] [Google Scholar]
- 42. Gelfand M. S., Koonin E. V. 1997, Avoidance of palindromic words in bacterial and archaeal genomes: a close connection with restriction enzymes. Nucleic Acids Res., 25, 2430–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cornet F., Hallet B., Sherratt D. J. 1997, Xer recombination in Escherichia coli. Site-specific DNA topoisomerase activity of the XerC and XerD recombinases. J. Biol. Chem., 272, 21927–31. [DOI] [PubMed] [Google Scholar]
- 44. Colloms S. D., McCulloch R., Grant K., Neilson L., Sherratt D. J. 1996, Xer-mediated site-specific recombination in vitro. EMBO J., 15, 1172–81. [PMC free article] [PubMed] [Google Scholar]
- 45. Roberts R. J., Belfort M., Bestor T., et al. 2003, A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res., 31, 1805–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wilson G. G., Murray N. E. 1991, Restriction and modification systems. Annu. Rev. Genet., 25, 585–627. [DOI] [PubMed] [Google Scholar]
- 47. Sugita C., Ogata K., Shikata M., et al. 2007, Complete nucleotide sequence of the freshwater unicellular cyanobacterium Synechococcus elongatus PCC 6301 chromosome: gene content and organization. Photosynth Res., 93, 55–67. [DOI] [PubMed] [Google Scholar]
- 48. Bandyopadhyay A., Elvitigala T., Welsh E., et al. 2011, Novel metabolic attributes of the genus cyanothece, comprising a group of unicellular nitrogen-fixing Cyanothece. MBio, 2, e00214–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Parkhill J., Sebaihia M., Preston A., et al. 2003, Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet., 35, 32–40. [DOI] [PubMed] [Google Scholar]
- 50. Rocha E. P. 2003, DNA repeats lead to the accelerated loss of gene order in bacteria. Trends Genet., 19, 600–3. [DOI] [PubMed] [Google Scholar]
- 51. van Belkum A., Scherer S., van Alphen L., Verbrugh H. 1998, Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev., 62, 275–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Moxon E. R., Rainey P. B., Nowak M. A., Lenski R. E. 1994, Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol., 4, 24–33. [DOI] [PubMed] [Google Scholar]
- 53. Rocha E. P. 2004, Order and disorder in bacterial genomes. Curr. Opin. Microbiol., 7, 519–27. [DOI] [PubMed] [Google Scholar]
- 54. Rebollo J. E., Francois V., Louarn J. M. 1988, Detection and possible role of two large nondivisible zones on the Escherichia coli chromosome. Proc. Natl. Acad. Sci. U S A, 85, 9391–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Campo N., Dias M. J., Daveran-Mingot M. L., Ritzenthaler P., Le Bourgeois P. 2004, Chromosomal constraints in Gram-positive bacteria revealed by artificial inversions. Mol. Microbiol., 51, 511–22. [DOI] [PubMed] [Google Scholar]
- 56. Garcia-Russell N., Harmon T. G., Le T. Q., Amaladas N. H., Mathewson R. D., Segall A. M. 2004, Unequal access of chromosomal regions to each other in Salmonella: probing chromosome structure with phage lambda integrase-mediated long-range rearrangements. Mol. Microbiol., 52, 329–44. [DOI] [PubMed] [Google Scholar]
- 57. Bench S. R., Ilikchyan I. N., Tripp H. J., Zehr J. P. 2011, Two strains of Crocosphaera watsonii with highly conserved genomes Eare distinguished by strain-specific features. Front. Microbiol., 2, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nakamura Y., Kaneko T., Sato S., et al. 2002, Complete genome structure of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. DNA Res., 9, 123–30. [DOI] [PubMed] [Google Scholar]
- 59. Frangeul L., Quillardet P., Castets A. M., et al. 2008, Highly plastic genome of Microcystis aeruginosa PCC 7806, a ubiquitous toxic freshwater cyanobacterium. BMC Genomics, 9, 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kaneko T., Nakajima N., Okamoto S., et al. 2007, Complete genomic structure of the bloom-forming toxic cyanobacterium Microcystis aeruginosa NIES-843. DNA Res., 14, 247–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. van Belkum A. 2007, Tracing isolates of bacterial species by multilocus variable number of tandem repeat analysis (MLVA). FEMS Immunol. Med. Microbiol., 49, 22–7. [DOI] [PubMed] [Google Scholar]
- 62. Lopes J., Ribeyre C., Nicolas A. 2006, Complex minisatellite rearrangements generated in the total or partial absence of Rad27/hFEN1 activity occur in a single generation and are Rad51 and Rad52 dependent. Mol. Cell Biol., 26, 6675–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bhaya D., Davison M., Barrangou R. 2011, CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet., 45, 273–297. [DOI] [PubMed] [Google Scholar]
- 64. Sander J. D., Joung J. K. 2014, CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol., 32, 347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.