Abstract
Transposable elements (TEs) have the capacity to replicate and insert into new genomic locations. This contributed significantly to evolution of genomes, but can also result in DNA breaks and illegitimate recombination, and therefore posing a significant threat to genomic integrity. Excess damage to the germ cell genome results in sterility. A specific RNA silencing pathway, termed the piRNA pathway operates in germ cells of animals to control TE activity. At the core of the piRNA pathway is a ribonucleo-protein complex consisting of a small RNA, called piRNA, and a protein from the PIWI subfamily of Argonaute nucleases. The piRNA pathway relies on the specificity provided by the piRNAs to recognize TEs targets, while effector functions are provided by the PIWI protein. PIWI-piRNA complexes silence TEs both at the transcriptional level – by attracting repressive chromatin modifications to genomic targets – and at the post-transcriptional level – by cleaving TE transcripts in the cytoplasm. Impairment of the piRNA pathway leads to overexpression of TEs, significantly compromised genome structure and, invariably, germ cells death and sterility.
The piRNA pathway is best understood in the fruit fly, Drosophila melanogaster, and in mouse. This Chapter gives an overview of current knowledge on piRNA biogenesis, and mechanistic details of both transcriptional and posttranscriptional TE silencing by the piRNA pathway. It further focuses on the importance of post-translational modifications and subcellular localization of the piRNA machinery. Finally, it provides a brief description of analogous pathways in other systems.
Keywords: piRNA, Small RNA, Argonautes, Piwi proteins, TE, Transposon, Transposable element, Tudor domain, Transcriptional silencing, Posttranscriptional silencing, Heterochromatin, H3K9me3, DNA methylation, Ping-Pong cycle, Germ granules, Nuage, Pole plasm, Pi-bodies, Intramitochondrial cement
4.1 Introduction
Of the 100 trillion cells in the body, germ cells are the only ones contributing to all future generations, and as such are essentially immortal. It is therefore of pivotal importance to preserve the integrity of the germ cell genome, as damage to it can result in an evolutionary dead end – sterility.
One of the greatest internal threats to genome stability is posed by transposable elements (TEs) – highly abundant and repetitive segments of DNA with the ability to replicate and insert into new locations, wreaking havoc in the process. New insertions cause breaks in the DNA, which lead to cell cycle arrest, disrupt genes and their regulatory regions, and increase the likelihood of non-homologous recombination (Slotkin and Martienssen 2007). Various systems have evolved in different organisms to combat and contain the expansion of TEs, but it is the elegant mechanism employed by the germ cells of metazoans that has attracted considerable interest in recent years: the piRNA pathway (Siomi et al. 2011).
The piRNA pathway is a germline-specific RNA silencing mechanism. The central effector complex of the pathway, called pi-RISC (piRNA-induced silencing complex) in analogy to canonical RNA interference pathways, consists of a protein from the PIWI subfamily of Argonaute nucleases, and a PlWI-interacting RNA, or piRNA. The piRNA pathway relies on the effector function of the Argonaute protein and the specificity provided by the piRNAs to restrict TE activity. PIWI proteins comprise a clade of the Argonaute protein family that is specific to metazoans and shows gonad-specific expression (Carmell et al. 2002; Cox et al. 1998; Kuramochi-Miyagawa et al. 2001, 2004; Lin and Spradling 1997; Houwing et al. 2007). PIWI, the protein that is the namesake of the PIWI clade of Argonautes, was discovered in a screen for factors affecting stem cell maintenance in the Drosophila germline. Mutants of this gene have very small gonads, and were therefore named P-element induced wimpy testis, or piwi mutants (Cox et al. 1998; Lin and Spradling 1997).
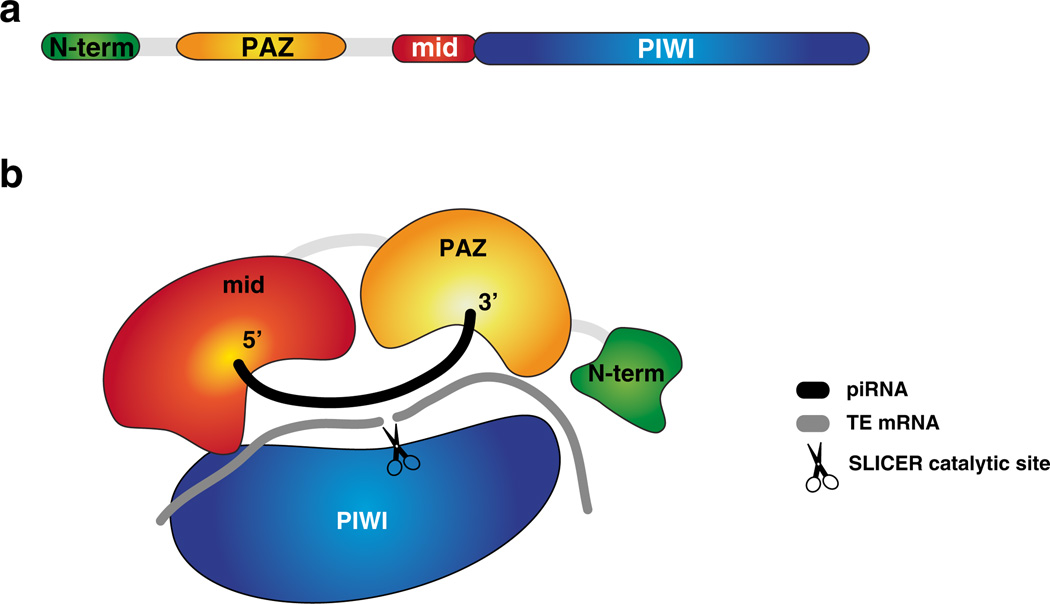
The domain structure of PIWI proteins is similar to that of Argonautes (Fig. 4.1) (Song et al. 2004; Wang et al. 2008). Like Argonautes, the mid domain anchors the 5′ end of a piRNA, and the PAZ domain lodges the 3′ end of the piRNA. The PIWI domain contains catalytic residues within an RNase H-like nucleolytic fold, providing catalytic cleavage of target transcripts. Unlike Argonautes however, PIWIs have Arginine-rich motifs near their N-termini. These residues are post-translationally dimethylated. Arginine methylation allows PIWI proteins to interact with components of the pathway from the Tudor family (Kirino et al. 2009; Liu et al. 2010a; Nishida et al. 2009; Vagin et al. 2009). Tudor family proteins containing the eponymous Tudor domain can bind methylated arginines in PIWI proteins, and this relationship appears to be conserved in many species. Many Tudor proteins have several Tudor domains and act as a scaffold for the formation of higher-order complexes (Huang et al. 2011a; Mathioudakis et al. 2012). In germ cells of Drosophila and in mouse testis, this mode of protein interaction translates into formation of distinct perinuclear granules similar to P-bodies, which harbour components of the piRNA pathway and have been known for a long time in cell biology as “nuage” (Aravin et al. 2008, 2009; Brennecke et al. 2007; Malone et al. 2009).
Fig. 4.1.
The domain structure of PlWI-clade argonaute proteins, (a) The domains of PIWI proteins are organized in a similar fashion to other Argonaute proteins, comprised of the N-terminal region, PAZ (Piwi-Argonaute-Zwille), mid and PIWI domains. The N-terminal region consists of a notional domain that is characterized by arginine rich motifs that are targeted for methylation and in the case of PIWI in flies and MIWI2 in mouse contains the NLS signal, (b) The organization of protein domains in space shows how the mid-domain anchors the piRNA at its 5′ end and the PAZ domain holds the 3′ end of the piRNA. PIWI is the largest domain and its catalytic site responsible for ‘slicer’ activity is positioned to cleave the backbone of annealed target RNA exactly 10 nt relative to the 5′ end of the piRNA
Based on shared domain structure with Argonaute proteins, it was inferred that PIWI proteins must also bind small RNAs. Indeed, at the emergence of high throughput sequencing techniques PIWI-associated small RNAs from several species were sequenced and characterized (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006). piRNAs are typically 23–31 nucleotides (nt) in length and are longer relatives of ubiquitous short interfering RNAs (siRNAs) and microRNAs (miRNAs). Based on their sequence, piRNAs and the conserved role of PIWI proteins in taming transposable elements in the germline of animals were elucidated.
piRNAs are generated either from RNA transcripts of active TE copies or from transcripts originating from specialized loci in the genome called piRNA clusters. Clusters are loci harbouring dysfunctional remnants of TEs and form the basis of immunity against TE propagation (Aravin et al. 2007, 2008; Brennecke et al. 2007). The piRNAs that are generated from piRNA clusters are mostly antisense to TE mRNA sequences and serve as guides for PIWI proteins to find TE transcripts by complementary base pairing. In the cytoplasm, the identification of target mRNAs by PIWIs leads to cleavage of TE transcripts resulting in destruction of the TE message and the concomitant amplification of defensive sequences targeting active TEs as the cleavage product itself is processed into piRNAs. The piRNA pathway has often been compared to an adaptive immune system, as it conveys memory of previous transposon invasions by storing TE sequence information in piRNA clusters. By amplifying piRNAs that are complementary to the active transposon sequence, the piRNA pathway can respond quickly and specifically to acute TE activation. In addition to targeted cleavage of TE mRNAs within the cytoplasm, PIWI proteins also function at the chromatin level to silence TE transcription by directing deposition of repressive histone marks and DNA methylation to copies of TEs (Le Thomas et al. 2013; Sienski et al. 2012). Thus, the piRNA pathway acts on two levels to limit germline transposable element activity.
In this Chapter we review the most important findings about this indispensable pathway, focusing mainly on discoveries in flies and mice. We first discuss sources of piRNAs and known mechanistic details of their biogenesis, followed by the two main ways in which the piRNA pathway acts to repress TEs. Furthermore, we address the localization and composition of the protein complexes that operate within the piRNA pathway. Lastly, we examine several mechanisms similar to the piRNA pathway that repress TE activity in organisms other than Drosophila and mouse.
4.2 piRNA Biogenesis
piRNAs are classified into two groups based on their biogenesis: primary and secondary piRNAs. Primary piRNAs are generated by yet poorly characterized biogenesis machinery, while secondary piRNAs arise in an amplification mechanism termed the Ping-Pong amplification loop to specifically enhance piRNA sequences targeting active elements. In this section we will concentrate on what is known about biogenesis of primary piRNAs focusing on their genomic origin, regulation of transcription and factors involved in their processing.
4.2.1 piRNA Cluster Architecture and Transcription
piRNA populations are highly complex: deep sequencing of piRNAs from mouse and fly revealed millions of individual, distinct piRNA molecules. Neither piRNAs nor their precursor sequences show any structural motif or sequence bias except for a preference for a Uracil as the first 5′ nucleotide (1U bias) of the piRNA. However, when mapped to the genome, this highly diverse population of piRNAs can be mapped to a few discrete genomic loci, called piRNA clusters (Brennecke et al. 2007; Aravin et al. 2006, 2007; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Ro et al. 2007). These clusters are transcriptional units of up to 200 kilobases long that, at least in flies, are mostly located in pericentromeric and subtelomeric beta-heterochromatin and are highly enriched in transposable element sequences (Brennecke et al. 2007). Primary piRNAs are generated almost exclusively from these piRNA clusters. It is believed that upon new transposon invasion the TE eventually “jumps” into one of the clusters leaving a memory of the invasion. Once a transposable element leaves a trace of its sequence in a cluster, it can be targeted by the piRNA machinery, which will limit further propagation of the TE. Currently it is not known what causes active transposons to preferentially integrate into clusters but the chromatin features of clusters have been proposed to play a role.
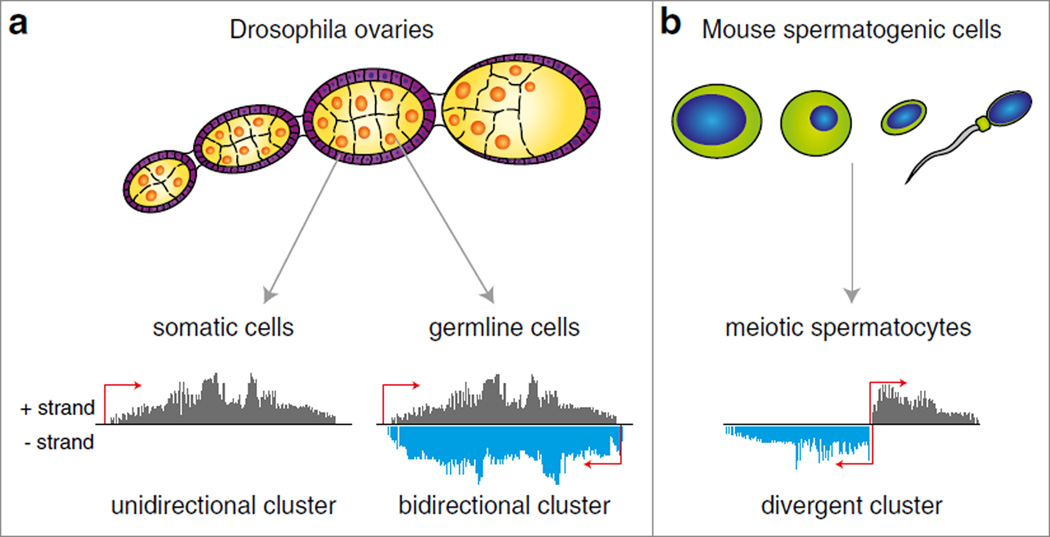
In fly ovaries, clusters are expressed in two cell types: cells of germline origin that include the developing oocyte and associated nurse cells in addition to somatic support cells called follicular cells. Interestingly, the structure of clusters seems to differ depending on where they are expressed: Germline clusters are transcribed bi-directionally, generating both sense and antisense piRNAs in relation to the transposon mRNA. Conversely, somatic clusters appear to be transcribed uni-directionally, producing mostly piRNAs that are antisense to TE coding regions (Fig. 4.2a) (Brennecke et al. 2007). Overall, the source of a piRNA dictates how it is processed.
Fig. 4.2.
The structure of piRNA clusters in Drosophila ovaries and mouse spermatogenic cells, (a) Clusters in Drosophila are divided into two groups based on their structure and they show differential expression in the two cell types of the ovary. Unidirectional clusters are expressed in follicular cells, which are the somatic support cells of the ovary. These clusters produce piRNAs from only one strand of their genomic locus, and are loaded into PIWI. Bidirectional clusters are expressed in germline cells of the ovary, and piRNAs from these clusters are preferentially loaded into AUB and PIWI. (b) Mouse spermatogenic cells contain unidirectional as well as divergent clusters. Divergent clusters are transcribed from a central promoter into both directions. Putative transcription start sites are shown with red arrows. piRNA density profiles derived from the + and − strands are indicated in grey and blue as they would appear in small RNA-seq data
In mouse testes, spermatogenic cells have two kinds of piRNA clusters: one class is transcribed during embryonic development and, just like in Drosophila, piRNAs derived from these clusters defend the germline against transposable elements. A second type of piRNA cluster is expressed in spermatogenic cells of adolescent mice during the first division of meiosis. These are called pachytene clusters, since piRNAs derived from these clusters are highly abundant in the pachytene stage of meiosis. Pachytene piRNAs are not enriched in transposon sequences and to date their function is unknown. An interesting feature of many pachytene clusters is that they are transcribed in both directions from a central promoter (Fig. 4.2b) (Aravin et al. 2006; Girard et al. 2006).
To date, the transcriptional regulation of piRNA clusters in flies and in embryonic stages of mouse spermatogenesis remains elusive. Chromatin immunoprecipitation (ChIP) analysis on histone modifications in the silkworm ovary-derived BmN4 cell line (this cell line contains a functional piRNA pathway and is used as an analogous model for the Drosophila piRNA pathway) revealed that piRNA clusters display features of euchromatin; they are enriched for histone modifications associated with transcriptional activity such as H3K4 di- and tri-methylation in addition to H3K9 acetylation and at the same time are devoid of repressive histone H3K9 di- and tri-methyl marks (Kawaoka et al. 2013). In ChIP performed on Drosophila ovaries, however, H3K9me3 marks are present on clusters and transposon loci (Rangan et al. 2011). The heterochromatin protein (HP1) homolog of Drosophila, Rhino, seems to bind those histone marks on piRNA clusters and is essential for piRNA cluster transcription (Klattenhoff et al. 2009). Still, it remains unresolved if these chromatin marks are a cause or consequence of cluster transcription, and transcription factors directly responsible for regulating cluster expression have not yet been identified. Taken together, data about piRNA clusters in Drosophila suggests the counterintuitive idea that piRNA clusters need to possess a repressive chromatin signature in order to generate piRNAs.
In mouse, a recent study has identified a transcription factor, A-MYB, which regulates expression of pachytene piRNA clusters. A-MYB also controls its own transcription and that of key genes in the piRNA pathway like Miwi, Maelstrom (Mael) and several Tudor domain-containing genes during the pachytene stage of spermatogenesis in mouse. The involvement of one transcription factor in the regulation of clusters and protein factors of the piRNA pathway suggests that a feed-forward loop orchestrates the transcription of piRNA clusters at the same time as the suite of proteins necessary to produce piRNAs from those cluster transcripts (Li et al. 2013). Whether a similar mechanism is at work in Drosophila and during embryonic stages of spermatogenesis remains to be elucidated.
4.2.2 Biogenesis of Primary piRNAs
piRNAs in flies are 23–25 nt in length, whereas in mouse they are slightly longer, averaging 25–28 nt. Similar to other small RNA classes, piRNAs are processed from longer precursors, however, in contrast to miRNAs and siRNAs, the precursors are single stranded transcripts without obvious hairpin structures. It is thought that piRNA biogenesis begins with the endonucleolytic cleavage of the long precursor transcript, generating shorter piRNA precursors. This cleavage event possibly specifies the 5′ end of the future piRNA. Genetic screens and structural studies suggest that the endonuclease, Zucchini (ZUC), might conduct this first cleavage of some piRNA precursors (Fig. 4.3a) (Czech et al. 2013; Handler et al. 2013; Muerdter et al. 2013; Olivieri et al. 2010; Ipsaro et al. 2012; Nishimasu et al. 2012; Voigt et al. 2012). After cleavage, the 5′ end of piRNA precursors gets loaded into a PIWI protein. In flies, primary piRNAs are loaded into two of the three PIWI proteins, PIWI and Aubergine (AUB). The factors that make up the piRNA loading machinery are currently unknown, however several studies have identified the proteins Shutdown (SHU), Vreteno (VRET), Brother-of-Yb (BoYb) and Sister-of-Yb (SoYb) as components that are necessary for loading of PIWI and AUB with primary piRNA (Handler et al. 2013; Olivieri et al. 2010) (Table 4.1). Loading of PIWI seems to require some additional factors including Armitage (ARMI) and Zucchini (ZUC) and in somatic cells, the Tudor domain protein, YB (Olivieri et al. 2010; Szakmary et al. 2009).
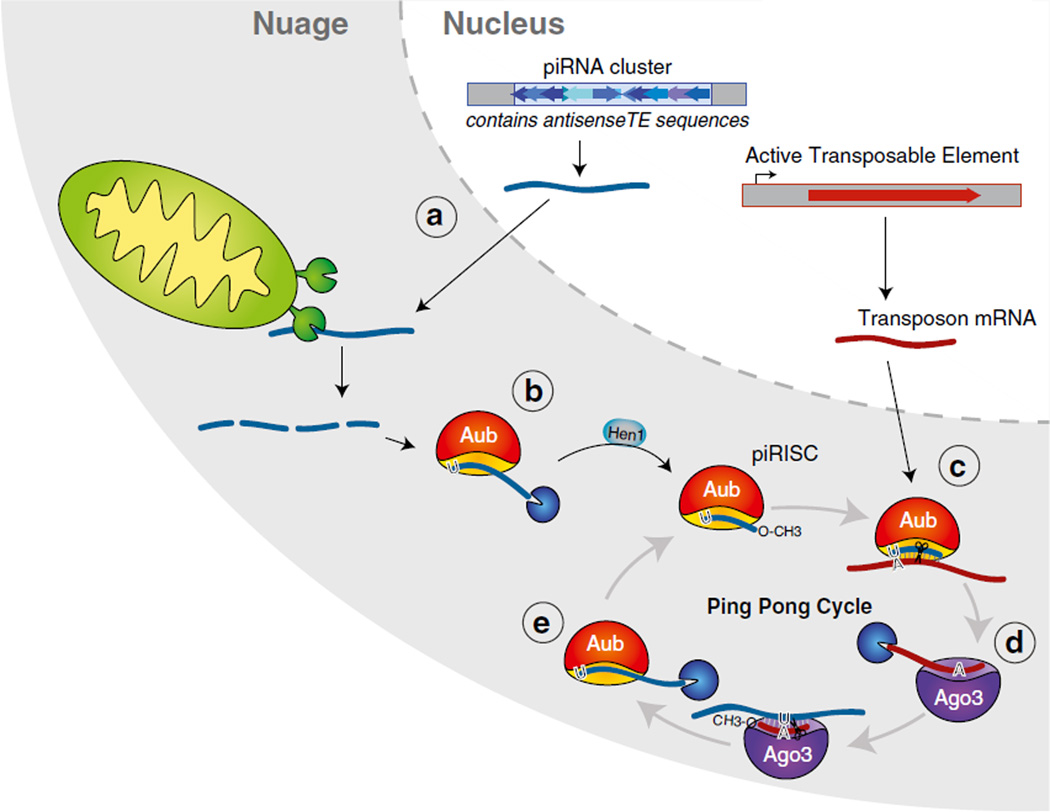
Fig. 4.3.
Biogenesis of piRNAs in flies begins in the nucleus and initiates the ping-ping cycle in the cytoplasm. (a) Precursor transcripts originating from piRNA clusters that contain anti-sense TE sequences are exported from the nucleus and processed into smaller fragments, possibly by the mitochondrial-associated endonuclease, Zucchini. (b) AUB binds precursor piRNA fragments with a preference for fragments that contain a 5′ terminal uracil (1U). Mature primary piRNAs are generated when an unknown 3′ ≥ 5′ exonuclease trims the precursor piRNA fragment bound to AUB and HEN1 catalyzes the 2-O methylation of the piRNA 3′ end. (c) When mRNA of active transposons is exported from the nucleus, AUB piRNA anneals to complementary sequences within the mRNA and cleaves the transcript. (d) The cleaved transcript is loaded into AGO3, followed by trimming and 2′ O-methylation of its 3′ end, to generate mature secondary piRNA. (e) AGO3 loaded with the secondary piRNA is believed to target and cleave newly generated precursor transcripts that are loaded into AUB to initiate a new round of Ping-Pong
Table 4.1.
| Gene | Synonyms | Tissue | Localization | Domains (M) | Function | Mouse | Ref. |
|---|---|---|---|---|---|---|---|
| Piwi proteins | |||||||
| Piwi | Piwi | Germ; Soma |
Nucleus | PAZ; PIWI | Argonaute; RISC effector |
Mili, Miwi, or Miwi2 |
Cox et al. (1998) and Lin and Spradling (1997) |
| Aubergine | Aub, Sting | Germ | Cyto.: nuage, pole plasm |
PAZ; PIWI | Argonaute; RISC effector |
Mili, Miwi, or Miwi2 |
Schmidt et al. (1999) and Harris and Macdonald (2001) |
| Argonaute-3 | Ago3 | Germ | Cyto.: cytoplasmic granules:germ granules in flies:nuagenuage |
PAZ; PIWI | Argonaute; RISC effector |
Mili, Miwi, or Miwi2 |
Li et al. (2009) |
| Nuclear factors | |||||||
| Rhino | Rhi; HP1D | Germ | Nucleus | Chromo; shadow | Chromatin factor | HP1a | Volpe et al. (2001) and Klattenhoff et al. (2009) |
| Eggless | Egg; SetDB1 | Germ | Nucleus | MDB, SET | H3K9me3 HMT | Rangan et al. (2011) | |
| Cutoff | Cuff | Germ | Nucleus | DOM3Z | DOM3Z | Pane et al. (2011) | |
| Deadlock | Del | Germ | Nucleus | – | Czech et al. (2013) | ||
| UAP56 | Hel25E | Germ; Soma |
Nucleus | DEXDc; Hel-C | RNA helicase | UAP56 | Zhang et al. (2012) |
| Asterix | Arx; CG3893 | Germ; Soma |
Nucleus | CHHC ZnF × 2 | HMT cofactor | Muerdter et al. (2013) | |
| Maelstrom | Mael | Germ | Nucleus and Cyto. | HMG | Mael | Findley et al. (2003), Sienski et al. (2012) and Sato et al. (2011) |
|
| Hsp90 | Hsp83 | Germ; Soma |
Nucleus and Cyto. | Heat-shock response; chaperone |
Hsp90 | Olivieri et al. (2012) and Zhang et al. (2012) |
|
| Cytoplasmic non-tudor proteins | |||||||
| Armitage | Armi | Germ; Soma |
Cyto. | SDE3 | Mov10L1 | Cook et al. (2004) | |
| Shutdown | Shu | Germ; Soma |
Cyto.: nuage | PPIASE; TPR | Co-chaperone; piRNA loading |
FKBP6 | Olivieri et al. (2012) and Preall et al. (2012) |
| Hen-1 | Hen1; PIMET | Germ; Soma |
Cyto. | MeT | RNA 2′ O-methyl transferase |
Saito et al. (2007) and Kirino and Mourelatos (2007) |
|
| Squash | Squ | Germ | Cyto.: Nuage | RNAase HII | RNA nuclease | Pane et al. (2007) | |
| Zucchini | Zuc | Germ; Soma |
Cyto.: Mito | HKD | RNA nuclease | PLD6 | Pane et al. (2007) and Ipsaro et al. (2012) |
| Minotaur | Mino; CG5508 | Germ; Soma |
Cyto.: Mito., ER, ring canals |
Glycerol-3-phosphate- O-acyltransferase (GPAT) |
Vagin et al. (2013) | ||
| GASZ | GASZ | Germ; Soma |
Cyto.: Mito. | Ankyrin × 3; SAM-2 |
GASZ |
Ma et al. (2009), Handler et al. (2013) and Czech et al. (2013) |
|
| Vasa | Vas | Germ | Cyto.: nuage, pole plasm |
DEXDc | RNA helicase | MVH | Liang et al. (1994) |
| Capsuléen |
Csul; dART5; PRMT5 |
Germ | Cytoplasm | PRMT5 | Argenine methyl transferase |
PRMT5 |
Anne and Mechler (2005) and Anne et al. (2007) |
| Valois | Vls | Germ | Cytoplasm | MEP50; WD × 4 | PRMT5 co-factor | Mep-50 | Cavey et al. (2004) |
| Polo | Cyto.: nuage | Kinase | Pek et al. (2012) | ||||
| Tudor proteins | |||||||
| Tudor | Tud | Germ | Cyto.: nuage, pole plasm |
Tud × 11 (Tud × 8) |
Tdrd6 |
Arkov et al. (2006) and Anne (2010) |
|
| Qin | Kumo | Germ | Cyto.: nuage | ZnF RING E3 Ligase; Tud × 5 |
E3 ligase | Tdrd4 | Annand et al. (2011) and Zhang et al. (2011) |
| Papi | Papi | Germ | Cyto.: nuage | KH × 2; Tud × 1 | Tdrd2 Siomi et al. (2009) | Liu et al. (2011) | |
| Tejas | Tej | Germ | Cyto.: nuage | LOTUS; Tud × 1 | Tdrd5 | Patil et al. (2010) | |
| CG8920 | Germ | LOTUS; DSRM; LOTUS; Tud × 3 |
Tdrd7 | Handler et al. (2011) | |||
| CG9925 | ZnF-MYND; Tud × 3 |
Tdrd1 | Handler et al. (2011) | ||||
| Spndle-E |
SpnE; Homeless; Hls |
Germ | Cyto.: nuage | DEXDc; Hel-C; HA2; OB; RRM; ZnF; Tud × 1 |
RNA helicase | Tdrd9 | Gillespie and Berg (1995) |
| Krimper |
Krimp; Montecristo |
Germ | Cyto.: nuage | Tud × 1 | Lim and Kai (2007) | ||
| Yb | fs(1)Yb | Soma | Cyto. | Hel-C like; DEAD; Hel-C; ZnF; Tud × 1 |
Szakmary et al. (2009) | ||
| Sister of Yb | SoYb | Germ; Soma |
Cyto.: nuage | Tud × 2; DEAD; Hel-C; ZnF; CS |
Tdrd12 | Handler et al. (2011) | |
| Brother of Yb | BoYb | Germ | Cyto.: nuage | Tud × 1; DEAD; Hel-C; ZnF; CS |
Handler et al. (2011) | ||
| Vreteno | Vret | Germ; Soma |
Cyto.: nuage | ZnF-MYND; RRM; Tud × 2 |
Zamparelli et al. (2011) and Handler et al. (2011) |
||
Primary piRNAs show a strong bias for a uridine as their 5′ terminal nucleotide. The step at which this bias is introduced is not clear, as there are two conceivable scenarios. First, cleavage of precursor transcripts could be random and the 5′ binding pocket of PIWI proteins could have a steric preference for binding piRNAs with a 5′ terminal 1U; or second, the nuclease conducting the first cleavage on precursor cluster transcripts could preferably cleave upstream of uridines, thereby creating the observed bias.
Once loaded into PIWI proteins, the piRNA precursor is trimmed at its 3′ end by an unknown 3′≥5′ exonuclease. This nuclease is tentatively named “Trimmer” (Fig. 4.3b). The final length of the piRNA is determined by the piRNA-binding pocket of the PIWI protein: PIWI associated piRNAs are 25 nt long, whereas piRNAs associated with AUB or AGO3 are 24 nt or 23 nt long, respectively. In mouse their length is slightly longer (25–31 nt) with an average of 26 nt, 28 nt and 30 nt for MILI, MIWI2 and MIWI-bound piRNAs respectively (Aravin et al. 2006, 2008; Grivna et al. 2006). However, piRNAs can vary in length by one to two nucleotides, possibly as a consequence of imprecise trimming. Finally, after trimming, piRNAs acquire a characteristic 2′ methyl modification at their 3′ end that is introduced by the piRNA methyl-transferase HEN1 and leads to stabilization of the small RNA (Kawaoka et al. 2011).
Overall, while the exact biochemistry and the involved factors are still not fully known, piRNA biogenesis consists of multiple consecutive steps from transcription of precursors, through precursor cleavage and loading, to trimming and 2′ O-methylation. Defects in any of these steps leads to diminished piRNAs, upregulation of transposons and sterility.
4.3 Nuclear and Cytoplasmic Function of the piRNA Pathway
Transposons are selfish genetic elements that can be classified into DNA or retrotransposons based on the mechanism by which they transpose in the genome. DNA transposons mobilize by a ‘cut and paste’ mechanism that is accomplished with the help of an encoded transposase protein. Retrotransposons propagate by reverse transcription of an RNA intermediate. Here too, the required enzymes are encoded by the transposable elements. Another class of TE is the semi-autonomous sequences, such as Alu repeats, that require the protein machinery of other transposons for their replication. In either case the transposon needs to be transcribed as an mRNA because it encodes the enzymes necessary for transposition, or is also required as a template for conversion to DNA by reverse transcription. Accordingly, transposition can be regulated on multiple levels, from the efficiency of transcription, to transcript stability and rate of translation of required factors, or by interfering with the process of re-integration into the genome. The piRNA pathway seems to target two steps that are required for all transposons: In the nucleus, piRNAs are implicated in the regulation of chromatin structure that can affect transcription of targeted loci. In the cytoplasm, the piRNA pathway directly targets and destroys RNAs of transposons that escaped transcriptional silencing. In this section we will describe the details of these two levels of defence against transposable elements.
4.3.1 The Role of piRNAs in Regulating Chromatin Structure
Transcriptional regulation of a specific locus is influenced by its contextual chromatin architecture. In mammals, regulation is achieved at two levels: by methylation of DNA and by modifications of histone proteins or even incorporation of special histone variants into nucleosomes. In Drosophila melanogaster, histone modifications and histone variants are mainly responsible for defining the properties of chromatin. In both flies and mice, one of the three PIWI proteins localizes to the nucleus, suggesting a nuclear role for the piRNA pathway possibly through transcriptional repression of targets (Cox et al. 1998; Aravin et al. 2008). Such a mechanism is further strengthened by the observation that in flies PIWI shows a banding pattern on polytene chromosomes of nurse cells, suggesting a direct interaction with chromatin (Le Thomas et al. 2013).
In mouse, DNA methylation plays an essential role in silencing of transposable elements (TEs). In mouse male germ cells, this silencing is established in a narrow developmental window during embryonic germ cell development, following global genome de-methylation. This is a critical point in the fate of spermatogenic cells, since the failure to methylate sequences of retrotransposons scattered throughout the genome leads to their activation and subsequent meiotic failure and sterility (Bourc’his and Bestor 2004). Several lines of evidence indicate that piRNAs are responsible for directing DNA methylation to their genomic targets. First, several proteins involved in the piRNA pathway have been described to localize to the nucleus: MIWI2, TDRD9, and MAEL (Aravin et al. 2008, 2009; Shoji et al. 2009; Soper et al. 2008). While TDRD9 and MAEL are expressed throughout male germ cell development, MIWI2 is expressed in a narrow developmental time window exactly overlapping the time of de novo DNA methylation in the embryonic spermatocyte. Additionally, genetic data implicate piRNAs in establishing de novo DNA methylation patterns in the mouse male germline on regulatory regions of TEs (Aravin et al. 2008; Carmell et al. 2007; Kuramochi-Miyagawa et al. 2008). DNMT3L is an accessory factor for the de novo DNA methyl-transferases DNMT3A and DNMT3B, and is essential for de novo DNA methylation. The defects observed in DNMT3L knockout animals are strikingly similar to those observed in animals deficient in many piRNA pathway components, including the two PIWI proteins, MILI and MIWI2: all result in meiotic arrest of spermatogenesis and apoptosis of germ cells. Furthermore, methylation patterns were not re-established on retrotransposon sequences in mice lacking MILI or MIWI2 (Kuramochi-Miyagawa et al. 2008). In contrast, piRNAs are still produced in DNMT3L mutants (Aravin et al. 2008). These observations indicate that the piRNA pathway directs DNA methylation of TEs and functions upstream of the methylation machinery.
A second PIWI protein, MILI is also expressed in the embryonic stage of spermatogenesis. While it localizes to the cytoplasm, its expression is necessary for piRNA loading and nuclear localization of MIWI2. The phenotype of Mili mutants is similar to that of Miwi2 suggesting that piRNAs serve as sequence-specific guides that direct MIWI2 to sites where DNA methylation is established (Kuramochi-Miyagawa et al. 2001, 2004, 2008; Aravin et al. 2008). Biochemically, this process remains unexplored due to the restriction of de novo DNA methylation to a small number of germ cells in a narrow developmental window during embryogenesis. It is not clear whether MIWI2 associates directly with chromatin and it seems not to interact directly with DNA methyltransferases. In fruit flies, the piRNA pathway participates in deposition of repressive histone marks on TEs (see below). It remains to be determined whether this mechanism is conserved in mice. If so, it will be interesting to see if histone modifications lead to subsequent DNA methylation, or vice versa (Fig. 4.4).
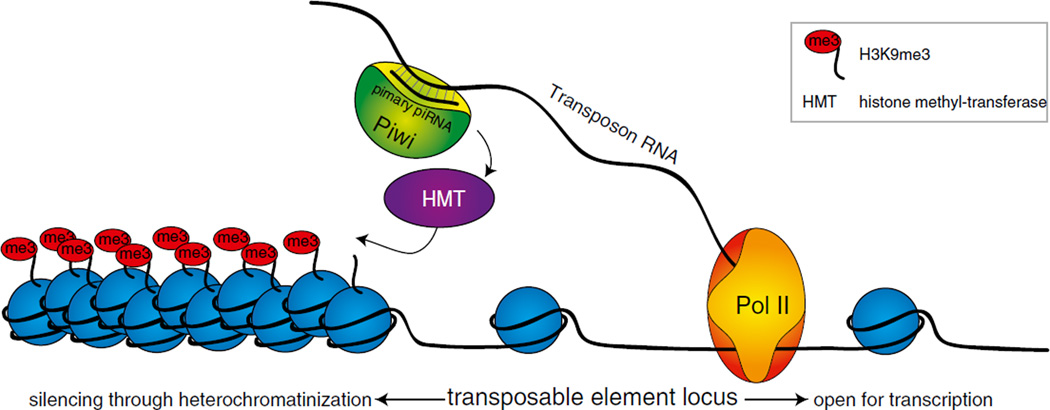
Fig. 4.4.
The nuclear function of the piRNA pathway in flies. PIWI loaded with cluster-derived primary piRNAs enters the nucleus and scans nascent transcripts for complementarity to its piRNA. If a transposon RNA transcript is encountered, PIWI is believed to recruit chromatin factors such as histone methyl-transferases (HMT) to deposit repressive H3K9-methyl marks. The modifications induced by PIWI binding create sites for the assembly of additional factors that repress transcription of transposon loci
Male germ cells also establish new methylation patterns at imprinted loci. Interestingly, the piRNA pathway is involved in methylation of at least one imprinted locus, Rasgrf1 (Watanabe et al. 2011a). This process depends on a solo-LTR transposon located in the Rasgrf1 locus and might be a consequence of piRNA-mediated retrotransposon methylation.
In contrast to mammals, DNA methylation in Drosophila does not play a major role in the regulation of chromatin structure and gene expression. In fruit flies it is believed that the piRNA pathway induces transcriptional repression by initiating deposition of repressive histone marks. Indeed, multiple studies have shown that the piRNA pathway transcriptionally represses at least some transposons. In cell culture experiments, deletion of PIWI’s nuclear localization signal leads to failure of transposon silencing, even though PIWI proteins are loaded with piRNAs in the cytoplasm (Saito et al. 2009a). Similarly, in flies the deletion of the N-terminus of PIWI leads to its delocalization from the nucleus and a slight increase in active histone marks (di-methylation of H3K79 and H3K4) and a decrease of repressive marks (di-/tri-methylation of H3K9) over several transposons (Klenov et al. 2007). Additionally, an increase in transcription and accumulation of several telomeric retrotransposons was observed in Drosophila ovaries upon mutation of piRNA pathway components (Chambeyron et al. 2008; Shpiz et al. 2009, 2011). The telomeric transposons, which had increased transcription showed slight changes in their chromatin marks (Shpiz et al. 2011). These observations all indirectly pointed towards an involvement of PIWI in transcriptional regulation of transposable elements. Recently, three genome-wide studies confirmed this assumption: knockdown of Piwi leads to transcriptional derepression of a significant fraction of TEs both in cell culture and in ovaries as assessed by Pol II ChIP-seq and by GRO-seq (Le Thomas et al. 2013; Sienski et al. 2012; Rozhkov et al. 2013). Upon Piwi knockdown transcript levels of transposable elements increase significantly, indicating that PIWI primarily, if not solely, represses transposons at the transcriptional level.
While it seems likely that transcriptional silencing of transposable elements is achieved through establishment of a repressive chromatin state, this correlation between repressive chromatin marks and silencing has not been shown directly yet. Likewise, factors involved in mediating PIWI’s repressive function are not known. Recently, two factors have been proposed to be involved in inducing transcriptional silencing. Maelstom (MAEL) is a conserved factor that has been implicated in TE silencing in both mouse and flies (Aravin et al. 2009; Sienski et al. 2012; Soper et al. 2008). Knockdown of Mael in flies leads to increased Pol II occupancy over TEs similar to changes observed upon Piwi knockdown (Sienski et al. 2012). Interestingly, while Piwi knockdown resulted in strong reduction of H3K9me3 signal at and downstream of target sequences Mael knockdown resulted in increased spreading and only a modest reduction in H3K9me3 signal, indicating that MAEL does not act in establishing the repressive H3K9me3 mark over targets. Another recent study identified Asterix (CG3893) as a factor required for PIWI-mediated establishment of H3K9me3 mark over at least a subset of TEs (Muerdter et al. 2013).
A study using a cell culture model showed that PIWI silences a significant number of novel TE insertions through tri-methylation of H3K9 (Sienski et al. 2012). This silencing mark does not only cover the site of the novel TE insertion, but spreads up to 15 kb downstream in the direction of transcription. This observation indicates that transcription plays a role in establishing repressive chromatin. The spreading of repressive marks upon insertion of a transposable element can even lead to repression of proximal protein coding genes. In fly, however, similar new insertions in proximity to protein coding genes were not observed (Le Thomas et al. 2013). It is conceivable, that while in cultured cells the silencing of nearby genes is tolerated and can be detected, in a living organism there is a strong selective pressure against insertions that would compromise functional gene expression.
The piRNA sequences bound to PIWI identify the sequences that are targeted for PIWI-mediated transcriptional repression. This is in agreement with the observation that host genes are generally unaffected by the pathway and that PIWI requires piRNAs to repress TEs (Le Thomas et al. 2013). Additionally, PIWI loaded with artificial sequences mapping to the lacZ transgene leads to silencing of lacZ-expressing loci in vivo. While the mechanism of target recognition remains unresolved, it is likely that piRNAs recognize nascent transcript targets through sequence complementarity. This is supported by the observation that Piwi knockdown results in decreased H3K9 tri-methylation of the genomic environment of active transposons, while non-transcribed fragments of transposable elements throughout the genome remained unaffected (Sienski et al. 2012).
Transcriptional repression by PIWI occurs in both germ cells and follicular cells of Drosophila ovaries. Follicular cells are the somatic support cells in fly ovaries, and while they are not germline cells, they do express a uniquely tailored version of the piRNA pathway. Out of the three PIWI proteins in Drosophila, only PIWI is expressed in follicular cells. In these cells it specifically loads with piRNAs generated from the unidirectional cluster, Flamenco (Malone et al. 2009). The Flamenco cluster appears to primarily target the LTR retrotransposon, Gypsy, whose expression can lead to the formation of viral particles that might infect the germ line (Song et al. 1997). Therefore, transcriptional repression of Gypsy loci by PIWI in follicular cells is crucial for germline survival.
In summary, while little is known about the mechanism of piRNA-mediated transcriptional silencing, it is apparent that both in mouse and in Drosophila, PIWI proteins play an essential role in inhibiting TE transcription and establishing a repressive genomic environment.
4.3.2 Ping-Pong: The Cytoplasmic Function of the piRNA Pathway
The beauty and power of the piRNA pathway lies in its ability to store information of previous TE invasions within piRNA clusters and to specifically respond to TE activation by distinctively amplifying sequences complementary to active elements. The latter function is achieved through the interplay of two cytoplasmic PIWI proteins – in Drosophila, Aubergine and Argonaute 3 – in a process termed the Ping-Pong cycle. Since these two proteins are only expressed in the germline, the Ping-Pong cycle is also restricted to germ cells (Brennecke et al. 2007).
In Drosophila, Aubergine (AUB) and Argonaute-3 (AGO3) act as partners in the defence against active transposable elements, with AUB binding to piRNAs mainly coming from piRNA cluster transcripts and AGO3 enriched in piRNAs from transposon mRNAs. Unlike PIWI, both AUB and AGO3 can catalytically cleave RNA targets. At the beginning of the pathway, AUB loads with primary piRNA sequences that are derived from piRNA clusters and are antisense to TE transcripts (see Sect. 2.2). These primary piRNAs guide AUB to scavenge the cytoplasm for complementary transcripts. When a TE mRNA with sequence complementarity to an AUB-bound piRNA is identified, AUB cleaves the target RNA ten nucleotides downstream of the piRNA 5′ end. This cleavage of transposon mRNA serves two purposes: first, it destroys the transposon transcript and second, it generates the 5′ end of a new piRNA precursor (Fig. 4.3c). This new precursor gets incorporated into AGO3 and is processed into a so-called secondary piRNA (Fig. 4.3d). Due to the fact that AGO3-bound piRNAs are sense to the transposable element, these piRNAs can promote the production of a new round of cluster-derived piRNAs by recognizing complementary cluster transcripts (which then get cleaved and incorporated into AUB) (Fig. 4.3e). As the cycle can only function if transposon mRNA is present, the Ping-Pong cycle shapes a specific piRNA repertoire against active TEs.
Since AUB-bound piRNAs have a strong bias for uracil at their 5′ ends, the piRNAs that are loaded into AGO3 have a bias for adenosine at position 10 (10A bias) and are complementary to the AUB-bound piRNAs over a 10-nucleotide stretch. This relationship between AUB piRNAs and AGO3 piRNAs is termed the Ping-Pong signature.
The precise mechanism of how AUB and AGO3 are coordinated in the Ping-Pong cycle is not yet fully understood. Protein components that are implicated in the Ping-Pong cycle include not only AUB and AGO3, but also the Tudor domain-containing proteins QIN, Krimper (KRIMP), and Spindle-E (SPNE) in addition to the essential germline helicase, VASA. SPNE and VASA are both putative DEAD-box RNA helicases, and QIN contains a RING E3 ligase domain with unknown function (Malone et al. 2009; Li et al. 2009; Anand and Kai 2012; Zhang et al. 2011). While the exact role of these additional proteins is not known, mutating them leads to aberrations in the Ping-Pong cycle and sterility. For example, in Qin mutant ovaries, Ping-Pong signatures between AUB and AGO3 are decreased and instead a homotypic Ping-Pong signature (i.e. Ping-Pong between sequences associated with the same PIWI protein) and AUB appears to predominate (Zhang et al. 2011). The biological function of homotypic Ping-Pong, if any, is currently unknown.
In spermatogenic cells of mouse embryos, Ping-Pong occurs between the two PIWI proteins MILI and MIWI2. MILI binds primary piRNAs and MIWI2 is bound to secondary piRNAs. A difference to Ping-Pong in Drosophila is that the MILI-bound primary piRNAs are mainly in sense orientation relative to TE coding sequences, which suggests that in mouse, active transposable elements initiate the cycle (Aravin et al. 2008). In embryonic mouse germline cells the PIWI proteins MILI and MIWI2 participate also in homotypic Ping-Pong.
4.3.3 Suppressor of Stellate Is a Specialized piRNA Mechanism in Drosophila Testis
Beyond transposon control, the piRNA pathway seems to have been adopted for alternative functions: Drosophila testes employ a unique piRNA pathway mechanism whose primary role is to provide “quality control” during spermatogenesis. The Stellate locus on chromosome X is comprised of a tandemly repeated gene that encodes a protein, Stellate, whose hyper-expression is responsible for the formation of large crystalline needle-like fibers of Stellate protein that are toxic to the cell. Due to their high copy number, unchecked expression of Stellate kills primary spermatocytes and results in male sterility. The antidote to Stellate toxicity is provided by the Crystal locus on chromosome Y, which encodes another set of tandem repeats named Suppressor-of-Stellate, Su(Ste) (Aravin et al. 2001; Bozzetti et al. 1995). Interestingly, the Su(Ste) gene shares 90 % homology to Stellate and produces both sense and antisense transcripts. The antisense transcripts play an integral role in silencing Stellate mRNA and the mechanism of silencing requires many of the same components as the piRNA pathway in ovaries.
Repression of Stellate in testes requires AUB and AGO3 while PIWI appears to be dispensable (Li et al. 2009; Aravin et al. 2001; Schmidt et al. 1999; Vagin et al. 2006). Additionally, the testes of mutants for piRNA pathway components SPNE, QIN, ARMI, TEJ, and VREt also express Stellate and/or appear to have meiotic abnormalities (Zhang et al. 2011; Vagin et al. 2006; Aravin et al. 2004; Handler et al. 2011; Patil and Kai 2010; Stapleton et al. 2001). The piRNA pathway in testes also directs TE repression through both AUB and PIWI, with PIWI being required for fertility in males (Lin and Spradling 1997; Vagin et al. 2006). It is unlikely that Ping-Pong plays a significant role in testis biology, since AGO3 mutant males are sub-fertile, even though crystals of Stellate do accumulate within spermatocytes, and male germ stem cell maintenance is only partially defective (Li et al. 2009; Kibanov et al. 2011). Many pathway components localize to perinuclear areas of spermatocytes forming piRNA nuage giant bodies, or piNG-bodies (Kibanov et al. 2011). These components likely have a similar structure to nuage in ovary, as discussed below.
The implied biological function of Stellate and Su(Ste) remains ambiguous. Certainly, this system does appear to ensure that spermatocytes develop with an intact piRNA pathway, in addition to preventing Y chromosome aneuploidy during spermatogenesis. Currently, a homologous “quality assurance” mechanism is not known to exist in the female germline of the fly, or in mouse. The existence of such alternative functions of the piRNA pathway raises the question whether it has unidentified functions in the germline or in other tissues. Although expression of the core components of the piRNA pathway – PIWI proteins – seems to be restricted to the germline, it can not be excluded that specified cells do express and employ the piRNA pathway for yet uncharacterized functions beyond transposon control.
4.4 Cytoplasmic Granules: Components, Structure and Function
4.4.1 Introduction to Germ Granules
Molecules involved in the same cellular process often assemble into granules – membraneless subcellular compartments – in order to increase the local concentration of factors and enhance the efficiency and specificity of processes. Germ granules are conserved germ cell components present in most if not all sexually reproducing metazoans, and are essential in ensuring the reproductive potential of the individual (Al-Mukhtar and Webb 1971; Eddy and Ito 1971; Mahowald 1968).
Germ granules were discovered more than a century ago, as darkly staining structures that could be traced from gametes of one generation into germ cells of the next in the fly (Hegner 1912). The importance of these granules for embryonic germ cell development and fertility was highlighted with experiments showing that their destruction by UV irradiation resulted in the so-called grandchild-less phenotype, i.e. sterility of the next generation (Hegner 1911; Hathaway and Selman 1961). Furthermore, transplanting germ plasm, which contains germ granules from a healthy donor strain into oocytes that had previously been treated with UV irradiation could rescue the fertility defect (Okada et al. 1974).
Detailed analysis of the constituents of germ granules reveals interesting relationships. In both mouse and fly, germ granules are frequently in close association with mitochondria and their constituent proteins have functions that are linked to the piRNA pathway (Aravin and Chan 2011). Interestingly, several mitochondrial proteins including Zucchini (ZUC) and GASZ are critical piRNA pathway components. Furthermore, many components that localize to germ granules contain Tudor domains (Ponting 1997). Tudor domains bind symmetrically di-methylated arginine (sDMA) residues, such as ones present in the N-terminal region of PIWI proteins. It is believed that Tudor domain proteins might serve as a scaffold to bring together PIWI proteins and other components of the piRNA pathway in germ granules.
4.4.2 Germ Granules in Flies
In flies, germ granules can be identified in almost all stages of germ cell development. Whether in testes or ovaries, granules have various names depending on their protein constituents, localization and stage of expression. Only recently has it been uncovered that many components of the granules operate to ensure successful transposon repression.
Nuage Are Composed of Many piRNA Pathway Components in Ovarian Nurse Cells
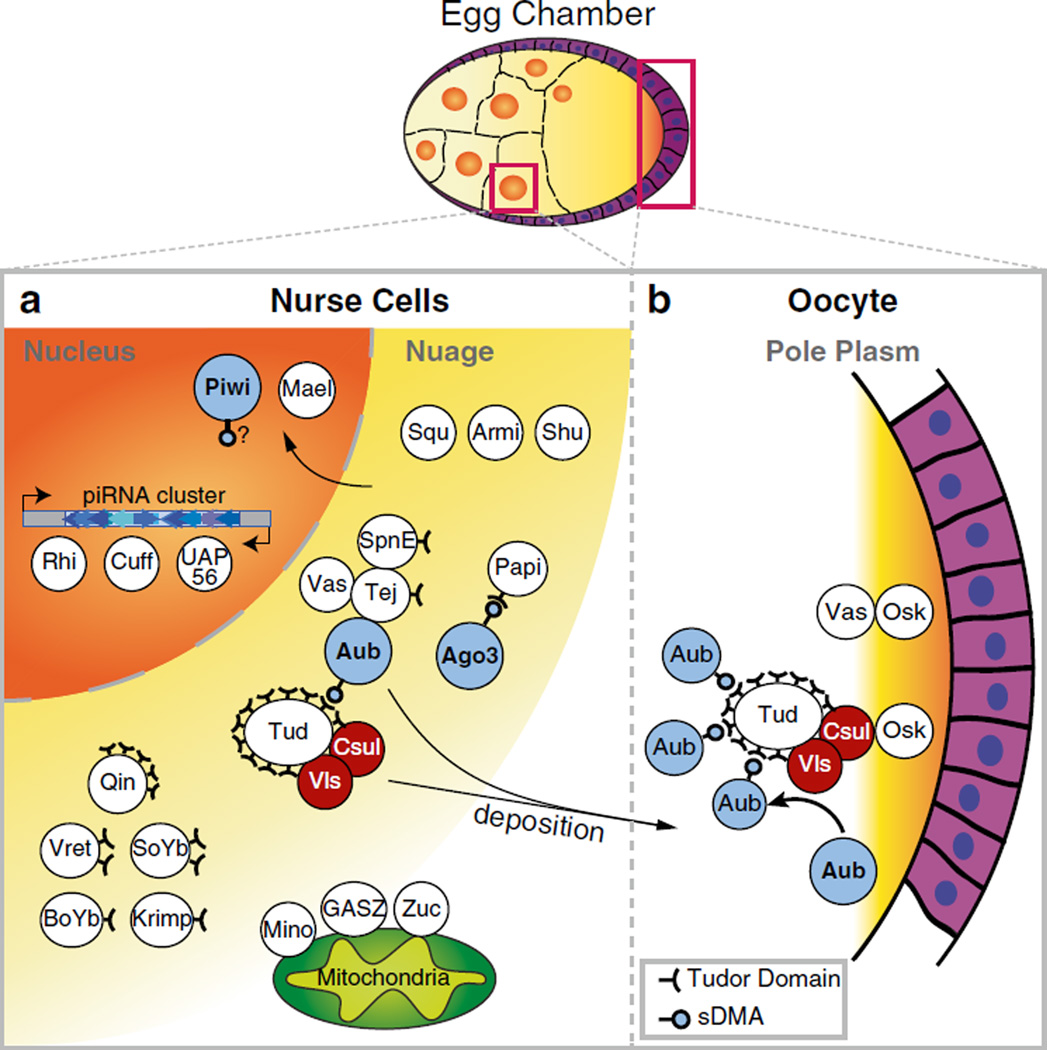
In nurse cells of the fruit fly ovary, nebulous perinuclear structures termed ‘nuage’ (after the French word for ‘cloud’) are visible in negative stained electron micrographs (Eddy and Ito 1971). The constituents of these granules include many cytoplasmic piRNA pathway components, including AUB and AGO3 (Fig. 4.5a; Table 4.1). The cytoplasmic functions of the piRNA pathway, such as piRNA loading and the Ping-Pong mechanism are believed to occur within these granules. Delocalization of any of the piRNA pathway components from nuage leads to impairment of the pathway, indicating the significance of these granules in proper TE silencing.
Fig. 4.5.
Many protein factors make up nuage granules in Drosophila nurse cells and some accumulate at the pole plasm of the developing oocyte. (a) Nuage granules surround the nucleus of nurse cells and consist of many factors involved in the piRNA pathway. Nuage also shows close proximity to mitochondria, which contain piRNA pathway components (ZUC, GASZ) in their outer membrane. Nuage integrity depends on nuclear piRNA biogenesis factors (such as RHI, CUFF, UAP56) and a number of nuage components of unknown function. Many nuage components contain Tudor domains, which are receptors for symmetrically di-methylated arginines (sDMA). These Tudor proteins are believed to form a scaffold for the factors involved in the pathway. AUB and AGO3 contain sDMA motifs at their N-terminal regions and can interact with Tudor proteins. The enzyme responsible for methylation, PRMT5, consisting of Capsuléen (CSUL) and Valois (VLS), is also present in nuage and associates with Tudor. Some nuclear proteins, such as PIWI and MAEL are believed to transiently visit the nuage and this is required for their function (Klenov et al. 2011; Saito et al. 2009b). Known interactions are indicated. (b) At later stages of oogenesis, nurse cells deposit their cytoplasmic contents, including piRNA pathway components, into the oocyte. Some components, including sDMA-modified AUB, accumulate at the posterior end of oocytes to form the pole plasm. The material that accumulates at the pole plasm becomes the cytoplasmic material of pole cells, which give rise to primordial germ cells of the embryo
The composition and requirements of nuage granule assembly is not yet fully understood. However, some general trends for nuage assembly have been worked out. First, the components of nuage granules seem to assemble in a fashion dependent on piRNA biogenesis. This is made evident by the fact that components including AUB and AGO3, which would normally localize to nuage, are cytoplasmically dispersed in ovaries mutant for piRNA biogenesis factors such as cluster-associated factors Rhino and Cutoff and the mRNA export protein UAP56 (Klattenhoff et al. 2009; Pane et al. 2011; Zhang et al. 2012). Second, the assembly of proteins that make up nuage granules appears to be hierarchical. For example, it is apparent from mapping pair-wise genetic interactions that some protein components only localize to nuage if other factors have already ‘built’ on the nuage complex. Some proteins may not localize to nuage in the absence of one or more upstream protein components, while the localization of these upstream components is unperturbed by mutations in downstream components (Malone et al. 2009; Anand and Kai 2012; Patil and Kai 2010; Lim and Kai 2007).
Lastly, arginine-rich motifs typically found near the N-termini of PIWI proteins are modified to symmetrical di-methyl arginines (sDMA) by the PRMT5 methylosome complex (Kirino et al. 2009, 2010a; Nishida et al. 2009; Anne and Mechler 2005). Methylated arginines can bind Tudor domains, which are a prevalent domain in piRNA pathway components (Table 4.1) (Kirino et al. 2010a, b; Liu et al. 2010b). It is believed that Tudor domain proteins might form a scaffold in nuage that helps bring together functional components of the pathway. While the role of arginine methylation in the function of the piRNA pathway is not entirely clear, AUB requires sDMA modifications to accumulate at the pole plasm during oogenesis indicating its requirement for proper localization. Additionally, sDMA modifications on AGO3 appear to be required for interactions with another component, PAPI (Liu et al. 2011). Further information regarding the ‘interactome’ of nuage is currently lacking, but will likely be required to understand how nuage components operate together to execute their biological functions.
The Pole Plasm: Germ Granules in the Oocyte
The pole plasm is defined by an accumulation of maternally deposited RNA, mitochondria and piRNA-associated granules at the posterior pole of the oocyte. Much of the material that adheres to the pole plasm will be inherited by pole cells, which are among the first cells to cellularize during embryogenesis (Hay et al. 1988). Pole cells migrate during gastrulation to the mid-gut and give rise to primordial germ cells of the developing larvae. AUB is among the hand-full of piRNA pathway proteins that localize to the pole plasm (Fig. 4.5b; Table 4.1). The enrichment of AUB at the pole plasm depends on sDMA modifications that allow it to bind Tudor, which itself is anchored to the pole plasm by the PRMT5 methylosome complex and the body-patterning-associated factor Oskar (Anne and Mechler 2005; Anne 2010; Arkov et al. 2006; Anne et al. 2007; Thomson and Lasko 2004; Kugler and Lasko 2009; Liang et al. 1994).
The accumulation of AUB at the pole plasm by a devoted mechanism, i.e. post-translational sDMA modification, suggests that AUB is required for the biology of primordial germ cells. Whether the role of AUB in primordial germ cells is solely palliative in destroying transposable element transcripts, as in the context of Ping-Pong, or whether its role in primordial germ cells serves a deeper purpose, one required for the establishment of the primordial gonad, remains to be understood. The idea that PIWI or AGO3 might also participate in germ cell development has been proposed, although direct evidence for their involvement in primordial gonads is still sparse (Nishida et al. 2009; Malone et al. 2009; Megosh et al. 2006).
4.4.3 Germ Granules in Mouse
Like in flies, mouse germ cells contain granular electron dense structures in their cytoplasm termed pi-bodies (Aravin et al. 2009). These structures were first cytologically described in rat germ cells and termed ‘intramitochondrial cement’ (IMC) (Eddy 1974). Studies using immunofluorescence microscopy showed that mouse piRNA pathway proteins, including MILI, MIWI2, MVH, and several Tudor proteins, accumulate in pibodies, which localize in the vicinity of mitochondria (Aravin et al. 2008, 2009; Shoji et al. 2009; Kuramochi-Miyagawa et al. 2010).
The functional importance of the integrity of pi-bodies and their spatial proximity to mitochondria was highlighted in studies that showed involvement of a mitochondrial protein mitoPLD in the piRNA pathway (Huang et al. 2011b; Watanabe et al. 2011b). MitoPLD is a mouse homolog of Drosophila protein ZUC, and was known to localize to the outer mitochondrial membrane and have a role in lipid signalling. In mitoPLD mutant, IMC formation is impaired, resulting in loss of localization of piRNA path-way components to the associated granules. As a consequence, TEs are upregulated, their regulatory regions lose DNA methylation, and the mice are sterile. A structural protein localized to IMC, GASZ, is also a conserved component of the piRNA pathway and its mutation leads to sterility. Loss of GASZ is detrimental for the integrity of the granules, resulting in loss of MILI and mislocalization of MVH and TDRD1 in embryonic testes (Ma et al. 2009).
The ‘second arm’ of the piRNA pathway, MIWI2 and its interacting partners TDRD9 and MAEL, occupy another discreet entity in the cytoplasm, partially co-localizing with the MILI-containing granules (Aravin et al. 2008, 2009; Shoji et al. 2009). These granules, which are larger and fewer than pi-bodies, also contain components of the ubiquitous P-bodies (such as GW182, XRN1 and DCP1A) and were termed piP-bodies. The integrity of these granules depends on the integrity of pi-bodies. Any disruption of the MILI-containing intramitochondrial cement results in loss of localization of MIWI2 and TDRD9 to piP-bodies and to the nucleus, downregulation of secondary piRNAs, and consequent TE overexpression.
Taken together, these data suggest that correct structure of these compartments and their spatial proximity heavily influences the efficiency of TE silencing, making them a pivotal subcellular compartment of embryonic male germ cells.
In postnatal germ cells, MILI, MIWI and their interacting proteins, such as TDRD6 and MAEL, are found in the chromatoid body, an entity thought to resemble the P-body of somatic cells. Chromatoid bodies are the proposed site of RNA storage and processing, but no specific function has been assigned to it yet. Given the localization of piRNA pathway components in this subcellular compartment, it has been proposed that pachytene piRNAs, in complex with MIWI and MILI, exert their function in the chromatoid body. This assumption still awaits experimental confirmation (Meikar et al. 2011).
4.5 Systems Similar to the piRNA Pathway in Other Organisms
A number of organisms contain small RNA pathways that share function with the germline-specific piRNA pathway of animals: protecting the host against harmful repetitive elements such as transposons by repressing their expression. In fact, small RNA pathways in yeast and in some single-cellular organisms are in many ways more similar to the germline-specific piRNA pathway than the ubiquitous miRNA or siRNA pathways of metazoans. In yeast and Tetrahymena the primary target seems to be transcriptional regulation of targets rather than the cytoplasmic posttran-scriptional control, with the extreme being elimination of undesired genomic regions. Transcriptional control is also the main mechanism of action for the siRNA pathway in plants, which is responsible for DNA-methylation and repression of TE transcription. Interestingly, in plants the germlines have an intriguing mechanism of small RNA-mediated TE silencing that seems to work non-cell autonomously. Finally, in C. elegans a system homologous to the piRNA pathway in flies and mouse closely interacts with the siRNA pathway to repress TEs in the germ-line. Here we briefly discuss some interesting aspects of small RNA pathways in single-celled organisms and in the germline of plants and C. elegans highlighting the similarities and differences to the Drosophila and mouse piRNA pathway.
4.5.1 RNAi-Dependent Heterochromatin Formation in Fission Yeast
In Schizosaccharomyces pombe, RNAi functions to maintain transcriptional silencing at constitutively heterochromatic regions of the genome. The importance of the RNA pathway is underlined in the observation that defects in RNAi pathway genes and pericentromeric heterochromatin formation lead to aneuploidy and genomic instability that result from defective kinetochore assembly and chromosome segregation during mitosis (reviewed in Reyes-Turcu and Grewal 2012).
While heterochromatic DNA is mostly transcriptionally silent, during S-phase transcription of centromeric loci, including pericentromeric repeats, is enhanced, while the HP1 homolog, SWI6 is removed (Kato et al. 2005; Zofall and Grewal 2006). Interestingly, this transcription is required for the recruitment of the silencing machinery in two ways: first, transcripts are converted into a double-stranded RNA by the RNA-dependent-RNA polymerase complex (RDRC), which are subsequently processed by Dicer (DCR1) and loaded into Argonaute 1 (AGO1). Secondly, the siRNA loaded into AGO1 directs it to complementary nascent transcripts originating from pericentromeric loci. The AGO1 complex recruits the histone methyltransferase CLR4, the homologue of SU(VAR)3–9, that installs repressive H3K9 trimethylation marks over the locus. The complex containing CLR4 in turn recruits the RNA-dependent RNA polymerase thereby ensuring production of more siRNAs from the locus (Zhang et al. 2008). This leads to a positive feedback loop that results in strong repression of the locus throughout other stages of the cell cycle.
The yeast system is similar to the siRNA pathway in higher eukaryotes in terms of small RNA biogenesis. However, the nature of the targets, namely repetitive elements, and the mechanism of action, that is, transcriptional silencing through establishment of a repressive chromatin over target loci strongly resemble the mechanism employed by the piRNA pathway in the Drosophila germline.
4.5.2 Transcriptional and Posttranscriptional Control of Repetitive Elements in Neurospora
Filamentous fungus, Neurospora crassa, has virtually no known active transposons. The presumed reason for this is the existence of three different mechanisms that defend the Neurospora genome from repetitive sequences. One, known as ‘quelling’, acts in the vegetative phase, whereas repeat-induced point mutation (RIP) and meiotic silencing of unpaired DNA (MSUD) operate upon fertilization, in pre-meiotic and meiotic stages, respectively (reviewed in Caterina et al. 2006). Unlike RIP, which does not employ RNA silencing components, Quelling and MSUD are post-transcriptional silencing mechanisms that rely on homologs of known RNA silencing factors Dicer, Argonaute, and RNA-directed RNA polymerase (RdRP) (Romano and Macino 1992; Shiu et al. 2001). During vegetative growth, presence of arrays of repetitive sequences, such as transgenes, results in transcription of so-called aberrant RNA. Aberrant RNA is converted into double stranded form by the RdRP QDE1, and processed into small RNAs by a Dicer-like protein (Cogoni and Macino 1999; Catalanotto et al. 2004). Small RNAs join the Argonaute protein QDE2 to form RISC (RNA-induced silencing complex) and target all homologous RNAs for degradation (Catalanotto et al. 2002). MSUD is triggered by regions of unpaired DNA during chromosome pairing in meiosis. These regions may arise upon insertion of foreign DNA, like a transgene, a virus, or a transposon. Unpaired regions are transcribed, and a different RdRP, SAD1, converts the transcripts into dsRNA. As in quelling, small RNAs are made by the Dicer-like protein and loaded into RISC, resulting in silencing of homologous RNA (Lee et al. 2003). The mechanism of quelling and MSUD strongly resemble siRNA biogenesis in yeast and plants in the requirement of RdRP for generation of dsRNA yet unlike in those organisms both silence targets posttranscriptionally similarly to RNAi in animals and the cytoplasmic piRNA pathway.
4.5.3 DNA Elimination in Ciliates
The most extreme form of gene silencing is seen in some unicellular organisms that literally eliminate substantial unnecessary genomic information from their “somatic” nuclei. Tetrahymena, Paramecium and other protozoans have an unconventional genetic configuration, possessing a germline genome, located in the micronucleus (mic), which is transcriptionally silent during vegetative growth, and a somatic genome, found in the macronucleus (mac) from which genes are expressed (Duharcourt et al. 1995, 1998; Jahn and Klobutcher 2002). The genomes of the two nuclei greatly differ both in content and in structure. The macronucleus contains multiple copies of fragmented chromosomes, which lack substantially repetitive sequences that are common in the micronucleus. During the sexual process of conjugation, ciliated protozoans undergo programmed excision of excess DNA – the so-called internal eliminated sequences (IES), which are believed to be derived from transposons. Interestingly, Tetrahymena uses RNA guides to target the heterochromatin modifications to the loci to be excised (reviewed in Mochizuki and Gorovsky 2004a; Yao and Chao 2005). During conjugation, the entire micronuclear genome, which contains IES, is transcribed into double-stranded RNA. Small RNAs called scan RNA (scnRNA) are derived from these transcripts and loaded into a PIWI protein, TWI1 (Mochizuki and Gorovsky 2004a, b, 2005; Chalker and Yao 2001; Malone et al. 2005). TWI1-bound scnRNAs travel to the old macronucleus, from which the IES have been removed in the previous conjugation, and ‘scan’ the entire genome. scnRNA matching the macronuclear genome are degraded, and the remaining ones, matching IES, shuttle to the developing new macronucleus, where they induce heterochromatinization and subsequent excision of IES (Malone et al. 2005; Liu et al. 2004; Mochizuki et al. 2002). This process ensures that TEs are removed, while genomic sequences required for somatic functions are maintained in the mac of the next generation.
4.5.4 RNA-Dependent DNA Methylation in Plants
Similar to S. pombe, plants also utilize siRNAs to establish repressive chromatin at repetitive regions. Contrary to yeast, heterochromatin is marked by DNA methylation. Plant DNA methylation occurs throughout the genome, mostly on repetitive sequences, but not restricted to them. Constitutive expression of dsRNA mapping to promoter regions results in production of corresponding siRNAs and in de novo DNA methylation and gene silencing (Mette et al. 2000; Matzke et al. 2004). Additionally, in plants repeats can influence development by nucleating RNAi-dependent methylation and silencing of surrounding protein-coding genes. This indicates a more general function of small RNA pathways in plant transcriptional control.
Plants also seem to utilize a small RNA pathway to repress TEs in the germline, similar to piRNAs in animal germ cells (Slotkin et al. 2009). Unlike small RNA system in animals, this process seems to function non-cell-autonomously and involves the supporter cells of the gonads. Pollen consists of two nuclei, one that will form the sperm and be transmitted to the offspring and an accompanying vegetative nucleus, which does not contribute its DNA to the progeny. Due to demethylation of TE sequences in the vegetative nucleus of the pollen, TEs are reactivated, transpose and contribute to the formation of mature siRNAs, which can freely diffuse into the sperm nucleus. It is thought, that this protects the sperm from activation of TEs and possibly directs establishment of the correct methylation pattern in the offspring (Slotkin et al. 2009). A similar process – albeit using slightly different machinery – seems to be involved in silencing of transposons in the female gametophyte as well. This function of small RNAs transmitted to the germ cells would possibly correspond to the maternally deposited piR-NAs observed in flies, which are thought to direct transposon repression in the offspring.
4.5.5 The Interplay of the piRNA and siRNA Pathways in C. elegans
C. elegans employs a remarkable system that uses trans-generational memory of gene expression to find and repress foreign sequences in germ cells (Ashe et al. 2012; Luteijn et al. 2012; Shirayama et al. 2012; Lee et al. 2012). The correct identification and silencing of non-self sequences in C. elegans requires the cooperation of three different small RNA pathways.
In the gonads of C. elegans a diverse population of so-called 22G-RNAs is generated from the transcriptome and loaded into the Argonaute CSR1 (Claycomb et al. 2009). CSR1 and the associated 22G-RNAs are transmitted into the progeny, where they recognize “self” transcripts that were present in the parental gonads. This recognition by CSR1 protects “self” transcripts against targeting by the second small RNA pathway, the C. elegans piRNA pathway, consisting of the C. elegans PIWI -clade protein PRG1 and the associated 21U-RNAs (the C. elegans piR-NAs). Accordingly, recognition of non-self sequences seems to be achieved by targeting of all sequences that are not protected by the CSR1 system (Ashe et al. 2012; Luteijn et al. 2012; Shirayama et al. 2012; Lee et al. 2012). 21U-RNAs are very diverse and can likely bind transcripts through incomplete sequence complementarity allowing for recognition of any sequence (Ruby et al. 2006; Bagijn et al. 2012). Binding of the PRG1/21U-RNA complex to a target transcript leads to production of cognate 22G-RNAs (similar to the 22G-RNAs in CSR1, but mapping to the foreign sequence), which get loaded into WAGO9, part of the third small RNA pathway (Ashe et al. 2012; Luteijn et al. 2012; Lee et al. 2012). WAGO9 enters the nucleus and, together with other factors including chromatin proteins, induces transcriptional silencing of the sequence, along with deposition of the H3K9me3 mark. Once 22G-RNAs that incorporate into WAGO9 are generated against non-self, the WAGO pathway can maintain silencing in the absence of 21U-RNAs, thereby keeping memory of previously identified foreign sequences. Similarly to 22G-RNAs associated with CSR1 these are also trans-generationally inherited.
Together, these three systems, in concert with chromatin factors, allow for stable transgenerational transmission of information regarding self and non-self, and ensure an immediate silencing of new foreign sequences. The promiscuity of targeting guided by 21U-RNAs associated with PRG1 allows the piRNA pathway to recognize and induce silencing of new foreign elements of any sequence.
Contributor Information
Katalin Fejes Tóth, Email: kft@caltech.edu.
Dubravka Pezic, Email: dubravka@caltech.edu.
Evelyn Stuwe, Email: estuwe@caltech.edu.
Alexandre Webster, Email: awebster@caltech.edu.
References
- Al-Mukhtar KA, Webb AC. An ultrastructural study of primordial germ cells, oogonia and early oocytes in Xenopus laevis. J Embryol Exp Morphol. 1971;26(2):195–217. [PubMed] [Google Scholar]
- Anand A, Kai T. The tudor domain protein kumo is required to assemble the nuage and to generate germ-line piRNAs in Drosophila. EMBO J. 2012;31(4):870–882. doi: 10.1038/emboj.2011.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anne J. Targeting and anchoring Tudor in the pole plasm of the Drosophila oocyte. PLoS One. 2010;5(12):e14362. doi: 10.1371/journal.pone.0014362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anne J, Mechler BM. Valois, a component of the nuage and pole plasm, is involved in assembly of these structures, and binds to Tudor and the methyltransferase Capsuleen. Development. 2005;132(9):2167–2177. doi: 10.1242/dev.01809. [DOI] [PubMed] [Google Scholar]
- Anne J, Ollo R, Ephrussi A, Mechler BM. Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development. 2007;134(1):137–146. doi: 10.1242/dev.02687. [DOI] [PubMed] [Google Scholar]
- Aravin A, Chan D. piRNAs meet mitochondria. Dev Cell. 2011;20(3):287–288. doi: 10.1016/j.devcel.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germ-line. Curr Biol: CB. 2001;11(13):1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Klenov MS, Vagin VV, Bantignies F, Cavalli G, Gvozdev VA. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol Cell Biol. 2004;24(15):6742–6750. doi: 10.1128/MCB.24.15.6742-6750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442(7099):203–207. doi: 10.1038/nature04916. doi: http://dx.doi.org/10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316(5825):744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Aravin A, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth K, Bestor T, Hannon G. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31(6):785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A, van der Heijden G, Castañeda J, Vagin V, Hannon G, Bortvin A. Cytoplasmic compart-mentalization of the fetal piRNA pathway in mice. PLoS Genet. 2009;5(12):e1000764. doi: 10.1371/journal.pgen.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkov AL, Wang JY, Ramos A, Lehmann R. The role of Tudor domains in germline development and polar granule architecture. Development. 2006;133(20):4053–4062. doi: 10.1242/dev.02572. [DOI] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, Pintacuda G, Sakaguchi A, Sarkies P, Ahmed S, Miska EA. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150(1):88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick EM, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science. 2012;337(6094):574–578. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc’his D, Bestor T. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431(7004):96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Bozzetti MP, Massari S, Finelli P, Meggio F, Pinna LA, Boldyreff B, Issinger OG, Palumbo G, Ciriaco C, Bonaccorsi S, et al. The Ste locus, a component of the parasitic cry-Ste system of Drosophila melanogaster, encodes a protein that forms crystals in primary spermatocytes and mimics properties of the beta sub-unit of casein kinase 2. Proc Natl Acad Sci U S A. 1995;92(13):6067–6071. doi: 10.1073/pnas.92.13.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin A, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon G. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128(6):1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Carmell M, Xuan Z, Zhang M, Hannon G. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16(21):2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Carmell M, Girard A, van de Kant H, Bourc’his D, Bestor T, de Rooij D, Hannon G. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12(4):503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Catalanotto C, Azzalin G, Macino G, Cogoni C. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev. 2002;16(7):790–795. doi: 10.1101/gad.222402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto C, Pallotta M, ReFalo P, Sachs MS, Vayssie L, Macino G, Cogoni C. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol Cell Biol. 2004;24(6):2536–2545. doi: 10.1128/MCB.24.6.2536-2545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterina C, Tony N, Carlo C. Homology effects in Neurospora crassa. FEMS Microbiol Lett. 2006;254:182. doi: 10.1111/j.1574-6968.2005.00037.x. [DOI] [PubMed] [Google Scholar]
- Chalker DL, Yao MC. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes Dev. 2001;15(10):1287–1298. doi: 10.1101/gad.884601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Popkova A, Payen-Groschêne G, Brun C, Laouini D, Pelisson A, Bucheton A. piRNA-mediated nuclear accumulation of retrotransposon transcripts in the Drosophila female germline. Proc Natl Acad Sci. 2008;105(39):14964–14969. doi: 10.1073/pnas.0805943105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, Conte D, Jr, Mello CC. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139(1):123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399(6732):166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- Cox D, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12(23):3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Preall J, McGinn J, Hannon G. A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol Cell. 2013;50(5):749–761. doi: 10.1016/j.molcel.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duharcourt S, Butler A, Meyer E. Epigenetic self-regulation of developmental excision of an internal eliminated sequence on Paramecium tetraurelia. Genes Dev. 1995;9(16):2065–2077. doi: 10.1101/gad.9.16.2065. [DOI] [PubMed] [Google Scholar]
- Duharcourt S, Keller AM, Meyer E. Homology-dependent maternal inhibition of developmental excision of internal eliminated sequences in Paramecium tetraurelia. Mol Cell Biol. 1998;18(12):7075–7085. doi: 10.1128/mcb.18.12.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy EM. Fine structural observations on the form and distribution of nuage in germ cells of the rat. Anat Rec. 1974;178(4):731–757. doi: 10.1002/ar.1091780406. [DOI] [PubMed] [Google Scholar]
- Eddy EM, Ito S. Fine structural and radioautographic observations on dense perinuclear cytoplasmic material in tadpole oocytes. J Cell Biol. 1971;49(1):90–108. doi: 10.1083/jcb.49.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon G, Carmell M. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442(7099):199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grivna S, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20(13):1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Olivieri D, Novatchkova M, Gruber FS, Meixner K, Mechtler K, Stark A, Sachidanandam R, Brennecke J. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J. 2011;30(19):3977–3993. doi: 10.1038/emboj.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Meixner K, Pizka M, Lauss K, Schmied C, Gruber F, Brennecke J. The genetic makeup of the Drosophila piRNA pathway. Mol Cell. 2013;50(5):762–777. doi: 10.1016/j.molcel.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hathaway DS, Selman GG. Certain aspects of cell lineage and morphogenesis studied in embryos of Drosophila melanogaster with an ultra-violet micro-beam. J Embryol Exp Morphol. 1961;9:310–325. [PubMed] [Google Scholar]
- Hay B, Ackerman L, Barbel S, Jan LY, Jan YN. Identification of a component of Drosophila polar granules. Development. 1988;103(4):625–640. doi: 10.1242/dev.103.4.625. [DOI] [PubMed] [Google Scholar]
- Hegner RW. The germ cell determinants in the eggs of chrysomelid beetles. Science. 1911;33(837):71–72. doi: 10.1126/science.33.837.71. [DOI] [PubMed] [Google Scholar]
- Hegner RW. The history of the germ cells in the paedogenetic larva of Miastor. Science. 1912;36(917):124–126. doi: 10.1126/science.36.917.124. [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga L, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov D, Blaser H, Raz E, Moens C, Plasterk R, Hannon G, Draper B, Ketting R. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129(1):69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Huang H-Y, Houwing S, Kaaij L, Meppelink A, Redl S, Gauci S, Vos H, Draper B, Moens C, Burgering B, Ladurner P, Krijgsveld J, Berezikov E, Ketting R. Tdrd1 acts as a molecular scaffold for Piwi proteins and piRNA targets in zebrafish. EMBO J. 2011a;30(16):3298–3308. doi: 10.1038/emboj.2011.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Gao Q, Peng X, Choi S-Y, Sarma K, Ren H, Morris A, Frohman M. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell. 2011b;20(3):376–387. doi: 10.1016/j.devcel.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipsaro J, Haase A, Knott S, Joshua-Tor L, Hannon G. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature. 2012;491(7423):279–283. doi: 10.1038/nature11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn CL, Klobutcher LA. Genome remodeling in ciliated protozoa. Annu Rev Microbiol. 2002;56:489–520. doi: 10.1146/annurev.micro.56.012302.160916. [DOI] [PubMed] [Google Scholar]
- Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309(5733):467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Izumi N, Katsuma S, Tomari Y. 3′ end formation of PIWI-interacting RNAs in vitro. Mol Cell. 2011;43:1015. doi: 10.1016/j.molcel.2011.07.029. [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Hara K, Shoji K, Kobayashi M, Shimada T, Sugano S, Tomari Y, Suzuki Y, Katsuma S. The comprehensive epigenome map of piRNA clusters. Nucleic Acids Res. 2013;41(3):1581–1590. doi: 10.1093/nar/gks1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibanov MV, Egorova KS, Ryazansky SS, Sokolova OA, Kotov AA, Olenkina OM, Stolyarenko AD, Gvozdev VA, Olenina LV. A novel organelle, the piNG-body, in the nuage of Drosophila male germ cells is associated with piRNA-mediated gene silencing. Mol Biol Cell. 2011;22(18):3410–3419. doi: 10.1091/mbc.E11-02-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein P, Rigoutsos I, Jongens T, Mourelatos Z. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol. 2009;11(5):652–658. doi: 10.1038/ncb1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Vourekas A, Sayed N, de Lima Alves F, Thomson T, Lasko P, Rappsilber J, Jongens TA, Mourelatos Z. Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA. 2010a;16(1):70–78. doi: 10.1261/rna.1869710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Vourekas A, Kim N, de Lima Alves F, Rappsilber J, Klein PS, Jongens TA, Mourelatos Z. Arginine methylation of vasa protein is conserved across phyla. J Biol Chem. 2010b;285(11):8148–8154. doi: 10.1074/jbc.M109.089821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, Seitz H, Zamore PD, Weng Z, Theurkauf WE. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138(6):1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov M, Lavrov S, Stolyarenko A, Ryazansky S, Aravin A, Tuschl T, Gvozdev V. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35(16):5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Sokolova OA, Yakushev EY, Stolyarenko AD, Mikhaleva EA, Lavrov SA, Gvozdev VA. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc Natl Acad Sci U S A. 2011;108(46):18760–18765. doi: 10.1073/pnas.1106676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler JM, Lasko P. Localization, anchoring and translational control of oskar, gurken, bicoid and nanos mRNA during Drosophila oogenesis. Fly. 2009;3(1):15–28. doi: 10.4161/fly.3.1.7751. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Yomogida K, Kuroiwa A, Tadokoro Y, Fujita Y, Sato M, Matsuda Y, Nakano T. Two mouse piwi-related genes: miwi and mili. Mech Dev. 2001;108(1-2):121–133. doi: 10.1016/s0925-4773(01)00499-3. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri T, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, Nakano T. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131(4):839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri T, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22(7):908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, Shiromoto Y, Asada N, Toyoda A, Fujiyama A, Totoki Y, Shibata T, Kimura T, Nakatsuji N, Noce T, Sasaki H, Nakano T. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 2010;24(9):887–892. doi: 10.1101/gad.1902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N, Seto A, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel D, Kingston R. Characterization of the piRNA complex from rat testes. Science (New York, NY) 2006;313(5785):363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Le Thomas A, Rogers A, Webster A, Marinov G, Liao S, Perkins E, Hur J, Aravin A, Tóth K. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013;27(4):390–399. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Pratt RJ, McLaughlin M, Aramayo R. An argonaute-like protein is required for meiotic silencing. Genetics. 2003;164(2):821–828. doi: 10.1093/genetics/164.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Gu W, Shirayama M, Youngman E, Conte D, Jr, Mello CC. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell. 2012;150(1):78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, Kittler EL, Zapp ML, Klattenhoff C, Schulz N, Theurkauf WE, Weng Z, Zamore PD. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137(3):509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Roy C, Dong X, Bolcun-Filas E, Wang J, Han B, Xu J, Moore M, Schimenti J, Weng Z, Zamore P. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell. 2013;50(1):67–81. doi: 10.1016/j.molcel.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Diehl-Jones W, Lasko P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development. 1994;120(5):1201–1211. doi: 10.1242/dev.120.5.1201. [DOI] [PubMed] [Google Scholar]
- Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104(16):6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Spradling A. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124(12):2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Liu Y, Mochizuki K, Gorovsky MA. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc Natl Acad Sci U S A. 2004;101(6):1679–1684. doi: 10.1073/pnas.0305421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Chen C, Guo Y, Lam R, Bian C, Xu C, Zhao D, Jin J, MacKenzie F, Pawson T, Min J. Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proc Natl Acad Sci U S A. 2010a;107(43):18398–18403. doi: 10.1073/pnas.1013106107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang JY, Huang Y, Li Z, Gong W, Lehmann R, Xu RM. Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes Dev. 2010b;24(17):1876–1881. doi: 10.1101/gad.1956010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Qi H, Wang J, Lin H. PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition. Development. 2011;138(9):1863–1873. doi: 10.1242/dev.059287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn MJ, van Bergeijk P, Kaaij LJ, Almeida MV, Roovers EF, Berezikov E, Ketting RF. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 2012;31(16):3422–3430. doi: 10.1038/emboj.2012.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Buchold G, Greenbaum M, Roy A, Burns K, Zhu H, Han D, Harris R, Coarfa C, Gunaratne P, Yan W, Matzuk M. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet. 2009;5(9):e1000635. doi: 10.1371/journal.pgen.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald AP. Polar granules of Drosophila. II. Ultrastructural changes during early embryogenesis. J Exp Zool. 1968;167(2):237–261. doi: 10.1002/jez.1401670211. [DOI] [PubMed] [Google Scholar]
- Malone CD, Anderson AM, Motl JA, Rexer CH, Chalker DL. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol Cell Biol. 2005;25(20):9151–9164. doi: 10.1128/MCB.25.20.9151-9164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137(3):522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathioudakis N, Palencia A, Kadlec J, Round A, Tripsianes K, Sattler M, Pillai R, Cusack S. The multiple Tudor domain-containing protein TDRD1 is a molecular scaffold for mouse Piwi proteins and piRNA biogenesis factors. RNA (New York, NY) 2012;18(11):2056–2072. doi: 10.1261/rna.034181.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M, Aufsatz W, Kanno T, Daxinger L, Papp I, Mette MF, Matzke AJ. Genetic analysis of RNA-mediated transcriptional gene silencing. Biochim Biophys Acta. 2004;1677(1-3):129–141. doi: 10.1016/j.bbaexp.2003.10.015. S0167478103002859 [pii] [DOI] [PubMed] [Google Scholar]
- Megosh HB, Cox DN, Campbell C, Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol: CB. 2006;16(19):1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- Meikar O, Da Ros M, Korhonen H, Kotaja N. Chromatoid body and small RNAs in male germ cells. Reproduction (Cambridge, England) 2011;142(2):195–209. doi: 10.1530/REP-11-0057. [DOI] [PubMed] [Google Scholar]
- Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19(19):5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky M. Small RNAs in genome rearrangement in Tetrahymena. Curr Opin Genet Dev. 2004a;14(2):181–187. doi: 10.1016/j.gde.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes Dev. 2004b;18(17):2068–2073. doi: 10.1101/gad.1219904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 2005;19(1):77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110(6):689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- Muerdter F, Guzzardo P, Gillis J, Luo Y, Yu Y, Chen C, Fekete R, Hannon G. A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila. Mol Cell. 2013;50(5):736–748. doi: 10.1016/j.molcel.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Okada T, Kawamura T, Mituyama T, Kawamura Y, Inagaki S, Huang H, Chen D, Kodama T, Siomi H, Siomi M. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 2009;28(24):3820–3831. doi: 10.1038/emboj.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani M, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, Ishitani R, Siomi H, Siomi M, Nureki O. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 2012;491(7423):284–287. doi: 10.1038/nature11509. [DOI] [PubMed] [Google Scholar]
- Okada M, Kleinman IA, Schneiderman HA. Restoration of fertility in sterilized Drosophila eggs by transplantation of polar cytoplasm. Dev Biol. 1974;37(1):43–54. doi: 10.1016/0012-1606(74)90168-7. [DOI] [PubMed] [Google Scholar]
- Olivieri D, Sykora M, Sachidanandam R, Mechtler K, Brennecke J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 2010;29(19):3301–3317. doi: 10.1038/emboj.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Jiang P, Zhao DY, Singh M, Schupbach T. The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. EMBO J. 2011;30(22):4601–4615. doi: 10.1038/emboj.2011.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil VS, Kai T. Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr Biol: CB. 2010;20(8):724–730. doi: 10.1016/j.cub.2010.02.046. [DOI] [PubMed] [Google Scholar]
- Ponting CP. Tudor domains in proteins that interact with RNA. Trends Biochem Sci. 1997;22(2):51–52. doi: 10.1016/s0968-0004(96)30049-2. [DOI] [PubMed] [Google Scholar]
- Rangan P, Malone CD, Navarro C, Newbold SP, Hayes PS, Sachidanandam R, Hannon GJ, Lehmann R. piRNA production requires heterochromatin formation in Drosophila. Curr Biol: CB. 2011;21(16):1373–1379. doi: 10.1016/j.cub.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Grewal SI. Different means, same end-heterochromatin formation by RNAi and RNAi-independent RNA processing factors in fission yeast. Curr Opin Genet Dev. 2012;22(2):156–163. doi: 10.1016/j.gde.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro S, Park C, Song R, Nguyen D, Jin J, Sanders K, McCarrey J, Yan W. Cloning and expression profiling of testis-expressed piRNA-like RNAs. RNA (New York, NY) 2007;13(10):1693–1702. doi: 10.1261/rna.640307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano N, Macino G. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol. 1992;6(22):3343–3353. doi: 10.1111/j.1365-2958.1992.tb02202.x. [DOI] [PubMed] [Google Scholar]
- Rozhkov N, Hammell M, Hannon G. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 2013;27(4):400–412. doi: 10.1101/gad.209767.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127(6):1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009a;461(7268):1296–1299. doi: 10.1038/nature08501. doi: http://dx.doi.org/10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009b;461(7268):1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Palumbo G, Bozzetti M, Tritto P, Pimpinelli S, Schäfer U. Genetic and molecular characterization of sting, a gene involved in crystal formation and meiotic drive in the male germ line of Drosophila melanogaster. Genetics. 1999;151(2):749–760. doi: 10.1093/genetics/151.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Jr, Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150(1):65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu PK, Raju NB, Zickler D, Metzenberg RL. Meiotic silencing by unpaired DNA. Cell. 2001;107(7):905–916. doi: 10.1016/s0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, Hata K, Martin SL, Noce T, Kuramochi-Miyagawa S, Nakano T, Sasaki H, Pillai RS, Nakatsuji N, Chuma S. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell. 2009;17(6):775–787. doi: 10.1016/j.devcel.2009.10.012. doi: http://dx.doi.org/10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- Shpiz S, Kwon D, Rozovsky Y, Kalmykova A. rasiRNA pathway controls antisense expression of Drosophila telomeric retrotransposons in the nucleus. Nucleic Acids Res. 2009;37(1):268–278. doi: 10.1093/nar/gkn960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpiz S, Olovnikov I, Sergeeva A, Lavrov S, Abramov Y, Savitsky M, Kalmykova A. Mechanism of the piRNA-mediated silencing of Drosophila telomeric retrotransposons. Nucleic Acids Res. 2011;39(20):8703–8711. doi: 10.1093/nar/gkr552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Dönertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151(5):964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi M, Sato K, Pezic D, Aravin A. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12(4):246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Slotkin R, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8(4):272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzic M, Becker JD, Feijo JA, Martienssen RA. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136(3):461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SU, Kurkulos M, Boeke JD, Corces VG. Infection of the germ line by retroviral particles produced in the follicle cells: a possible mechanism for the mobilization of the gypsy retroelement of Drosophila. Development. 1997;124(14):2789–2798. doi: 10.1242/dev.124.14.2789. [DOI] [PubMed] [Google Scholar]
- Song J-J, Smith S, Hannon G, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science (New York, NY) 2004;305(5689):1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Soper S, van der Heijden G, Hardiman T, Goodheart M, Martin S, de Boer P, Bortvin A. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15(2):285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton W, Das S, McKee BD. A role of the Drosophila homeless gene in repression of Stellate in male meiosis. Chromosoma. 2001;110(3):228–240. doi: 10.1007/s004120100136. [DOI] [PubMed] [Google Scholar]
- Szakmary A, Reedy M, Qi H, Lin H. The Yb protein defines a novel organelle and regulates male germline stem cell self-renewal in Drosophila melanogaster. J Cell Biol. 2009;185:613. doi: 10.1083/jcb.200903034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson T, Lasko P. Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis. 2004;40(3):164–170. doi: 10.1002/gene.20079. [DOI] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313(5785):320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Vagin V, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon G, Aravin A. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23(15):1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt F, Reuter M, Kasaruho A, Schulz E, Pillai R, Barabas O. Crystal structure of the primary piRNA biogenesis factor Zucchini reveals similarity to the bacterial PLD endonuclease Nuc. RNA (New York, NY) 2012;18(12):2128–2134. doi: 10.1261/rna.034967.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel D. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456(7224):921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Tomizawa S-i, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy P, Toyoda A, Gotoh K, Hiura H, Arima T, Fujiyama A, Sado T, Shibata T, Nakano T, Lin H, Ichiyanagi K, Soloway P, Sasaki H. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science (New York, NY) 2011a;332(6031):848–852. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, Noce T, Nakano T, Nakatsuji N, Lin H, Sasaki H. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell. 2011b;20(3):364–375. doi: 10.1016/j.devcel.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao MC, Chao JL. RNA-guided DNA deletion in Tetrahymena: an RNAi-based mechanism for programmed genome rearrangements. Annu Rev Genet. 2005;39:537–559. doi: 10.1146/annurev.genet.39.073003.095906. [DOI] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15(4):381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Xu J, Koppetsch BS, Wang J, Tipping C, Ma S, Weng Z, Theurkauf WE, Zamore PD. Heterotypic piRNA Ping-Pong requires qin, a protein with both E3 ligase and Tudor domains. Mol Cell. 2011;44(4):572–584. doi: 10.1016/j.molcel.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD, Weng Z, Theurkauf WE. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell. 2012;151(4):871–884. doi: 10.1016/j.cell.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Grewal SI. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of hetero-chromatic repeats. Mol Cell. 2006;2(5):681. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]