Abstract
Context
Our genome adapts to environmental influences, in part through epigenetic mechanisms, including DNA methylation. Variations in the quality of the early environment are associated with alterations in DNA methylation in rodents, and recent data suggest similar processes in humans in response to early-life adversity.
Objective
To determine genome-wide DNA methylation alterations induced by early-life trauma.
Design
Genome-wide study of promoter methylation in individuals with severe abuse during childhood.
Patients, Setting, and Main Outcome Measures
Promoter DNA methylation levels were profiled using methylated DNA immunoprecipitation followed by microarray hybridization in hippocampal tissue from 41 French-Canadian men (25 with a history of severe childhood abuse and 16 control subjects). Methylation profiles were compared with corresponding genome-wide gene expression profiles obtained by messenger RNA microarrays. Methylation differences between groups were validated on neuronal and nonneuronal DNA fractions isolated by fluorescence-assisted cell sorting. Functional consequences of site-specific promoter methylation were assessed by luciferase assays.
Results
We identified 362 differentially methylated promoters in individuals with a history of abuse compared with controls. Among these promoters, 248 showed hypermethylation and 114 demonstrated hypomethylation. Validation and site-specific quantification of DNA methylation in the 5 most hypermethylated gene promoters indicated that methylation differences occurred mainly in the neuronal cellular fraction. Genes involved in cellular/neuronal plasticity were among the most significantly differentially methylated, and, among these, Alsin (ALS2) was the most significant finding. Methylated ALS2 constructs mimicking the methylation state in samples from abused suicide completers showed decreased promoter transcriptional activity associated with decreased hippocampal expression of ALS2 variants.
Conclusion
Childhood adversity is associated with epigenetic alterations in the promoters of several genes in hippocampal neurons.
Childhood adversity, characterized by the presence of sexual and physical abuse, is a global problem of significant proportions.1,2 Epidemiologic studies3–5 indicate prevalence rates for all forms of childhood sexual abuse and physical abuse ranging from 11% to 35%. Childhood sexual abuse and childhood physical abuse are among the strongest predictors of psychiatric pathology and severity of clinical course, including suicide.2,4–14 The influence of childhood sexual abuse and childhood physical abuse on psychological development is thought to be mediated directly by changes in cognitive processing of threatening stimuli,15–18 resulting in enhanced negative affect to daily life stressors.19 Although there is a clear link between early-life adversity and psychopathology, very little is known about the molecular mechanisms responsible for the long-lasting behavioral consequences of childhood abuse. Significant insight into this critical issue comes primarily from animal studies and from recent translation research in humans investigating epigenetic mechanisms.20,21
Early studies20,22 have shown that variations in the quality of postnatal parent-offspring interactions directly alter intracellular signals that regulate epigenetic states, with sustained effects on gene transcription. For instance, in rats, adult offspring of low licking and grooming mothers show decreased hippocampal expression of glucocorticoid receptor variant 7, glutamic acid decarboxylase 1, and estrogen receptor α associated with overall promoter hypermethylation that constrains the binding of transcription factors, such as nerve growth factor–induced protein A.23–25 Similarly, site-specific hypermethylation in brain-derived neurotropic factor promoter associates with lower expression of this neurotrophin in the prefrontal cortex of offspring of chronically stressed mothers.26 Prolonged periods of maternal separation in the mouse regulate the methylation of an arginine vasopressin enhancer, arginine vasopressin expression, and hypothalamic-pituitary-adrenal responses to stress.27 Chronic social stress in adult mice decreases methylation levels in the promoter of the corticotrophin-releasing factor and increases expression in the paraventricular nucleus of the hypothalamus.28 In humans, suicide completers with a history of childhood abuse (SAs) show hypermethylation in the nerve growth factor–induced protein A binding site within glucocorticoid receptor variant 1F promoter that is associated with decreased glucocorticoid receptor variant 1F receptor expression in the hippocampus (HPC).21 Abused suicide completers also have overall hypermethylation in the ribosomal RNA gene promoters associated with lower hippocampal expression, suggesting widespread effects across the genome.29
These studies suggest that early-life adversity induces epigenetic alterations in gene regulatory regions. An important question is whether childhood abuse affects only a restricted number of candidate genes or whether it has a broader effect on the epigenome and, as a consequence, on a large spectrum of functional pathways. To date, no large-scale genome-wide study has been performed to identify epigenetic alterations found in the brains of individuals who experienced childhood abuse. We report results from a comprehensive genome-wide screening of promoter DNA methylation modifications found in the HPC of SAs compared with control individuals. Results from this study support the hypothesis that child abuse induces a coordinated DNA methylation response in multiple promoters throughout the genome.
METHODS
Complete methods are described in the Author eAppendix (available at http://www.douglas.qc.ca/page/mgss-supplementary).
SAMPLE SELECTION
The project was approved by the research ethics board at the Douglas University Mental Health Institute. Brain tissue was obtained from the Quebec Suicide Brain Bank (Douglas Mental Health University Institute, Verdun, Québec, Canada). The sample for this study consisted of brain tissue from 41 individuals (25 SAs and 16 controls [Author eTable 1]). An additional group composed of 20 nonabused suicide completers (SNAs) was included in the validation experiments. All subjects were white males of French-Canadian descent, a population with a well-identified founder effect,30 and were group matched for age, pH, and postmortem intervals. Presence of severe early-life abuse was based on adapted Childhood Experience of Care and Abuse interviews assessing various dimensions of the childhood experience, including abuse.31
METHYLATED DNA IMMUNOPRECIPITATION, LABELING, AND HYBRIDIZATION
Methylated DNA was extracted following an adaptation of a methylated DNA immunoprecipitation (meDIP) method developed32 using 5′ methylcytosine antibody bound to sepharose beads. Input, unmethylated, and methylated fractions were purified by phenol-chloroform and precipitated in ethanol. Labeling, hybridization, and data extraction were performed following the manufacturer’s (Agilent Technologies) instructions. Every subject was hybridized on a separate microarray. Microarrays were scanned (High-Resolution C Scanner; Agilent Technologies), and data were extracted using commercial software (Feature Extraction; Agilent Technologies).
MICROARRAY DESIGN AND ANALYSIS
A custom-designed 400K promoter tiling array was used for this study (Agilent Technologies). The array was designed using the manufacturer’s array design platform (eArray) in July 2009. Probes were selected to tile all known gene promoters, ie, intervals approximately 2000 base pairs (bp) upstream and 400 bp downstream of the transcription start sites of genes were described in Ensembl software (version 55; http://www.ensembl.org) at 100-bp spacing. Extracted microarray intensities were processed and analyzed using the R software environment for statistical computing.33
EXPRESSION MICROARRAY DATA
Whole-genome gene expression data were obtained from gene expression microarrays previously generated in our laboratory on the HU 133 plus2 microarrays (Affymetrix Inc).34 Methylation and expression data were compared in a subset of samples for which we added expression and methylation profiles (13 SAs and 9 controls). Expression data were normalized as previously described.35
NEURONAL AND NONNEURONAL ISOLATION AND MICROARRAY VALIDATION
Nuclei were isolated from hippocampal tissue by fluorescence-assisted cell sorting using human anti-NeuN antibody conjugated to a fluorescent marker (Alexa Fluor 488; Life Technologies Corp) as described previously.36 Nuclei were filtered and sorted (FACSVantage SE system; BD Bioscience). Microarray validation was performed by EpiTYPER (Sequenom) at the Génome Québec Innovation Centre. Every sample used in the microarray experiments was used in the validation experiments. Results were analyzed by 2-way mixed-model analysis of variance (ANOVA), with groups as a fixed factor and CpG dinucleotides as a repeated measure followed by Fisher least significant difference (LSD) post hoc tests. The level of significance was fixed at P = .05.
LUCIFERASE ASSAYS
Alsin gene (ALS2 [HGNC 443]) full-length and truncated promoters were amplified by polymerase chain reaction (PCR) from human genomic DNA. A methylated full-length construct was obtained by means of inverse PCR using a methylated primer and ligated into a pGL3 vector before transfection into Be(2)c cells. Firefly renilla plasmid was used as a control for transfection efficiency and to normalize luciferase activity. All experiments were performed in 6 replicates. Results were analyzed by independent sample t test, and the significance level was fixed at P = .05.
QUANTIFICATION OF GENE EXPRESSION USING QUANTITATIVE REAL-TIME PCR
Total RNA was extracted from the same samples used in the microarray and validation experiments using a lipid tissue extraction kit (RNeasy; Qiagen) and was followed by Dnase I treatment, and cDNA conversion was performed using oligo(dT) primers. Expression of ALS2 was quantified using custom-designed probes (ABI 7900HT Taqman; Applied Biosystems). Mean quantities from all samples were normalized to the reference gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH [HGNC 4141]). Results were analyzed by 1-way ANOVA followed by LSD post hoc tests. The level of significance was fixed at P = .05.
RESULTS
GENOME-WIDE PROMOTER METHYLATION DIFFERENCES
Using custom-designed Agilent high-density microarrays, we investigated genome-wide methylation profiles in gene promoter sequences in the HPC of SAs compared with controls. A total of 330 600 probes were distributed in the promoter regions of 23 551 genes. The SAs and controls did not differ significantly with respect to mean (SEM) age (SAs, 37.3[2.1] years; controls, 40.9 [3.6] years; t = −0.92, P = .36), pH (SAs, 6.5 [0.1]; controls, 6.5 [0.1]; t = −0.06, P = .95), and postmortem interval (SAs, 28.5 [2.6] hours; controls, 32.6 [3.8] hours; t = −0.95, P = .35).
We first examined promoter methylation levels and distribution across the genome in SA compared with control samples, adjusting for confounders that showed evidence of a significant effect on the total amount of methylation variance in the microarray data or for variables known to have an effect on methylation. Specifically, we controlled for substance disorders, age, and postmortem interval. A total of 362 probe sets mapping to 307 different promoters were significantly differentially methylated between groups. (See the Author eAppendix for the methods used in the analyses of the arrays.) Our analyses revealed an overrepresentation of hypermethylated probes in the promoters of the SA group. Thus, 68.5% of the probes (n = 248) showed higher methylation and 31.5% (n = 114) showed lower methylation in SA compared with control samples after false discovery rate correction (P ≤ 1.2E−6, hypergeometric). Interestingly, our analyses revealed that CpG density is more than 3.2 times lower in hypomethylated regions compared with hyper-methylated regions. The CpG density in hypermethylated regions was identical to density in all regions tiled by the microarray (P <8.2E−38, Wilcoxon rank sum test).
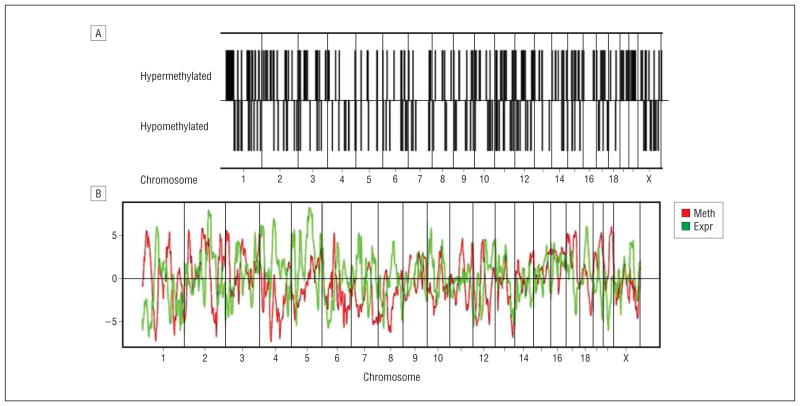
Analysis of chromosomal distribution (Figure 1A) of differentially methylated promoters across the genome showed that no chromosome or chromosomal region was overrepresented. As evidence of this, we noted that only 20% of the differentially methylated promoters were within 500 kilobases (kb) of another differentially methylated promoter. A permutation test revealed that this level (20%) was in the expected range for the random distribution of 307 differentially methylated promoters (P = .65). Thus, epigenetic alterations associated with early-life adversity were equally distributed throughout the genome.
Figure 1.
Chromosomal location and expression. A, Chromosomal distribution of differentially methylated probes in abused suicide completers (SAs). The first row represents the chromosomal location of probes with increased methylation (hypermethylated) in the SAs and the second row indicates the chromosomal location of probes with decreased methylation (hypomethylated) in the SAs. B, Inverse correlation between whole-genome expression and promoter methylation differences between SAs and controls. The mean promoter methylation differences (Meth) and gene expression (Expr) differences show that differential methylation is inversely correlated with differential gene expression across the genome (Pearson r= −0.19, P≤4.0E−6). Data were obtained by summarizing promoter methylation and gene expression differences across regions of 1 megabase throughout the whole genome.
Promoter hypermethylation has classically been associated with gene silencing, whereas hypomethylation has been observed with increased gene expression.37 We assessed the transcriptional consequences of methylation changes in gene promoters by comparing the methylation data with messenger RNA (mRNA) gene expression data generated in an overlapping sample using Affymetrix HU 133 plus2 microarrays. We first compared expression and methylation data independent of group differences. As expected, genome-wide expression levels were inversely correlated with estimated genome-wide promoter methylation levels (Pearson r = −0.14, P ≤4.6E−247). Furthermore, this inverse correlation became more pronounced when summarizing promoter methylation and gene expression differences across 1-megabase regions (Pearson r = −0.19, P ≤4.0E−6) (Figure 1B). These findings are consistent with an overall negative effect of promoter methylation on the regulation of gene expression.
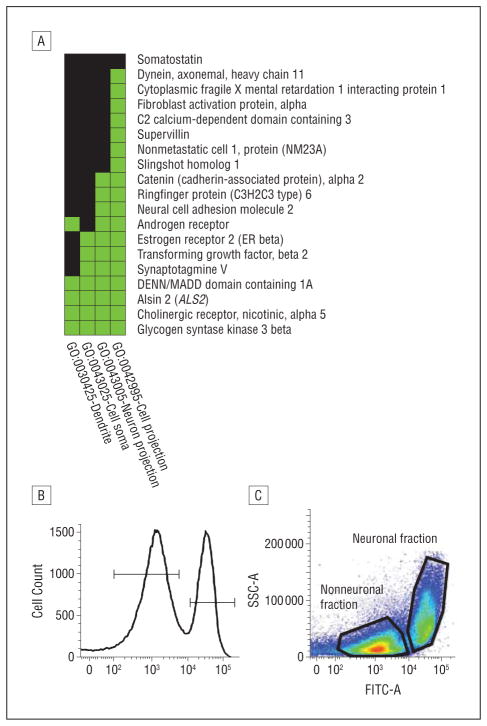
The Table provides a list of the most significantly differentially methylated gene promoters ranked by corrected (false discovery rate) P values and log2-fold change (see Author eTable 2 for a complete list). Genomic regions in the promoter of the most significantly differentially methylated genes in the SA group (DGKZ [HGNC 2857], HIST2H2AB [HGNC 20508], NR1D1 [HGNC 7962], RGS3 [HGNC 9999], and TAF5L [HGNC 17304]) were selected for validation (eFigure; http://www.archgenpsychiatry.com). We validated our findings in neuronal and nonneuronal cell fractions from the same hippocampal samples used in the promoter methylation array study to investigate whether significant differential methylation between groups resulted from methylation differences in neurons or glial cells. We first isolated nuclei from both cellular fractions using a fluorescence-assisted cell-sorting method with the neuronal nuclei marker NeuN.38 Sorting total nuclei stained by NeuN generated a bimodal fluorescence intensity distribution (Figure 2B and C) composed of approximately 30% of neuronal nuclei and 70% of nonneuronal nuclei. The sorting procedure was assessed under fluorescence microscopy and revealed a high level of purity (Author eFigure 2). With the exception of RGS3, all promoters investigated confirmed significant methylation differences specific to the neuronal fraction (eFigure).
Table.
List of the 25 Most Significantly Differentially Methylated Gene Promoters Subjected to Multiple Testing Correction (FDR)
| Chromosomea | P Value FDR | LFC | Gene | TSS, bp |
|---|---|---|---|---|
| Hypermethylated in SAs | ||||
| 11 | 1.48E−3 | 0.97 | DGKZ | 642 |
| 1 | 1.48E−3 | 0.97 | HIST2H2AB | 836 |
| 17 | 5.24E−3 | 1.27 | NR1D1 | 314 |
| 17 | 5.24E−3 | 1.08 | NR1D1 | 358 |
| 9 | 5.24E−3 | 0.98 | RGS3 | 878 |
| 1 | 5.49E−3 | 0.90 | TAF5L | −968 |
| 1 | 5.49E−3 | 0.85 | TAF5L | −518 |
| 1 | 5.49E−3 | 0.82 | TAF5L | −957 |
| 11 | 5.59E−3 | 0.98 | ABCG4 | 208 |
| 20 | 5.72E−3 | 1.34 | HCK | 564 |
| 2 | 5.72E−3 | 1.10 | MIR10B | −96 |
| 20 | 5.72E−3 | 0.81 | HCK | 383 |
| 16 | 5.73E−3 | 0.89 | HYDIN | 377 |
| 11 | 6.07E−3 | 0.85 | C2CD3 | −548 |
| 14 | 6.63E−3 | 1.17 | C14orf174 | −739 |
| 3 | 6.63E−3 | 0.82 | FHIT | 294 |
| 1 | 6.79E−3 | 1.06 | C1orf51 | 1199 |
| 2 | 6.79E−3 | 1.22 | ALS2 | 596 |
| 2 | 6.79E−3 | 0.97 | ALS2 | 352 |
| 1 | 6.79E−3 | 1.20 | PPFIA4 | 68 |
| 1 | 6.79E−3 | 0.98 | PPFIA4 | 84 |
| Hypomethylated in SAs | ||||
| 14 | 8.05E−4 | 0.81 | SNORD114-14 | 884 |
| 14 | 8.05E−4 | 0.78 | SNORD114-14 | 950 |
| 9 | 1.48E−3 | 1.03 | AL449083.1 | 505 |
| 15 | 3.13E−3 | 0.96 | SNRPN | 535 |
Abbreviations: bp, base pairs; FDR, false discovery rate; LFC, log-fold change; SAs, suicide completers who were abused as children; TSS, distance from the transcription start site (positive, upstream; negative, downstream).
Multiple probes were differentially methylated on the same chromosome.
Figure 2.
Pathway analysis and fluorescence-assisted cell-sorting summary. A, Functional annotation chart of the neuronal plasticity cluster in the abused suicide completer group determined from the differentially methylated gene sets after false discovery rate correction. Shown are the genes and associated annotations for functional annotation cluster 4 related to neuronal plasticity. Green represents corresponding gene-term associations previously reported, and black represents corresponding gene-term associations not yet reported. B, Total nuclei stained with NeuN generated a bimodal fluorescence intensity distribution. The intensity of fluorescence in the nonneuronal fraction (left peak) is less than in the neuronal fraction (right peak). C, Density plot representing the neuronal and nonneuronal cell fractions as a function of the intensity of fluorescence (FITC-A). SSC-A indicates side scatter (internal granulosity).
We explored the functional significance of the differentially methylated genes with a functional annotation-clustering analysis using DAVID 6.7 (http://david.abcc.ncifcrf.gov). The 5 functional clusters most significantly enriched with differentially methylated genes are listed in Author eTable 3. The list suggests that genes associated with neuronal plasticity, including both cell adhesion and cell plasticity, were significantly enriched categories. We focused on differentially methylated genes within this functional cluster to further understand the relationship between early-life adversity and hippocampal function. Figure 2A shows the cell-plasticity functional annotation cluster and the genes it contains. This annotation was enriched with 19 genes, among which ALS2 was the only gene appearing among our top candidates (Table) and found in all ontologic terms. We thus further investigated ALS2 to define the functional effect of hypermethylation in its promoter.
CHARACTERIZATION OF METHYLATION ALTERATIONS IN ALS2 PROMOTER
The ALS2 (Alsin) gene is located on chromosome 2q and is composed of 34 exons spanning a region of 80 kb of DNA. It has 2 major transcripts predicted to encode 2 protein isoforms39,40 composed of multiple guanine-nucleotide exchange factor motifs.41 Interestingly, ALS2 is thought to be expressed specifically in neurons.41
We quantified individual CpG methylation levels in a region of 409 bp, including 14 CpG sites in the promoter of ALS2 (Figure 3A). A group of SNAs matched with SAs and controls for age, pH, and postmortem interval was included to control for the confounding effect of suicide on methylation associated with childhood adversity. Consistent with the microarray results and with a neuronal pattern of ALS2 expression, a significant main effect of group was found in the neuronal (2-way ANOVA, F2,54 = 5.1, P=.007) (Figure 4A) but not in the nonneuronal cell fraction (Figure 4B). A post hoc test in the neuronal fraction revealed significant hypermethylation in SAs compared with SNAs (LSD, P=.001) and controls (LSD, P=.05). A 2-way ANOVA also revealed a significant main effect of CpG site (F9,54=59.9, P =1.58E-67) and a significant group × CpG site interaction (F29,54=2.1, P =.006) in the neuronal cell fraction. Post hoc analysis indicated significant hypermethylation at CpG4 in SA compared with SNA (LSD, P=7.07E−5) and control samples (LSD, P=9.59E−9) and between SNA and control samples (LSD, P=.03) (Figure 4C). There was also a trend toward a significant hypermethylation at CpGs 16 and 17 between SAs and controls (LSD, P = .07) (Figure 4C). These results suggest that child abuse is associated with differential ALS2 promoter methylation levels in the neuronal cell fraction, specifically at CpG site 4.
Figure 3.
In vitro analysis of ALS2 promoter sequence methylation. A, ALS2 promoter sequence, showing the location of the CpG dinucleotides. The full-length (FL) (1050 basepairs [bp], black triangle, solid underline) and the truncated (Trunc) (543 bp, white triangle, broken underline) constructs are shown, along with the specific CpG dinucleotide that was hypermethylated as shown by EpiTYPER (circled, CpG4). The solid-lined box represents an upstream stimulating factor 1 putative binding site with the blue area indicating area investigated with EpiTYPER. Capitalized letters represent the beginning of exon 1. B, Mean (SE) levels of luciferase expression in Be(2)c cells for the FL and Trunc constructs. Results are expressed as mean luciferase expression normalized by renilla expression. Experiments were run with 6 replicates. C, Mean (SE) levels of luciferase expression in Be(2)c cells for the FL ALS2 promoter that was either unmethylated (FL No CH3) or artificially methylated at CpG4 (FL CH3). Results are expressed as mean luciferase expression normalized by renilla expression. Experiments were run with 6 replicates. D, Mean (SE) levels of ALS2 isoform 1 expression in the hippocampus. Results are expressed as mean ALS2 expression normalized with the glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH) expression. Abused suicide completer (SA) (n = 22), nonabused suicide completer (SNA) (n = 19), and control subjects (n = 12). E, Mean (SE) levels of ALS2 isoform 2 expression in the hippocampus. Results are expressed as mean ALS2 expression normalized with GAPDH expression. SA (n = 22), SNA (n = 17), and controls (n = 13). *P<.05. †P<.0001.
Figure 4.
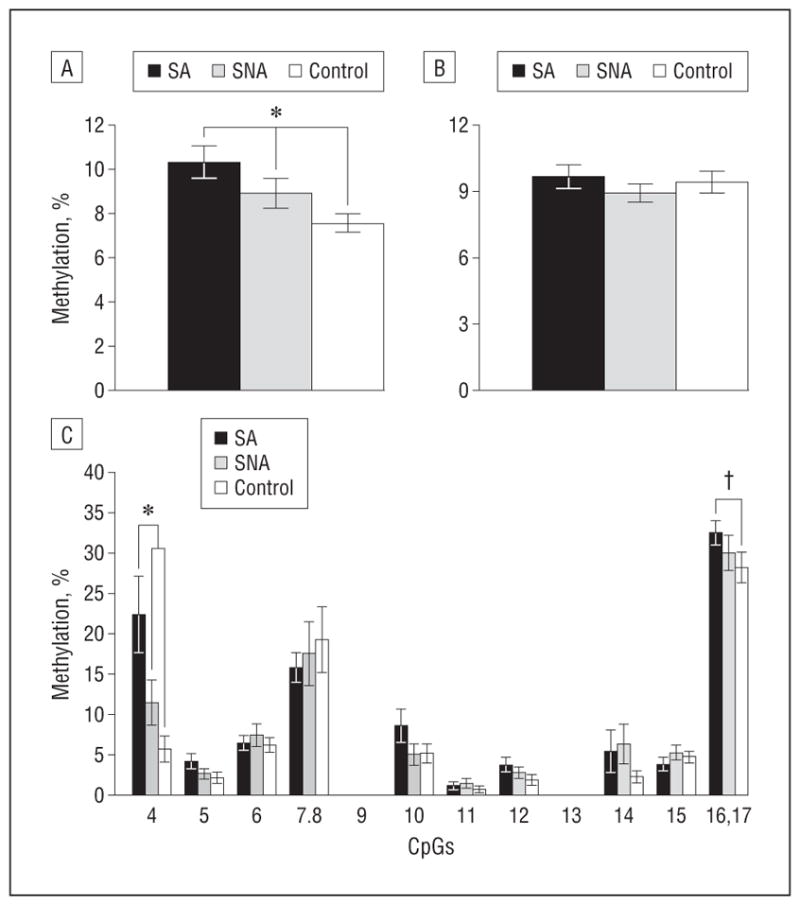
Methylation levels in ALS2 promoters in abused suicide completer (SA), nonabused suicide completer (SNA), and control groups. A, Total percentage of methylation in all CpGs for ALS2 in the neuronal cell fraction. B, Total percentage of methylation in all CpGs in ALS2 promoter in the nonneuronal cell fraction. C, Individual CpG methylation levels in the promoter of ALS2 in the neuronal cell fraction. Abused suicide completer (n = 24), SNA (n = 19), and controls (n = 16). For ALS2, methylation values for CpGs 7 and 8, as well as 16 and 17, are pooled. Values are given as the mean (SE) percentage of methylation.*P<.05. †P<.10.
FUNCTIONAL EFFECTS OF ALS2 PROMOTER HYPERMETHYLATION
We used a dual-reporter cell assay using the neuroblastoma Be(2)c cell line to investigate the functional effects of hypermethylation at CpG site 4 in the ALS2 promoter. We first assessed transcriptional activity of the ALS2 promoter. Two deletion constructs of the ALS2 promoter were generated: a full-length construct composed of a 1000-bp region upstream of the distance from the transcription start site (TSS) of ALS2, including the site of hypermethylation identified at CpG4, and a truncated construct of 543-bp upstream ALS2 TSS, excluding the site of hypermethylation (Figure 3A). The luciferase assays showed significantly higher transcriptional activity for the full-length compared with the truncated construct (t = 2.8, P = .02) (Figure 3B), suggesting that the region identified as hypermethylated in ALS2 participates in the regulation of ALS2 expression.
We then methylated CpG site 4 in the full-length construct to assess whether methylation at this site decreases transcriptional activity. Site-specific methylation (see the Author eAppendix for the complete procedure) significantly decreased ALS2 promoter activity compared with the unmethylated, full-length construct (t = 11.3, P = 3.38E−6) (Figure 3C). Given the transcriptional effect of methylation in the ALS2 promoter, we then used quantitative real-time PCR to examine the hippocampal expression of the 2 major ALS2 transcripts. A 1-way ANOVA performed on ALS2 isoform 1 transcript expression levels revealed no significant group effect (Figure 3D). However, a significant group effect was found for ALS2 isoform 2 (F2,51 = 3.2, P = .049) with post hoc analysis revealing lower levels of ALS2 isoform 2 expression in SA (LSD, P = .02) and SNA (LSD, P = .048) samples compared with control samples (Figure 3E). Together, these results suggest that hypermethylation in the ALS2 promoter decreases its transcriptional activity, leading to decreased expression of isoform 2 in the HPC of all suicide completers.
COMMENT
The results of a genome-wide methylation analysis of HPC suggest that early-life trauma alters methylation levels in several gene promoters. This finding is in accordance with studies23–26,42 performed in rats showing that variability in early-life social environment alters promoter methylation in the brain of offspring. Previous candidate-gene studies in humans, such as the study of the GR gene,21,43 also show increased promoter methylation associated with early-life adversity. Our results indicated that early-life adversity is associated with both hypomethylated and hypermethylated promoters, suggesting that active DNA methylation and demethylation may result from social stressors during early development. Interestingly, prolonged periods of maternal separation during early post-natal development in the mouse are associated with hypomethylation of the Avp gene.27 Likewise, a recent study28 reported hypomethylation in the promoter of the corticotrophin-releasing factor (Crf) gene in socially stressed mice. Taken together with previous studies in rats, these findings suggest dynamic, bidirectional alterations in methylation as a function of early-life adversity.
Clustering analyses of the annotated gene terms suggested that differential methylation associated with early-life adversity occurs across a number of biological processes. It was noteworthy that terms related to cellular/neuronal plasticity were among the most significantly enriched functions. Multiple studies with human and non-human models suggest that adult hippocampal neurogenesis and synaptic architecture are altered by stress. For instance, acute psychosocial stress decreases the number of proliferating cells in nonhuman primates44 and the survival of adult-born hippocampal cells in rats,45 whereas chronic stress decreases hippocampal cell proliferation and neurogenesis, as well as hippocampal volume, in tree shrews, rats, and mice.46–55 Furthermore, chronic pharmacologic antidepressant treatment increases adult hippocampal neurogenesis in rats,49,56 mice,57,58 primates,53,54 and humans,59 whereas suppressing adult hippocampal neurogenesis abolishes the therapeutic effects of antidepressants.58,60,61 Moreover, epigenetic mechanisms have been involved in the regulation of adult neurogenesis in mice.62 Importantly, variations in maternal care regulate both neuronal survival and synaptic density in the rat63–65 as well as synaptic plasticity.65,66 Therefore, our results suggest that early-life stress induces molecular changes regulating methylation patterns in genes involved in neuroplasticity. Given the retrospective design of our study, we cannot directly validate this observation. However, as mentioned previously, these results are consistent with experimental data in rodents, suggesting that adult hippocampal neurogenesis is affected by early-life stress.44–66
Our follow-up analyses focused on the ALS2 gene, the most differentially methylated gene in the cellular/neuronal plasticity cluster and among the most significantly affected genes in the array. The ALS2 gene encodes 2 major mRNA variants generating 2 functional protein isoforms39,40 that regulate small GTPase activity.67,68 The ALS2 gene contains several guanine nucleotide exchange factor domains39,40 stimulating the exchange of GDP to GTP and generating the active form of GTPase68 by interacting with the small GTPase Rab5.67 Small GTPases control a broad spectrum of cellular and molecular processes, including chromatin condensation,69 regulation of actin cytoskeleton organization,70 signaling cascades,71 neuronal morphogenesis,72,73 axonal/neurite growth, and neuroprotection processes.74–76 Although deletions and mutations in the coding sequence of ALS2 are involved in a juvenile recessive form of amyotrophic lateral sclerosis,39,40 recent data suggest that this gene is also associated with psychiatric phenotypes.77 Indeed, loss of ALS2 function is associated with behavioral alterations related to anxietylike phenotypes. For instance, ALS2−/− mice exhibit more freezing episodes in the open field compared with wild-type mice, which is an index of anxiety behavior.78 The ALS2−/− mice also show fewer visits into the open arm and spend less time in both the central area and the open arms of an elevated plus maze compared with wild-type mice.79 Thus, beside its well-characterized peripheral function, ALS2 appears to have important central functions and more work will be required to elucidate the involvement of ALS2 in the regulation of behavior.
Differences in both ALS2 expression and promoter methylation were specific to the neuronal cell fraction. Our results suggest that methylation of the distal region of the ALS2 promoter, and CpG4 in particular, is increased as a result of childhood adversity. Transfection studies with CpG4 methylated constructs indicated that methylation of this site decreases ALS2 promoter activity, an observation that is consistent with the evidence for significantly lower hippocampal expression of isoform 2 in all suicides. Interestingly, CpG4 is located in the binding site for the upstream stimulating factor 1 (USF1). Decreasing binding of USF1 to the ALS2 promoter through increased CpG4 methylation provides a possible mechanism for the in vitro effect on promoter activity as well as the postmortem expression of ALS2 observed in our study. However, given that the expression of ALS2 isoform 2 was decreased in both SAs and SNAs compared with controls, other mechanisms, such as histone modifications, should be considered.
Further experiments will be required to demonstrate the role of USF on ALS2 expression and the possible effects of promoter methylation preventing its action. Interestingly, the transcriptional dynamic of ALS2, generating 2 alternative transcripts, suggests the use of alternative promoters and/or different transcription factors. Moreover, protein levels, as well as neuronal morphologic characteristics, are other important features that need to be investigated.
One limitation of this study was the absence of a group of controls who did not die by suicide and who experienced early-life adversity. Such a group would have allowed us to fully separate the methylation changes associated with childhood adversity from the methylation changes associated with suicide. However, obtaining samples from an age- and sex-matched group of individuals who died suddenly by causes other than suicide and had a history of early-life adversity comparable to the suicide group is logistically challenging because of the very low frequency of severe early-life adversity among controls. In this study, we analyzed only samples of SAs with a history of severe early-life adversity. However, to control for the effect of suicide, all validation studies included a group of suicide completers with no history of childhood adversity. In general, SNAs showed intermediary results, falling between SAs and controls. This finding suggests that, at least for the differentially methylated genes, the effect of early-life adversity on promoter methylation is independent of suicide.
Another important point to consider in this study is the method used to isolate methylated DNA. The me-DIP method is highly sensitive, allowing the enrichment of methylated genomic DNA,32,80 and its advantage over other methods is that it is not limited by the sequence context of methylation-specific enzymes81 and does not require extensive bisulfite treatment.82 This method enriches DNA sequences with both low and high CpG density, although it is known to have a bias toward CpG-rich regions and CpG islands.83 Consequently, it is possible that some differences in gene promoters with low CpG density may not be well represented in this study and that our results represent only a subset of even larger methylation changes that are taking place throughout the genome.
CONCLUSIONS
In summary, this study assessed genome-wide promoter methylation patterns in the HPC of SAs. Our findings indicate that early-life adversity induces a pattern of alterations, including both hypermethylation and hypomethylation, in gene promoters that inversely correlate with gene expression throughout the whole genome. Our data suggest that methylation alterations associated with early-life adversity are significantly enriched in the promoter of genes functionally related to neuronal plasticity. Validation experiments revealed that alterations in methylation of the ALS2 promoter were specific to the neuronal cell fraction. Functional assays revealed that site-specific hypermethylation in the promoter of ALS2 reduces transcriptional activity. This effect was also associated with decreased expression of 1 major mRNA variant in the HPC of suicide completers. These results highlight the importance of the molecular modifications induced by early-life adversity in brain functions. Taken together, this study suggests that early-life adversity induces sustained modifications in DNA methylation across the genome that associate with alterations in transcriptional patterns that may be relevant to help understand suicide risk among individuals who were abused during childhood.
Acknowledgments
Funding/Support: This study was supported by grant MOP84291 from the Canadian Institutes of Health Research (CIHR) and by a National Alliance for Research on Schizophrenia and Depression (NARSAD) Independent Investigator Award to Dr Turecki and by support to the Brain Bank from the Réseau Québécois de Recherche sur le Suicide. Dr Turecki is a chercheur boursier from the FRSQ (Fond de Recherche du Québec). Mr Labonté is supported by a CIHR Frederick Banting and Charles Best doctoral fellowship.
Footnotes
Author Contributions: Drs Szyf, Meaney, and Turecki contributed equally to the study. Mr Labonté has full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Financial Disclosure: None reported.
Online-Only Material: The eFigure is available at http://www.archgenpsychiatry.com. An eAppendix, eTables, and eFigures are available on the authors’ website at http://www.douglas.qc.ca/page/mgss-supplementary.
Additional Contributions: Danièle Gagné, MSc, from the University of Montreal and Simon Young, MSc, from the Lady Davis Institute assisted with flow cytometry experiments. Alexandre Belisle, MSc, from Génome Québec helped with the sequenom platform.
References
- 1.Green JG, McLaughlin KA, Berglund PA, Gruber MJ, Sampson NA, Zaslavsky AM, Kessler RC. Childhood adversities and adult psychiatric disorders in the National Comorbidity Survey replication, I: associations with first onset of DSM-IV disorders. Arch Gen Psychiatry. 2010;67(2):113–123. doi: 10.1001/archgenpsychiatry.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McLaughlin KA, Green JG, Gruber MJ, Sampson NA, Zaslavsky AM, Kessler RC. Childhood adversities and adult psychiatric disorders in the National Comorbidity survey replication, II: associations with persistence of DSM-IV disorders. Arch Gen Psychiatry. 2010;67(2):124–132. doi: 10.1001/archgenpsychiatry.2009.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmes WC, Slap GB. Sexual abuse of boys: definition, prevalence, correlates, sequelae, and management. JAMA. 1998;280(21):1855–1862. doi: 10.1001/jama.280.21.1855. [DOI] [PubMed] [Google Scholar]
- 4.Kessler RC, Davis CG, Kendler KS. Childhood adversity and adult psychiatric disorder in the US National Comorbidity Survey. Psychol Med. 1997;27(5):1101–1119. doi: 10.1017/s0033291797005588. [DOI] [PubMed] [Google Scholar]
- 5.Edwards VJ, Holden GW, Felitti VJ, Anda RF. Relationship between multiple forms of childhood maltreatment and adult mental health in community respondents: results from the Adverse Childhood Experiences study. Am J Psychiatry. 2003;160(8):1453–1460. doi: 10.1176/appi.ajp.160.8.1453. [DOI] [PubMed] [Google Scholar]
- 6.Afifi TO, Enns MW, Cox BJ, Asmundson GJ, Stein MB, Sareen J. Population-attributable fractions of psychiatric disorders and suicide ideation and attempts associated with adverse childhood experiences. Am J Public Health. 2008;98(5):946–952. doi: 10.2105/AJPH.2007.120253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collishaw S, Pickles A, Messer J, Rutter M, Shearer C, Maughan B. Resilience to adult psychopathology following childhood maltreatment: evidence from a community sample. Child Abuse Negl. 2007;31(3):211–229. doi: 10.1016/j.chiabu.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Dinwiddie S, Heath AC, Dunne MP, Bucholz KK, Madden PA, Slutske WS, Bierut LJ, Statham DB, Martin NG. Early sexual abuse and lifetime psychopathology: a co-twin–control study. Psychol Med. 2000;30(1):41–52. doi: 10.1017/s0033291799001373. [DOI] [PubMed] [Google Scholar]
- 9.Evans E, Hawton K, Rodham K. Suicidal phenomena and abuse in adolescents: a review of epidemiological studies. Child Abuse Negl. 2005;29(1):45–58. doi: 10.1016/j.chiabu.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 10.Fergusson DM, Horwood LJ, Lynskey MT. Childhood sexual abuse and psychiatric disorder in young adulthood, II: psychiatric outcomes of childhood sexual abuse. J Am Acad Child Adolesc Psychiatry. 1996;35(10):1365–1374. doi: 10.1097/00004583-199610000-00024. [DOI] [PubMed] [Google Scholar]
- 11.Gladstone GL, Parker GB, Mitchell PB, Malhi GS, Wilhelm K, Austin MP. Implications of childhood trauma for depressed women: an analysis of pathways from childhood sexual abuse to deliberate self-harm and revictimization. Am J Psychiatry. 2004;161(8):1417–1425. doi: 10.1176/appi.ajp.161.8.1417. [DOI] [PubMed] [Google Scholar]
- 12.Mullen PE, Martin JL, Anderson JC, Romans SE, Herbison GP. The long-term impact of the physical, emotional, and sexual abuse of children: a community study. Child Abuse Negl. 1996;20(1):7–21. doi: 10.1016/0145-2134(95)00112-3. [DOI] [PubMed] [Google Scholar]
- 13.Widom CS, DuMont K, Czaja SJ. A prospective investigation of major depressive disorder and comorbidity in abused and neglected children grown up. Arch Gen Psychiatry. 2007;64(1):49–56. doi: 10.1001/archpsyc.64.1.49. [DOI] [PubMed] [Google Scholar]
- 14.Ystgaard M, Hestetun I, Loeb M, Mehlum L. Is there a specific relationship between childhood sexual and physical abuse and repeated suicidal behavior? Child Abuse Negl. 2004;28(8):863–875. doi: 10.1016/j.chiabu.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 15.Heim C, Young LJ, Newport DJ, Mletzko T, Miller AH, Nemeroff CB. Lower CSF oxytocin concentrations in women with a history of childhood abuse. Mol Psychiatry. 2009;14(10):954–958. doi: 10.1038/mp.2008.112. [DOI] [PubMed] [Google Scholar]
- 16.Lara ME, Klein DN. Psychosocial processes underlying the maintenance and persistence of depression: implications for understanding chronic depression. Clin Psychol Rev. 1999;19(5):553–570. doi: 10.1016/s0272-7358(98)00066-x. [DOI] [PubMed] [Google Scholar]
- 17.Lupien SJ, McEwen BS, Gunnar MR, Heim C. Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nat Rev Neurosci. 2009;10(6):434–445. doi: 10.1038/nrn2639. [DOI] [PubMed] [Google Scholar]
- 18.Goodman M, New A, Siever L. Trauma, genes, and the neurobiology of personality disorders. Ann N Y Acad Sci. 2004;1032:104–116. doi: 10.1196/annals.1314.008. [DOI] [PubMed] [Google Scholar]
- 19.Wichers M, Myin-Germeys I, Jacobs N, Peeters F, Kenis G, Derom C, Vlietinck R, Delespaul P, Van Os J. Genetic risk of depression and stress-induced negative affect in daily life. Br J Psychiatry. 2007;191:218–223. doi: 10.1192/bjp.bp.106.032201. [DOI] [PubMed] [Google Scholar]
- 20.Meaney MJ, Szyf M. Maternal care as a model for experience-dependent chromatin plasticity? Trends Neurosci. 2005;28(9):456–463. doi: 10.1016/j.tins.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 21.McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonté B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12(3):342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kappeler L, Meaney MJ. Epigenetics and parental effects. Bioessays. 2010;32(9):818–827. doi: 10.1002/bies.201000015. [DOI] [PubMed] [Google Scholar]
- 23.Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 24.Zhang TY, Hellstrom IC, Bagot RC, Wen X, Diorio J, Meaney MJ. Maternal care and DNA methylation of a glutamic acid decarboxylase 1 promoter in rat hippocampus. J Neurosci. 2010;30(39):13130–13137. doi: 10.1523/JNEUROSCI.1039-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Champagne FA, Weaver IC, Diorio J, Dymov S, Szyf M, Meaney MJ. Maternal care associated with methylation of the estrogen receptor-α1b promoter and estrogen receptor-α expression in the medial preoptic area of female offspring. Endocrinology. 2006;147(6):2909–2915. doi: 10.1210/en.2005-1119. [DOI] [PubMed] [Google Scholar]
- 26.Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65(9):760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmühl Y, Fischer D, Holsboer F, Wotjak CT, Almeida OF, Spengler D. Dynamic DNA methylation programs persistent adverse effects of early-life stress [published correction appears in Nat Neurosci. 2010;13(5):649] Nat Neurosci. 2009;12(12):1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- 28.Elliott E, Ezra-Nevo G, Regev L, Neufeld-Cohen A, Chen A. Resilience to social stress coincides with functional DNA methylation of the Crf gene in adult mice. Nat Neurosci. 2010;13(11):1351–1353. doi: 10.1038/nn.2642. [DOI] [PubMed] [Google Scholar]
- 29.McGowan PO, Meaney MJ, Szyf M. Diet and the epigenetic (re)programming of phenotypic differences in behavior. Brain Res. 2008;1237:12–24. doi: 10.1016/j.brainres.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labuda M, Labuda D, Korab-Laskowska M, Cole DE, Zietkiewicz E, Weissen-bach J, Popowska E, Pronicka E, Root AW, Glorieux FH. Linkage disequilibrium analysis in young populations: pseudo-vitamin D–deficiency rickets and the founder effect in French Canadians. Am J Hum Genet. 1996;59(3):633–643. [PMC free article] [PubMed] [Google Scholar]
- 31.Bifulco A, Brown GW, Harris TO. Childhood Experience of Care and Abuse (CECA): a retrospective interview measure. J Child Psychol Psychiatry. 1994;35(8):1419–1435. doi: 10.1111/j.1469-7610.1994.tb01284.x. [DOI] [PubMed] [Google Scholar]
- 32.Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Pikarski E, Young RA, Niveleau A, Cedar H, Simon I. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38(2):149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 33.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2007. [Google Scholar]
- 34.Klempan TA, Rujescu D, Mérette C, Himmelman C, Sequeira A, Canetti L, Fiori LM, Schneider B, Bureau A, Turecki G. Profiling brain expression of the spermidine/spermine N1-acetyltransferase 1 (SAT1) gene in suicide. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(7):934–943. doi: 10.1002/ajmg.b.30920. [DOI] [PubMed] [Google Scholar]
- 35.Sequeira A, Klempan T, Canetti L, Ffrench-Mullen J, Benkelfat C, Rouleau GA, Turecki G. Patterns of gene expression in the limbic system of suicides with and without major depression. Mol Psychiatry. 2007;12(7):640–655. doi: 10.1038/sj.mp.4001969. [DOI] [PubMed] [Google Scholar]
- 36.Matevossian A, Akbarian S. Neuronal nuclei isolation from human postmortem brain tissue. J Vis Exp. 2008;(20 pt ii):914. doi: 10.3791/914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31(2):89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 38.Mullen RJ, Buck CR, Smith AM. NeuN, a neuronal-specific nuclear protein in vertebrates. Development. 1992;116(1):201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- 39.Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Devon RS, Miyamoto N, Showguchi-Miyata J, Okada Y, Singaraja R, Figlewicz DA, Kwiatkowski T, Hosler BA, Sagie T, Skaug J, Nasir J, Brown RH, Jr, Scherer SW, Rouleau GA, Hayden MR, Ikeda JE. A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet. 2001;29(2):166–173. doi: 10.1038/ng1001-166. [DOI] [PubMed] [Google Scholar]
- 40.Yang Y, Hentati A, Deng HX, Dabbagh O, Sasaki T, Hirano M, Hung WY, Ouahchi K, Yan J, Azim AC, Cole N, Gascon G, Yagmour A, Ben-Hamida M, Pericak-Vance M, Hentati F, Siddique T. The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet. 2001;29(2):160–165. doi: 10.1038/ng1001-160. [DOI] [PubMed] [Google Scholar]
- 41.Gros-Louis F, Gaspar C, Rouleau GA. Genetics of familial and sporadic amyotrophic lateral sclerosis. Biochim Biophys Acta. 2006;1762(11–12):956–972. doi: 10.1016/j.bbadis.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 42.Roth TL, Zoladz PR, Sweatt JD, Diamond DM. Epigenetic modification of hippocampal Bdnf DNA in adult rats in an animal model of post-traumatic stress disorder. J Psychiatr Res. 2011;45(7):919–926. doi: 10.1016/j.jpsychires.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGowan PO, Sasaki A, Huang TC, Unterberger A, Suderman M, Ernst C, Meaney MJ, Turecki G, Szyf M. Promoter-wide hypermethylation of the ribosomal RNA gene promoter in the suicide brain. PLoS One. 2008;3(5):e2085. doi: 10.1371/journal.pone.0002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gould E, Tanapat P, McEwen BS, Flügge G, Fuchs E. Proliferation of granule cell precursors in the dentate gyrus of adult monkeys is diminished by stress. Proc Natl Acad Sci U S A. 1998;95(6):3168–3171. doi: 10.1073/pnas.95.6.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas RM, Hotsenpiller G, Peterson DA. Acute psychosocial stress reduces cell survival in adult hippocampal neurogenesis without altering proliferation. J Neurosci. 2007;27(11):2734–2743. doi: 10.1523/JNEUROSCI.3849-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanapat P, Hastings NB, Rydel TA, Galea LA, Gould E. Exposure to fox odor inhibits cell proliferation in the hippocampus of adult rats via an adrenal hormone-dependent mechanism. J Comp Neurol. 2001;437(4):496–504. doi: 10.1002/cne.1297. [DOI] [PubMed] [Google Scholar]
- 47.Czéh B, Welt T, Fischer AK, Erhardt A, Schmitt W, Müller MB, Toschi N, Fuchs E, Keck ME. Chronic psychosocial stress and concomitant repetitive transcranial magnetic stimulation: effects on stress hormone levels and adult hippocampal neurogenesis. Biol Psychiatry. 2002;52(11):1057–1065. doi: 10.1016/s0006-3223(02)01457-9. [DOI] [PubMed] [Google Scholar]
- 48.Pham K, Nacher J, Hof PR, McEwen BS. Repeated restraint stress suppresses neurogenesis and induces biphasic PSA-NCAM expression in the adult rat dentate gyrus. Eur J Neurosci. 2003;17(4):879–886. doi: 10.1046/j.1460-9568.2003.02513.x. [DOI] [PubMed] [Google Scholar]
- 49.Malberg JE, Duman RS. Cell proliferation in adult hippocampus is decreased by inescapable stress: reversal by fluoxetine treatment. Neuropsychopharmacology. 2003;28(9):1562–1571. doi: 10.1038/sj.npp.1300234. [DOI] [PubMed] [Google Scholar]
- 50.Alonso R, Griebel G, Pavone G, Stemmelin J, Le Fur G, Soubrie P. Blockade of CRF(1) or V(1b) receptors reverses stress-induced suppression of neurogenesis in a mouse model of depression. Mol Psychiatry. 2004;9(3):278–286. doi: 10.1038/sj.mp.4001464. [DOI] [PubMed] [Google Scholar]
- 51.Toth E, Gersner R, Wilf-Yarkoni A, Raizel H, Dar DE, Richter-Levin G, Levit O, Zangen A. Age-dependent effects of chronic stress on brain plasticity and depressive behavior. J Neurochem. 2008;107(2):522–532. doi: 10.1111/j.1471-4159.2008.05642.x. [DOI] [PubMed] [Google Scholar]
- 52.Mineur YS, Belzung C, Crusio WE. Functional implications of decreases in neurogenesis following chronic mild stress in mice. Neuroscience. 2007;150(2):251–259. doi: 10.1016/j.neuroscience.2007.09.045. [DOI] [PubMed] [Google Scholar]
- 53.Czéh B, Michaelis T, Watanabe T, Frahm J, de Biurrun G, van Kampen M, Bartolomucci A, Fuchs E. Stress-induced changes in cerebral metabolites, hippocampal volume, and cell proliferation are prevented by antidepressant treatment with tianeptine. Proc Natl Acad Sci U S A. 2001;98(22):12796–12801. doi: 10.1073/pnas.211427898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gould E, McEwen BS, Tanapat P, Galea LA, Fuchs E. Neurogenesis in the dentate gyrus of the adult tree shrew is regulated by psychosocial stress and NMDA receptor activation. J Neurosci. 1997;17(7):2492–2498. doi: 10.1523/JNEUROSCI.17-07-02492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van der Hart MG, Czéh B, de Biurrun G, Michaelis T, Watanabe T, Natt O, Frahm J, Fuchs E. Substance P receptor antagonist and clomipramine prevent stress-induced alterations in cerebral metabolites, cytogenesis in the dentate gyrus and hippocampal volume. Mol Psychiatry. 2002;7(9):933–941. doi: 10.1038/sj.mp.4001130. [DOI] [PubMed] [Google Scholar]
- 56.Malberg JE, Eisch AJ, Nestler EJ, Duman RS. Chronic antidepressant treatment increases neurogenesis in adult rat hippocampus. J Neurosci. 2000;20(24):9104–9110. doi: 10.1523/JNEUROSCI.20-24-09104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Couillard-Despres S, Wuertinger C, Kandasamy M, Caioni M, Stadler K, Aigner R, Bogdahn U, Aigner L. Ageing abolishes the effects of fluoxetine on neurogenesis. Mol Psychiatry. 2009;14(9):856–864. doi: 10.1038/mp.2008.147. [DOI] [PubMed] [Google Scholar]
- 58.David DJ, Samuels BA, Rainer Q, Wang JW, Marsteller D, Mendez I, Drew M, Craig DA, Guiard BP, Guilloux JP, Artymyshyn RP, Gardier AM, Gerald C, Antonijevic IA, Leonardo ED, Hen R. Neurogenesis-dependent and -independent effects of fluoxetine in an animal model of anxiety/depression. Neuron. 2009;62(4):479–493. doi: 10.1016/j.neuron.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boldrini M, Underwood MD, Hen R, Rosoklija GB, Dwork AJ, John Mann J, Arango V. Antidepressants increase neural progenitor cells in the human hippocampus. Neuropsychopharmacology. 2009;34(11):2376–2389. doi: 10.1038/npp.2009.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang JW, David DJ, Monckton JE, Battaglia F, Hen R. Chronic fluoxetine stimulates maturation and synaptic plasticity of adult-born hippocampal granule cells. J Neurosci. 2008;28(6):1374–1384. doi: 10.1523/JNEUROSCI.3632-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Surget A, Saxe M, Leman S, Ibarguen-Vargas Y, Chalon S, Griebel G, Hen R, Belzung C. Drug-dependent requirement of hippocampal neurogenesis in a model of depression and of antidepressant reversal. Biol Psychiatry. 2008;64(4):293–301. doi: 10.1016/j.biopsych.2008.02.022. [DOI] [PubMed] [Google Scholar]
- 62.Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323(5917):1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu D, Diorio J, Day JC, Francis DD, Meaney MJ. Maternal care, hippocampal synaptogenesis and cognitive development in rats. Nat Neurosci. 2000;3(8):799–806. doi: 10.1038/77702. [DOI] [PubMed] [Google Scholar]
- 64.Bredy TW, Grant RJ, Champagne DL, Meaney MJ. Maternal care influences neuronal survival in the hippocampus of the rat. Eur J Neurosci. 2003;18(10):2903–2909. doi: 10.1111/j.1460-9568.2003.02965.x. [DOI] [PubMed] [Google Scholar]
- 65.Champagne DL, Bagot RC, van Hasselt F, Ramakers G, Meaney MJ, de Kloet ER, Joëls M, Krugers H. Maternal care and hippocampal plasticity: evidence for experience-dependent structural plasticity, altered synaptic functioning, and differential responsiveness to glucocorticoids and stress. J Neurosci. 2008;28(23):6037–6045. doi: 10.1523/JNEUROSCI.0526-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bagot RC, Meaney MJ. Epigenetics and the biological basis of gene×environment interactions. J Am Acad Child Adolesc Psychiatry. 2010;49(8):752–771. doi: 10.1016/j.jaac.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 67.Otomo A, Hadano S, Okada T, Mizumura H, Kunita R, Nishijima H, Showguchi-Miyata J, Yanagisawa Y, Kohiki E, Suga E, Yasuda M, Osuga H, Nishimoto T, Narumiya S, Ikeda JE. ALS2, a novel guanine nucleotide exchange factor for the small GTPase Rab5, is implicated in endosomal dynamics. Hum Mol Genet. 2003;12(14):1671–1687. doi: 10.1093/hmg/ddg184. [DOI] [PubMed] [Google Scholar]
- 68.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294(5545):1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 69.Dasso M. Running on Ran: nuclear transport and the mitotic spindle. Cell. 2001;104(3):321–324. doi: 10.1016/s0092-8674(01)00218-5. [DOI] [PubMed] [Google Scholar]
- 70.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420(6916):629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 71.Snider WD, Zhou FQ, Zhong J, Markus A. Signaling the pathway to regeneration. Neuron. 2002;35(1):13–16. doi: 10.1016/s0896-6273(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 72.Luo L. Rho GTPases in neuronal morphogenesis. Nat Rev Neurosci. 2000;1(3):173–180. doi: 10.1038/35044547. [DOI] [PubMed] [Google Scholar]
- 73.da Silva JS, Dotti CG. Breaking the neuronal sphere: regulation of the actin cytoskeleton in neuritogenesis. Nat Rev Neurosci. 2002;3(9):694–704. doi: 10.1038/nrn918. [DOI] [PubMed] [Google Scholar]
- 74.Otomo A, Kunita R, Suzuki-Utsunomiya K, Mizumura H, Onoe K, Osuga H, Hadano S, Ikeda JE. ALS2/alsin deficiency in neurons leads to mild defects in macropinocytosis and axonal growth. Biochem Biophys Res Commun. 2008;370(1):87–92. doi: 10.1016/j.bbrc.2008.01.177. [DOI] [PubMed] [Google Scholar]
- 75.Lai C, Xie C, McCormack SG, Chiang HC, Michalak MK, Lin X, Chandran J, Shim H, Shimoji M, Cookson MR, Huganir RL, Rothstein JD, Price DL, Wong PC, Martin LJ, Zhu JJ, Cai H. Amyotrophic lateral sclerosis 2–deficiency leads to neuronal degeneration in amyotrophic lateral sclerosis through altered AMPA receptor trafficking. J Neurosci. 2006;26(45):11798–11806. doi: 10.1523/JNEUROSCI.2084-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tudor EL, Perkinton MS, Schmidt A, Ackerley S, Brownlees J, Jacobsen NJ, Byers HL, Ward M, Hall A, Leigh PN, Shaw CE, McLoughlin DM, Miller CC. ALS2/Alsin regulates Rac-PAK signaling and neurite outgrowth. J Biol Chem. 2005;280(41):34735–34740. doi: 10.1074/jbc.M506216200. [DOI] [PubMed] [Google Scholar]
- 77.Pedrosa E, Shah A, Tenore C, Capogna M, Villa C, Guo X, Zheng D, Lachman HM. β-Catenin promoter ChIP-chip reveals potential schizophrenia and bipolar disorder gene network. J Neurogenet. 2010;24(4):182–193. doi: 10.3109/01677063.2010.495182. [DOI] [PubMed] [Google Scholar]
- 78.Devon RS, Orban PC, Gerrow K, Barbieri MA, Schwab C, Cao LP, Helm JR, Bissada N, Cruz-Aguado R, Davidson TL, Witmer J, Metzler M, Lam CK, Tetzlaff W, Simpson EM, McCaffery JM, El-Husseini AE, Leavitt BR, Hayden MR. ALS2-deficient mice exhibit disturbances in endosome trafficking associated with motor behavioral abnormalities. Proc Natl Acad Sci U S A. 2006;103(25):9595–9600. doi: 10.1073/pnas.0510197103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cai H, Lin X, Xie C, Laird FM, Lai C, Wen H, Chiang HC, Shim H, Farah MH, Hoke A, Price DL, Wong PC. Loss of ALS2 function is insufficient to trigger motor neuron degeneration in knock-out mice but predisposes neurons to oxidative stress. J Neurosci. 2005;25(33):7567–7574. doi: 10.1523/JNEUROSCI.1645-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schübeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37(8):853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 81.Fazzari MJ, Greally JM. Epigenomics: beyond CpG islands. Nat Rev Genet. 2004;5(6):446–455. doi: 10.1038/nrg1349. [DOI] [PubMed] [Google Scholar]
- 82.Rakyan VK, Hildmann T, Novik KL, Lewin J, Tost J, Cox AV, Andrews TD, Howe KL, Otto T, Olek A, Fischer J, Gut IG, Berlin K, Beck S. DNA methylation profiling of the human major histocompatibility complex: a pilot study for the Human Epigenome Project. PLoS Biol. 2004;2(12):e405. doi: 10.1371/journal.pbio.0020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, Olshen A, Ballinger T, Zhou X, Forsberg KJ, Gu J, Echipare L, O’Geen H, Lister R, Pelizzola M, Xi Y, Epstein CB, Bernstein BE, Hawkins RD, Ren B, Chung WY, Gu H, Bock C, Gnirke A, Zhang MQ, Haussler D, Ecker JR, Li W, Farnham PJ, Waterland RA, Meissner A, Marra MA, Hirst M, Milosavljevic A, Costello JF. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28(10):1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]