Abstract
Background
Protein kinases catalyze the transfer of a phosphate moiety from a phosphate donor to the substrate molecule, thus playing critical roles in cell signaling and metabolism. Although plant genomes contain more than 1000 genes that encode kinases, knowledge is limited about the function of each of these kinases. A major obstacle that hinders progress towards kinase characterization is functional redundancy. To address this challenge, we previously developed the rice kinase database (RKD) that integrated omics-scale data within a phylogenetics context.
Results
An updated version of rice kinase database (RKD) that contains metadata derived from NCBI GEO expression datasets has been developed. RKD 2.0 facilitates in-depth transcriptomic analyses of kinase-encoding genes in diverse rice tissues and in response to biotic and abiotic stresses and hormone treatments. We identified 261 kinases specifically expressed in particular tissues, 130 that are significantly up- regulated in response to biotic stress, 296 in response to abiotic stress, and 260 in response to hormones. Based on this update and Pearson correlation coefficient (PCC) analysis, we estimated that 19 out of 26 genes characterized through loss-of-function studies confer dominant functions. These were selected because they either had paralogous members with PCC values of <0.5 or had no paralog.
Conclusion
Compared with the previous version of RKD, RKD 2.0 enables more effective estimations of functional redundancy or dominance because it uses comprehensive expression profiles rather than individual profiles. The integrated analysis of RKD with PCC establishes a single platform for researchers to select rice kinases for functional analyses.
Electronic supplementary material
The online version of this article (doi:10.1186/s12284-016-0106-5) contains supplementary material, which is available to authorized users.
Keywords: Rice kinase database, Phylogenomics, Functional redundancy, Mutant analysis, Meta-analysis, Pearson correlation coefficient, Transcriptome
Background
Protein kinases are involved in diverse cellular and biological processes. Elucidation of their roles is limited; functions for only 61 kinases in rice (Oryza sativa) have been ascribed (Yamamoto et al. 2012). This subset represents approximately 4.1 % of all kinase genes in that genome. Since sequencing of the entire rice genome was completed in 2005 (IRGSP 2005), diverse omics datasets have been generated that include genome-wide expression analysis using microarrays as well as RNA-seq, re-sequencing of 3000 rice accessions, protein–protein interactomes, gene regulatory networks, and metabolic pathways. Because these databases use very different platforms for formatting, it has been a challenge to integrate publicly available datasets to facilitate functional genomics studies (Chandran and Jung 2014; Alexandrov et al. 2015).
Functional redundancy is a significant obstacle to the identification of gene functions. Completed whole-genome sequences enable researchers to estimate the extent of functional redundancy due to gene duplications. From 20 to 60 % of the rice genome is tandemly or segmentally duplicated (Itoh et al. 2007; Lin et al. 2008). In addition, 21,403 proteins in rice have been assigned to 3856 paralogous protein family groups, thus demonstrating that more than 50 % of all rice genes annotated to non-transposable element (non-TE) gene models have functional redundancy because of their paralogous relationships (Lin et al. 2008). However, not all paralogous genes have such redundancy. Knock-out mutant analysis has identified light response-defective phenotypes in T-DNA insertional mutant lines for nine gene family members that are predominantly expressed under high illumination (Jung et al. 2008). In addition, 79 of 127 ubiquitously expressed genes which have been functionally characterized through loss-of-function studies belong to gene families (Jung et al. 2015). In both cases, functional dominance of the characterized genes within a particular family has been readily estimated through phylogenomics analyses that integrate metadata from diverse expression datasets within a phylogenetics context (Jung et al. 2015).
To date, six phylogenomics databases have been developed for rice. These include kinase, glycosyltransferase, glycoside hydrolase, transcription factor, transporter, and cytochrome p450 databases (Jung et al. 2015). Rice kinase database (RKD) was the first of these to be established, and it has provided a basic framework for all others (Dardick et al. 2007; Jung et al. 2010). Application of RKD in facilitating functional genomic studies was previously demonstrated. For example, phylogenomics approach played a major role in the identification of a set of MAPK, MAPKK, and MAPKKK genes that are part of same signaling cascades, and in predicting the regulatory model of a light-inducible kinase gene (Jung et al. 2010).
One unique feature of RKD compared with other databases is an option to integrate protein–protein interaction network data based on yeast two-hybrid or tandem affinity purification tagging analyses (Dardick et al. 2007). A large set of this integrated transcriptomic data, retrieved from the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO; www.ncbi.nlm.nih.gov/geo/. accessed on November 21, 2015), is a main provider of diverse biological information about interesting kinase genes for further functional studies (Cao et al. 2008; Jung et al. 2010). The current format of those databases for integrating transcriptome data has proven cumbersome because users must mine data from a list of datasets within NCBI GEO. However, Genevestigator, RiceXPro, and Rice Oligonucleotide Array Database (ROAD; www.ricearray.org/, accessed on December 2, 2015) have recently integrated these meta-expression data through classification and reconstruction to enhance accessibility (Zimmermann et al. 2008; Cao et al. 2012; Sato et al. 2013).
Here, we describe RKD 2.0, an updated RKD that integrates metadata from genes expressed in particular tissues and in response to abiotic/biotic stresses and hormone treatments. Using these data, a genome-wide meta-analysis of expression patterns for all predicted rice kinases have been performed and identified kinase-encoding genes with distinct expression patterns. Also, steps involved in the construction and application of RKD 2.0 has been discussed.
Results and discussion
Updated features of RKD 2.0
The earlier version, RKD, presented phylogenomics data for 1508 kinases comprising 65 subfamilies, and a group of kinase genes that could not be assigned to any of those subfamilies (Dardick et al. 2007; Jung et al. 2010). Because that version utilized all gene models (splicing variants), the integrated transcriptome information showed redundant expression data for multiple models produced from a particular locus. However, for the current version, we selected one representative model per locus based on the information available from rice genome annotation project (RGAP) database (Ouyang et al. 2007) that could provide a simplified and comprehensive visualization of the integrated data. Likewise, instead of taking individual datasets from NCBI GEO (Barrett et al. 2011), Affymetrix-based anatomical metadata (ROAD; http://www.ricearray.org/expression/experiment_search.php, accessed on December 2, 2015), used in ROAD (Cao et al. 2012) have been integrated within the phylogenic tree context. Furthermore, meta-analysis with diverse datasets of genes that are expressed in response to biotic and abiotic stresses as well as hormones treatment was performed. Unlike the average normalized intensity data used for an anatomical meta-expression database, log2 fold-change data for treatment versus control have been introduced. Differential expression in response to five pathogens (Magnaporthe grisea, MG; Magnaporthe oryzae, MO; rice stripe virus, RSV; Xanthomonas oryzae pv. oryzae, Xoo; and the brown plant hopper, BPH), in addition to five abiotic stresses (drought, salt, cold, heat, and submergence) and six hormones (abscisic acid, ABA; brassinolide, BL; gibberellic acid, GA; indole-3-acetic acid, IAA; trans-zeatin, tZ; and jasmonic acid, JA) were also analyzed. The results are presented in Additional files 1, 2, 3, 4 and 5: Tables S1-S5. The expression data used for meta-analysis from anatomical tissues and in stress responses were based on Affymetrix array data downloaded from the NCBI GEO (Cao et al. 2012; Zimmermann et al. 2008). For hormone responses, those expression data were based on Agilent 44 K arrays (GSE39429) (NCBI GEO; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39429, accessed on November 21, 2015) (Sato et al. 2013). As an alternative platform for meta-analysis of microarray data, RNASeq data of anatomical and abiotic stress treated samples were integrated to the database (Secco et al. 2013; Wang et al. 2015). All of these updated features are available in the new rice kinase database (RKD 2.0; http://ricephylogenomics-khu.org/kinase/index.php, accessed on February 20, 2016).
Meta-analysis of expression data available for kinase genes with known functions
Using information from OGRO, the Overview of Functionally Characterized Genes in Rice online database (OGRO; http://qtaro.abr.affrc.go.jp/ogro/table. Accessed 20 October 2015) (Yamamoto et al. 2012), 61 RGAP loci related to kinases that have been functionally characterized were retrieved. Among them, function of a subset of kinase genes were related to more than one trait. Thus, 61 kinases are associated with 124 traits. Although these genes were broadly classified as conferring morphological traits (35 genes), physiological traits (27), stress responses resistance (59), or other biological functions (3) (Table 1), 63 of them are associated with multiple functions. Among those for morphological traits, 10 genes each target for culm leaves and dwarfism. Another three genes each are related to panicle/flower formation and seeds, four to roots and five to seedling development. The major physiological effects of rice mutants lead to sterility (6 genes), indicating that the genes in this category are important for fertilization and embryo development. The physiological-trait classification also includes four genes related to source activity, six for flowering, two for eating quality, and one for seed production. In that group, mutations of three genes are attributed to germination dormancy. In addition to the panicle-dependent effect, physiological effects can be observed in the roots (two genes) and culm leaves (one gene). Mutation of one gene causes lethality. The stress-response classification is subdivided into genes that function in biotic stresses, abiotic stresses, and resistance to mechanical stresses such as lodging. With regard to biotic stresses, 15 genes have roles in blast resistance, eight including Xa21 gene involved in bacterial blight resistance (Song et al. 1995) , function of a gene associated with insect resistance and one in resistance to other diseases. For abiotic stresses, eight genes are associated with drought, 15 with salinity, six with chilling, and another five with other types of stresses. Analysis of previously characterized genes indicates that kinases plays diverse role in physiology and growth of rice plants.
Table 1.
Functional information for kinase genes with known functions
| Type of meta-expression | Character_major | LOC_id | Gene name | Gene symbol | Method | character_minor | DOI reference |
|---|---|---|---|---|---|---|---|
| IAA | Morphological trait | LOC_Os01g52050.1 | dwarf 61 | d61 | Mutant | Culm leaf | 10.1105/tpc.12.9.1591 |
| Anther/Pollen | Morphological trait | LOC_Os02g58610.1 | N-acetylglucosaminyltransferase I | GnTI | Mutant | Culm leaf | 10.1111/tpj.12087 |
| Root, BPH | Morphological trait | LOC_Os06g50920.1 | increased leaf angle1 | ila1 | Mutant | Culm leaf | 10.1105/tpc.111.093419 |
| Morphological trait | LOC_Os02g05980.1 | leucine-rich repeat receptor-like kinase1 | LRK1 | Overexpression | Culm leaf | 10.1111/j.1467-7652.2009.00428.x | |
| Morphological trait | LOC_Os02g05980.1 | Leucine-rich repeat receptor-like kinase 1 | LRK1 | Overexpression | Culm leaf | 10.1007/s10529-012-1054-9 | |
| Morphological trait | LOC_Os02g40860.1 | Low Temperature Growth 1 | LTG1 | Others | Culm leaf | 10.1111/tpj.12487 | |
| Morphological trait | LOC_Os08g07760.1 | OsBAK1 | OsBAK1 | Knockdown | Culm leaf | 10.1111/j.1467-7652.2009.00444.x | |
| Morphological trait | LOC_Os05g40770.1 | Leucine-rich-repeat receptor-like kinases | OsRPK1 | Knockdown Overexpression | Culm leaf | 10.1016/j.bbagen.2014.01.003 | |
| Morphological trait | LOC_Os02g32610.2 | CONSTITUTIVE TRIPLE-RESPONSE2 | OsCTR2 | Mutant | Culm leaf | 10.1093/jxb/ert272 | |
| Morphological trait | LOC_Os04g48760.1 | xiao | xiao | Mutant | Culm leaf | 10.1111/j.1365-313X.2011.04877.x | |
| IAA | Morphological trait | LOC_Os01g52050.1 | dwarf 61 | d61 | Mutant | Dwarf | 10.1105/tpc.12.9.1591 |
| Anther/Pollen | Morphological trait | LOC_Os02g58610.1 | N-acetylglucosaminyltransferase I | GnTI | Mutant | Dwarf | 10.1111/tpj.12087 |
| Morphological trait | LOC_Os02g05980.1 | leucine-rich repeat receptor-like kinase1 | LRK1 | Overexpression | Dwarf | 10.1111/j.1467-7652.2009.00428.x | |
| Morphological trait | LOC_Os02g05980.1 | Leucine-rich repeat receptor-like kinase 1 | LRK1 | Overexpression | Dwarf | 10.1007/s10529-012-1054-9 | |
| Morphological trait | LOC_Os02g40860.1 | Low Temperature Growth 1 | LTG1 | Others | Dwarf | 10.1111/tpj.12487 | |
| Leaf/Flag leaf/Shoot, ABA | Morphological trait | LOC_Os02g36570.1 | activity of bc1 complex 1–2 | OsABC1-2 | Mutant | Dwarf | 10.1016/j.gene.2012.02.017 |
| Morphological trait | LOC_Os08g07760.1 | OsBAK1 | OsBAK1 | Knockdown | Dwarf | 10.1111/j.1467-7652.2009.00444.x | |
| Morphological trait | LOC_Os02g03410.1 | Ca2 + −dependent protein kinase 4 | OsCPK4 | Knockdown | Dwarf | 10.1104/pp.113.230268 | |
| Morphological trait | LOC_Os05g40770.1 | Leucine-rich-repeat receptor-like kinases | OsRPK1 | Knockdown Overexpression | Dwarf | 10.1016/j.bbagen.2014.01.003 | |
| Morphological trait | LOC_Os04g48760.1 | xiao | xiao | Mutant | Dwarf | 10.1111/j.1365-313X.2011.04877.x | |
| Morphological trait | LOC_Os06g50340.1 | FLORAL ORGAN NUMBER1 | fon1 | Mutant | Panicle flower | 10.1242/dev.01441 | |
| Morphological trait | LOC_Os02g05980.1 | leucine-rich repeat receptor-like kinase1 | LRK1 | Overexpression | Panicle flower | 10.1111/j.1467-7652.2009.00428.x | |
| SAM/Panicle, IAA | Morphological trait | LOC_Os12g42020.1 | pinoid | OsPID | Overexpression | Panicle flower | 10.1093/pcp/pcm024 |
| Morphological trait | LOC_Os02g14120.1 | defective in outer cell layer specification 1 | Docs1 | Mutant | Root | 10.1111/j.1365-313X.2011.04824.x | |
| Morphological trait | LOC_Os02g40860.1 | Casein Kinase I | OsCKI1 | Knockdown | Root | 10.1046/j.1365-313X.2003.01866.x | |
| SAM/Panicle, IAA | Morphological trait | LOC_Os12g42020.1 | pinoid | OsPID | Overexpression | Root | 10.1093/pcp/pcm024 |
| Morphological trait | LOC_Os05g40770.1 | Leucine-rich-repeat receptor-like kinases | OsRPK1 | Knockdown Overexpression | Root | 10.1016/j.bbagen.2014.01.003 | |
| Leaf/Flag leaf/Shoot, ABA | Morphological trait | LOC_Os02g36570.1 | activity of bc1 complex 1–2 | OsABC1-2 | Mutant | Seed | 10.1016/j.gene.2012.02.017 |
| Ubiquitous, JA | Morphological trait | LOC_Os03g03660.1 | calcium-dependent protein kinase 1 | OsCDPK1 | Knockdown Overexpression | Seed | 10.1007/s11103-012-0006-z |
| Morphological trait | LOC_Os04g48760.1 | xiao | xiao | Mutant | Seed | 10.1111/j.1365-313X.2011.04877.x | |
| Ubiquitous, JA | Morphological trait | LOC_Os03g03660.1 | calcium-dependent protein kinase 1 | OsCDPK1 | Knockdown Overexpression | Shoot seedling | 10.1007/s11103-012-0006-z |
| tZ | Morphological trait | LOC_Os03g20380.1 | Calcineurin B-like protein-interacting protein kinases 31 | oscipk31 | Mutant | Shoot seedling | 10.1007/s10059-010-0084-1 |
| JA | Morphological trait | LOC_Os08g42750.1 | Calcium-dependent protein kinase21 | OsCPK21 | Overexpression | Shoot seedling | 10.1007/s11103-010-9717-1 |
| Morphological trait | LOC_Os02g32610.2 | CONSTITUTIVE TRIPLE-RESPONSE2 | OsCTR2 | Mutant | Shoot seedling | 10.1093/jxb/ert272 | |
| Morphological trait | LOC_Os05g45420.1 | Snf1-related protein kinases 1a | snrk1a | Mutant | Shoot seedling | 10.1105/tpc.105.037887 | |
| SAM/Panicle, IAA | Physiological trait | LOC_Os07g32480.1 | Bub1-Related Kinase 1 | BRK1 | Mutant | Sterility | 10.1105/tpc.112.105874 |
| RSV, IAA | Physiological trait | LOC_Os01g68870.1 | MULTIPLE SPOROCYTE 1 | msp1 | Mutant | Sterility | 10.1105/tpc.012401 |
| Root | Physiological trait | LOC_Os07g05620.1 | CBL (Calcineurin B-Like) Interacting Protein Kinase 23 | OsCIPK23 | Knockdown Overexpression | Sterility | 10.1016/S1673-8527(08)60073-9 |
| JA | Physiological trait | LOC_Os09g38850.1 | DEFECT IN EARLY EMBRYO SAC1 | OsDEES1 | Knockdown | Sterility | 10.1104/pp.112.203943 |
| Physiological trait | LOC_Os07g08000.1 | NIMA-related kinase 3 | OsNek3 | Overexpression | Sterility | 10.1093/pcp/pcp026 | |
| Physiological trait | LOC_Os04g48760.1 | xiao | xiao | Mutant | Sterility | 10.1111/j.1365-313X.2011.04877.x | |
| Anther/Pollen | Physiological trait | LOC_Os02g58610.1 | N-acetylglucosaminyltransferase I | GnTI | Mutant | Source activity | 10.1111/tpj.12087 |
| Leaf/Flag leaf/Shoot, ABA | Physiological trait | LOC_Os02g36570.1 | activity of bc1 complex 1–2 | OsABC1-2 | Mutant | Source activity | 10.1016/j.gene.2012.02.017 |
| Leaf/Flag leaf/Shoot | Physiological trait | LOC_Os05g40180.1 | A serine/threo- nine protein kinase 8 | OsSTN8 | Mutant | Source activity | 10.1111/tpj.12331 |
| Physiological trait | LOC_Os11g01140.1 | Phototropin1a | phot1a | Mutant | Source activity | 10.1007/s11103-008-9442-1 | |
| Physiological trait | LOC_Os03g57940.1 | earlier flowering1 | el1 | Mutant | Flowering | 10.1038/emboj.2010.75 | |
| Physiological trait | LOC_Os03g57940.1 | Heading date 16 | Hd16 | Natural variation | Flowering | 10.1111/tpj.12268 | |
| Physiological trait | LOC_Os03g55389.1 | heading date 6 | Hd6 | Natural variation | Flowering | 10.1073/pnas.111136798 | |
| Physiological trait | LOC_Os02g40860.1 | Low Temperature Growth 1 | LTG1 | Others | Flowering | 10.1111/tpj.12487 | |
| Physiological trait | LOC_Os03g56270.1 | NUTRITION RESPONSE AND ROOT GROWTH b | NRRb | Knockdown | Flowering | 10.1093/mp/sss157 | |
| Physiological trait | LOC_Os02g32610.2 | CONSTITUTIVE TRIPLE-RESPONSE2 | OsCTR2 | Mutant | Flowering | 10.1093/jxb/ert272 | |
| Physiological trait | LOC_Os10g39420.1 | seed-specific protein kinase | SPK | Knockdown | Eating quality | 10.1093/pcp/pch122 | |
| Physiological trait | LOC_Os10g39420.1 | seed-specific protein kinase | SPK | Knockdown | Eating quality | 10.1105/tpc.010454 | |
| tZ | Physiological trait | LOC_Os03g20380.1 | Calcineurin B-like protein-interacting protein kinases 31 | oscipk31 | Mutant | Germination dormancy | 10.1007/s10059-010-0084-1 |
| Physiological trait | LOC_Os04g12540.1 | lectin receptor kinase | OslecRK | Knockdown | Germination dormancy | 10.1111/tpj.12328 | |
| Physiological trait | LOC_Os05g45420.1 | Snf1-related protein kinases 1a | snrk1a | Mutant | Germination dormancy | 10.1105/tpc.105.037887 | |
| Physiological trait | LOC_Os05g40770.1 | Leucine-rich-repeat receptor-like kinases | OsRPK1 | Knockdown Overexpression | Seed | 10.1016/j.bbagen.2014.01.003 | |
| Physiological trait | LOC_Os05g41090.1 | Ca2 + calmodulin-de-pendent protein kinase | OsCCaMK | Mutant | Root | 10.1128/AEM.03646-13. | |
| Physiological trait | LOC_Os02g03410.1 | Ca2 + −dependent protein kinase 4 | OsCPK4 | Overexpression | Root | 10.1104/pp.113.230268 | |
| Root | Physiological trait | LOC_Os05g40770.1 | Leucine-rich-repeat receptor-like kinases | OsRPK1 | Knockdown Overexpression | Culm leaf | 10.1016/j.bbagen.2014.01.003 |
| Physiological trait | LOC_Os02g40860.1 | hybrid breakdown 2 | hbd2 | Natural variation | Lethality | 10.1007/s00438-010-0514-y | |
| Physiological trait | LOC_Os02g05480.1 | TEY-type rice mitogen-activated protein kinase 3 | OsMPK3 | Others | Others | 10.1007/s00299-014-1620-9 | |
| Cold | Resistance or Tolerance | LOC_Os09g36320.1 | BROAD-SPECTRUM RESISTANCE 1 | BSR1 | Overexpression | Blast resistance | 10.1111/j.1467-7652.2010.00568.x |
| Heat, JA | Resistance or Tolerance | LOC_Os07g22710.1 | calcium-dependent protein kinase (CDPK) 18 | CPK18 | Knockdown | Blast resistance | 10.1105/tpc.114.126441 |
| Resistance or Tolerance | LOC_Os10g40100.1 | receptor-like cytoplasmic kinase gene | NRRB | Knockdown | Blast resistance | 10.1007/s11033-014-3069-x | |
| BPH | Resistance or Tolerance | LOC_Os03g06410.1 | accelerated cell death and resistance 1 | OsACDR1 | Knockdown Overexpression | Blast resistance | 10.1007/s10059-009-0161-5 |
| Resistance or Tolerance | LOC_Os03g12730.1 | blast resistance-related 1 | OsBRR1 | Knockdown Overexpression | Blast resistance | 10.1007/s00425-009-0951-1 | |
| JA | Resistance or Tolerance | LOC_Os08g42580.1 | chitin elicitor receptor kinase1 | OsCERK1 | Knockdown | Blast resistance | 10.1111/j.1365-313X.2010.04324.x |
| Cold | Resistance or Tolerance | LOC_Os03g57450.1 | Calcium-dependent protein kinase 10 | OsCPK10 | Overexpression | Blast resistance | 10.1016/j.plaphy.2013.10.004 |
| Resistance or Tolerance | LOC_Os04g47300.1 | Calcium-dependent protein kinase 12 | OsCPK12 | Mutant | Blast resistance | 10.1111/j.1365-313X.2011.04766.x | |
| Resistance or Tolerance | LOC_Os04g12540.1 | lectin receptor kinase | OslecRK | Knockdown | Blast resistance | 10.1111/tpj.12328 | |
| Resistance or Tolerance | LOC_Os03g17700.1 | Mitogen-activated protein kinase5 | OsMAPK5 | Knockdown Overexpression | Blast resistance | 10.1105/tpc.008714 | |
| Resistance or Tolerance | LOC_Os02g05480.1 | TEY-type rice mitogen-activated protein kinase 3 | OsMPK3 | Others | Blast resistance | 10.1007/s00299-014-1620-9 | |
| Cold | Resistance or Tolerance | LOC_Os04g41160.1 | oxidative signal inducible 1 | OsOxi1 | Overexpression | Blast resistance | 10.1093/pcp/pcq132 |
| Resistance or Tolerance | LOC_Os01g65230.1 | 3-phosphoinositide-dependent protein kinase 1 | OsPdk1 | Mutant | Blast resistance | 10.1093/pcp/pcq167 | |
| Cold | Resistance or Tolerance | LOC_Os04g38480.1 | somatic embryogenesis receptor-like kinase 1 | OsSERK1 | Overexpression | Blast resistance | 10.1007/s00425-005-1534-4 |
| Resistance or Tolerance | LOC_Os06g29810.1 | Pi-d2 | Pi-d2 | Natural variation | Blast resistance | 10.1111/j.1365-313X.2006.02739.x | |
| Cold | Resistance or Tolerance | LOC_Os09g36320.1 | BROAD-SPECTRUM RESISTANCE 1 | BSR1 | Overexpression | Bacterial blight resistance | 10.1111/j.1467-7652.2010.00568.x |
| BPH | Resistance or Tolerance | LOC_Os03g06410.1 | enhanced disease resistance 1 | OsEDR1 | Mutant | Bacterial blight resistance | 10.1111/j.1365-3040.2010.02219.x |
| Resistance or Tolerance | LOC_Os04g12540.1 | lectin receptor kinase | OslecRK | Knockdown | Bacterial blight resistance | 10.1111/tpj.12328 | |
| Resistance or Tolerance | LOC_Os03g17700.1 | Mitogen-activated protein kinase5 | OsMAPK5 | Knockdown Overexpression | Bacterial blight resistance | 10.1105/tpc.008714 | |
| Resistance or Tolerance | LOC_Os10g38950.1 | Mitogen-Activated Protein Kinase Phosphatase6 | OsMPK6 | Mutant | Bacterial blight resistance | 10.1007/s00425-007-0541-z | |
| Cold, JA | Resistance or Tolerance | LOC_Os04g41160.1 | oxidative signal inducible 1 | OsOxi1 | Overexpression | Bacterial blight resistance | 10.1093/pcp/pcq132 |
| Resistance or Tolerance | LOC_Os01g65230.1 | 3-phosphoinositide-dependent protein kinase 1 | OsPdk1 | Mutant | Bacterial blight resistance | 10.1093/pcp/pcq167 | |
| Resistance or Tolerance | LOC_Os11g35500.1 | Xa21 | Xa21 | Natural variation | Bacterial blight resistance | 10.1126/science.270.5243.1804 | |
| Resistance or Tolerance | LOC_Os03g17700.1 | Mitogen-activated protein kinase5 | OsMAPK5 | Knockdown Overexpression | Other disease resistance | 10.1105/tpc.008714 | |
| Resistance or Tolerance | LOC_Os04g12540.1 | lectin receptor kinase | OslecRK | Knockdown | Insect resistance | 10.1111/tpj.12328 | |
| Cold | Resistance or Tolerance | LOC_Os02g50970.1 | drought-hypersensitive mutant1 | dsm1 | Mutant | Drought tolerance | 10.1104/pp.109.149856 |
| JA | Resistance or Tolerance | LOC_Os03g03660.1 | calcium-dependent protein kinase 1 | OsCDPK1 | Knockdown Overexpression | Drought tolerance | 10.1007/s11103-012-0006-z |
| Cold | Resistance or Tolerance | LOC_Os04g49510.1 | calcium-dependent protein kinase 7 | OsCDPK7 | Overexpression | Drought tolerance | 10.1046/j.1365-313x.2000.00787.x |
| Drought/Salt, M. grisea, ABA | Resistance or Tolerance | LOC_Os01g55450.1 | calcineurin B-like protein-interacting protein kinase12 | OsCIPK12 | Overexpression | Drought tolerance | 10.1104/pp.107.101295 |
| Cold, M. oryzae | Resistance or Tolerance | LOC_Os02g03410.1 | Ca2 + −dependent protein kinase 4 | OsCPK4 | Knockdown | Drought tolerance | 10.1104/pp.113.230268 |
| Resistance or Tolerance | LOC_Os01g10840.1 | glycogen synthase kinase3-like gene 1 | OsGSK1 | Mutant | Drought tolerance | 10.1007/s11103-007-9213-4 | |
| Resistance or Tolerance | LOC_Os03g17700.1 | Mitogen-activated protein kinase5 | OsMAPK5 | Knockdown Overexpression | Drought tolerance | 10.1105/tpc.008714 | |
| Resistance or Tolerance | LOC_Os06g03970.1 | stress-induced protein kinase gene 1 | sik1 | Mutant | Drought tolerance | 10.1111/j.1365-313X.2010.04146.x | |
| Cold | Resistance or Tolerance | LOC_Os04g49510.1 | calcium-dependent protein kinase 7 | OsCDPK7 | Overexpression | Salinity tolerance | 10.1046/j.1365-313x.2000.00787.x |
| BPH | Resistance or Tolerance | LOC_Os11g02240.1 | calcineurin B-like protein-interacting protein kinase15 | OsCIPK15 | Overexpression | Salinity tolerance | 10.1104/pp.107.101295 |
| Resistance or Tolerance | LOC_Os07g05620.1 | CBL (Calcineurin B-Like) Interacting Protein Kinase 23 | OsCIPK23 | Knockdown Overexpression | Salinity tolerance | 10.1016/S1673-8527(08)60073-9 | |
| tZ | Resistance or Tolerance | LOC_Os03g20380.1 | Calcineurin B-like protein-interacting protein kinases 31 | oscipk31 | Mutant | Salinity tolerance | 10.1007/s10059-010-0084-1 |
| Resistance or Tolerance | LOC_Os04g47300.1 | Calcium-dependent protein kinase 12 | OsCPK12 | Mutant | Salinity tolerance | 10.1111/j.1365-313X.2011.04766.x | |
| JA | Resistance or Tolerance | LOC_Os08g42750.1 | Calcium-dependent protein kinase21 | OsCPK21 | Overexpression | Salinity tolerance | 10.1007/s11103-010-9717-1 |
| Cold, M. oryzae | Resistance or Tolerance | LOC_Os02g03410.1 | Ca2 + −dependent protein kinase 4 | OsCPK4 | Knockdown | Salinity tolerance | 10.1104/pp.113.230268 |
| Resistance or Tolerance | LOC_Os01g10840.1 | glycogen synthase kinase3-like gene 1 | OsGSK1 | Mutant | Salinity tolerance | 10.1007/s11103-007-9213-4 | |
| Resistance or Tolerance | LOC_Os02g05480.1 | Mitogen-activated protein kinase33 | OsMAPK33 | Overexpression | Salinity tolerance | 10.1007/s12038-011-9002-8 | |
| Resistance or Tolerance | LOC_Os03g17700.1 | Mitogen-activated protein kinase5 | OsMAPK5 | Knockdown Overexpression | Salinity tolerance | 10.1105/tpc.008714 | |
| Resistance or Tolerance | LOC_Os06g05520.1 | mitogen-activated protein kinase kinase | OsMKK1 (MAPKK) | Knockdown | Salinity tolerance | 10.1016/j.plantsci.2014.08.007 | |
| ABA | Resistance or Tolerance | LOC_Os01g64970.1 | SNF1-type serine-threonine protein kinase4 | SAPK4 | Overexpression | Salinity tolerance | 10.1186/1471-2229-8-49 |
| Resistance or Tolerance | LOC_Os06g03970.1 | stress-induced protein kinase gene 1 | sik1 | Mutant | Salinity tolerance | 10.1111/j.1365-313X.2010.04146.x | |
| JA | Resistance or Tolerance | LOC_Os02g42780.1 | Salt Intolerance 1 | SIT1 | Knockdown Overexpression | Salinity tolerance | 10.1105/tpc.114.125187 |
| Cold | Resistance or Tolerance | LOC_Os04g44900.1 | Salt Intolerance 2 | SIT2 | Knockdown | Salinity tolerance | 10.1105/tpc.114.125187 |
| JA | Resistance or Tolerance | LOC_Os03g03660.1 | calcium-dependent protein kinase 13 | CDPK13 | Overexpression | Cold tolerance | 10.1007/s00438-007-0220-6 |
| JA | Resistance or Tolerance | LOC_Os03g03660.1 | calcium-dependent protein kinase 13 | OsCDPK13 | Overexpression | Cold tolerance | 10.1007/s11103-004-1178-y |
| tZ | Resistance or Tolerance | LOC_Os03g20380.1 | calcineurin B-like protein-interacting protein kinase03 | OsCIPK03 | Overexpression | Cold tolerance | 10.1104/pp.107.101295 |
| Resistance or Tolerance | LOC_Os01g10840.1 | glycogen synthase kinase3-like gene 1 | OsGSK1 | Mutant | Cold tolerance | 10.1007/s11103-007-9213-4 | |
| Cold | Resistance or Tolerance | LOC_Os03g17700.1 | Mitogen-activated protein kinase5 | OsMAPK5 | Knockdown Overexpression | Cold tolerance | 10.1105/tpc.008714 |
| Cold, M. oryzae, IAA | Resistance or Tolerance | LOC_Os01g32660.1 | Oryza sativa MAPK kinase 6 | OsMKK6 | Overexpression | Cold tolerance | 10.1016/j.jplph.2011.11.012 |
| Cold | Resistance or Tolerance | LOC_Os02g50970.1 | drought-hypersensitive mutant1 | dsm1 | Mutant | Other stress resistance | 10.1104/pp.109.149856 |
| ABA | Resistance or Tolerance | LOC_Os02g36570.1 | activity of bc1 complex 1–2 | OsABC1-2 | Mutant | Other stress resistance | 10.1016/j.gene.2012.02.017 |
| Heat | Resistance or Tolerance | LOC_Os05g41090.1 | Doesn’t make infection 3 | OsDMI3 | Mutant | Other stress resistance | 10.1093/mp/sss068 |
| Resistance or Tolerance | LOC_Os01g10840.1 | glycogen synthase kinase3-like gene 1 | OsGSK1 | Mutant | Other stress resistance | 10.1007/s11103-007-9213-4 | |
| Cold | Resistance or Tolerance | LOC_Os04g41160.1 | oxidative signal inducible 1 | OsOxi1 | Overexpression | Other stress resistance | 10.1093/pcp/pcq132 |
| tZ | Others | LOC_Os08g34380.1 | commissural vein excessive1 | coe1 | Mutant | Others | 10.1111/j.1365-313X.2010.04250.x |
| Cold | Others | LOC_Os08g40170.1 | plant-specific cyclin-dependent kinase2;1 | Orysa;CDKB2;1 | Knockdown | Others | 10.1111/j.1365-313X.2011.04847.x |
| Cold, JA | Others | LOC_Os06g06090.1 | Mitogen-activated protein kinase6 | OsMAPK6 | Knockdown | Others | 10.1104/pp.104.057414 |
Functional assignment of rice kinase genes using expression data
To assess the biological functions of rice kinases, meta-expression profiles generated from a large collection of microarray-based expression datasets available in NCBI GEO have been used. Global analysis with anatomical data suggested that 55 kinases are predominantly expressed in the roots, 41 in the leaves/shoots, 58 in the callus/panicles, 36 in anthers/pollen and 71 genes are ubiquitously expressed (Fig. 1; Additional file 1: Table S1). Of these, functions for 16 genes have been elucidated through genetics and molecular studies (Table 1). For example, LOC_Os05g40770 is categorized under root preferred featured anatomical expression group in the current investigation. Previous study revealed that OsRPK1/LOC_Os05g40770 is expressed in the root tips and encodes a Ca2 +-independent leucine-rich-repeat (LRR) receptor-like kinase (RLK). Transgenic rice plants over-expressing OsRPK1 showed undeveloped adventitious and lateral roots (Zou et al. 2014).
Fig. 1.
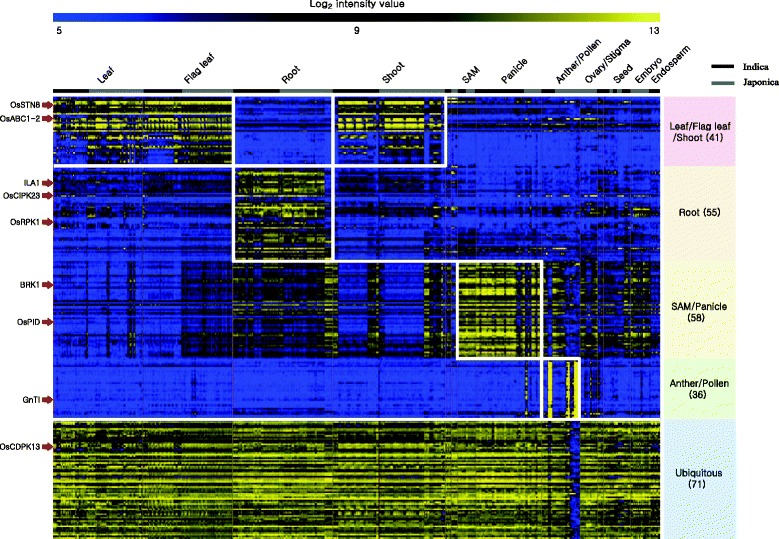
Heatmap analysis of kinase genes showing anatomical tissue-preferred expression pattern. Gene expression pattern reveals five tissue-preferential groups and the total genes in each group is shown in parenthesis. Affymetrix dataset includes the gene expression profiles from both indica and japonica subspecies plant samples. Sample profiling from subspecies are represented using black and grey bars for indica and japonica respectively. Log2-transformed intensity values were taken for generating heatmap and the scale is given above the heatmap. Genes with red arrow pointing the heatmap are the known genes
Biotic stress-related meta-expression data revealed significant upregulation in response to MG (29 kinases), MO (21 kinases), RSV (13 kinases), Xoo (18 kinases), and BPH (49 kinases) (Fig. 2; Additional file 2: Table S2). Of these, functions have been elucidated for eight genes (Table 1). OsACDR1/LOC_Os03g06410, for which present meta-analysis revealed induction under BPH infection, encodes a putative Raf-like mitogen-activated protein kinase kinase kinase (MAPKKK) and plays a positive regulatory role against fungal infection by modulating defense related gene expression (Kim et al. 2009).
Fig. 2.
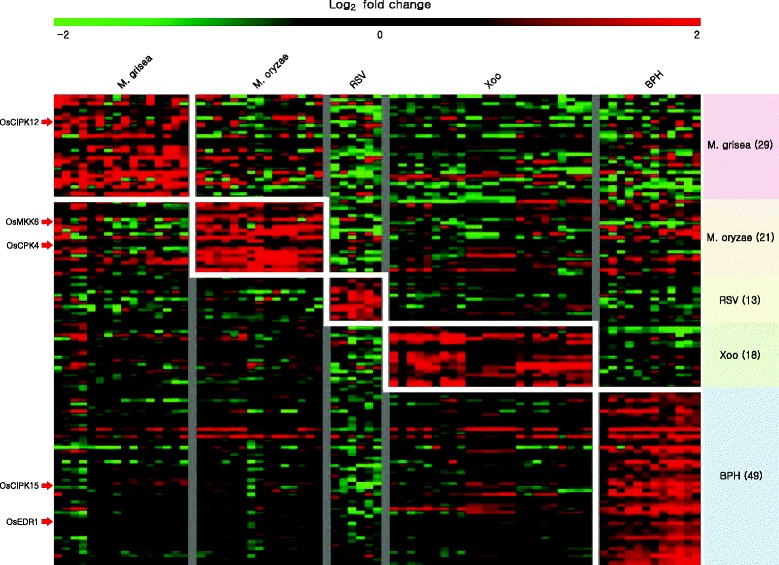
Heatmap analysis of expression patterns for kinase genes related to biotic-stress responses. Biotic stress responsive kinase genes heatmap is generated using log2-transformed fold change values under four stress conditions. Number of stress responsive genes in each condition is shown inside parenthesis after the applied biotic stress. Genes with red arrow pointing the heatmap are the known genes
Abiotic stress-related meta-expression data showed that 43 kinases are induced by drought or salt stress, 166 by low temperature, 44 by heat, and 43 by submergence (Fig. 3; Additional file 3: Table S3). Of these, functions of 23 kinase genes have been elucidated (Table 1). They include calcium-dependent protein kinase OsCDPK7/ LOC_Os04g49510 which acts as positive regulator for stress conditions and confers cold, salt and drought resistance on overexpression (Saijo et al. 2000). Interestingly, we also noted the cold depend induction of OsCDPK7 in the abiotic stress dataset based analysis and provides a hint about more potential candidates in the featured groups. Furthermore, the genes that are differentially expressed (log2 fold change >2 and P < 0.05) in root under heavy metal treatment were analyzed to reveal the potential kinase genes involved in soil toxicity. It was revealed that 21 genes were upregulated whereas expressions of 92 genes were declined under arsenic treatment in soil. Similarly, 44 and 79 genes are induced and declined by cadmium treatment, respectively. More numbers of kinase genes were responsive to chromium: expressions of 142 genes are induced; and that of 206 genes were reduced. In case of lead treatment in soil, 36 genes are upregulated and 54 genes are downregulated (Additional file 4: Table S4).
Fig. 3.
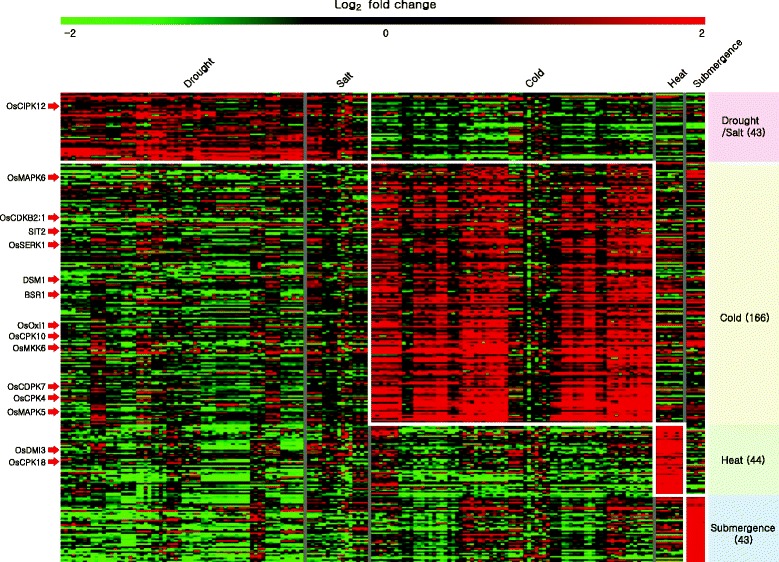
Heatmap analysis of kinase genes related to abiotic-stress responses. Abiotic stress responsive kinase genes heatmap is generated using log2-transformed fold change values under four stress conditions. Number of stress responsive genes in each condition is shown inside parenthesis after the applied abiotic stress. Genes with red arrow pointing the heatmap are the known genes
The meta-expression data for hormone responses indicated that 67 kinases are up-regulated upon ABA treatment, 27 by IAA, 32 by tZ, and 134 by JA treatment (Fig. 4; Additional file 5: Table S5). Among them, functions of 18 genes have been characterized previously.
Fig. 4.
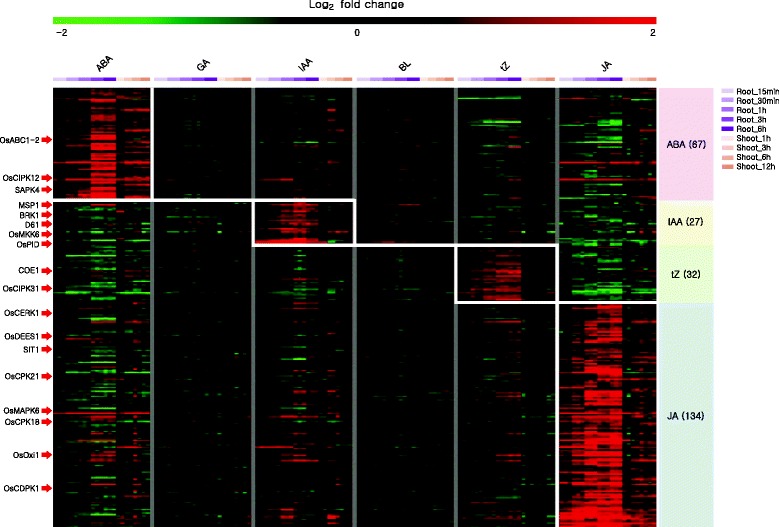
Heatmap analysis of expression patterns for kinase genes in response to hormone treatments. Hormone responsive kinase genes are generated from Agilent log2-transformed fold change hormone dataset, which includes responses from six major hormone treatments at different time intervals in root and shoot. Group of genes induced under specific hormones treatment along with their total number is indicated in parenthesis. Genes with red arrow pointing the heatmap are the known genes
Based on the featured expression data, it is evident that these 947 kinases may serve as potential primary targets for further functional studies, with 261 related to anatomy; 130, to biotic stresses; 296, to abiotic stresses; and 260, to hormone responses.
Evaluation of functional redundancy or functional dominance for rice kinases with known functions via PCC analysis
Regarding morphological or physiological traits, the roles played by 26 kinases have been identified through loss-of-function studies. Pearson correlation coefficient (PCC) can be used to measure the linear dependence between two genes, with a value of ‘1’ representing a perfect, positive correlation. Among paralogous kinase members, PCC values are useful indicators when evaluating functional similarities. PCC values for one to three family members closely linked to each kinase have been generated based on anatomical meta-expression data. These data, summarized in Table 2 and Fig. 5, indicated that 10 kinases exist independently from a clade and another nine have no closely linked members with PCC values >0.5. This explains functional dominancy for the 73 % of kinases with known functions. Therefore, the PCC approach can provide useful estimates of functional redundancy or dominance among family members. In addition, integrating anatomical expression data within the phylogenetics context serves as an effective platform for examining functional dormancy or redundancy within a gene family. Examples of integrated anatomical expression data for subgroups of three kinase families are shown in Fig. 6. Compared with other members, the dominant expression of OsCDPK1 (LOC_Os03g03660) has a PCC value of 0 to 0.5 (Fig. 6a) and predominant expression of OsBAK1 (LOC_Os08g07760) produces a PCC value below zero (Fig. 6B), while OsABC1-2 (LOC_Os02g36570) has redundant expression patterns with PCC values above 0.5 with LCO_Os09g07660 (Fig. 6c). In the case of OsABC1-2, functional studies have used RNAi to repress the expression of OsABC1-2 to overcome the functional redundancy. The observed phenotype might have been caused by suppression of multiple targets closely linked with LOC_Os09g07660 in that tree. Further investigation is needed to clarify this possibility.
Table 2.
Rice kinase paralogs with known roles based on loss-of-function studies, and PCC values calculated for paralog pairings
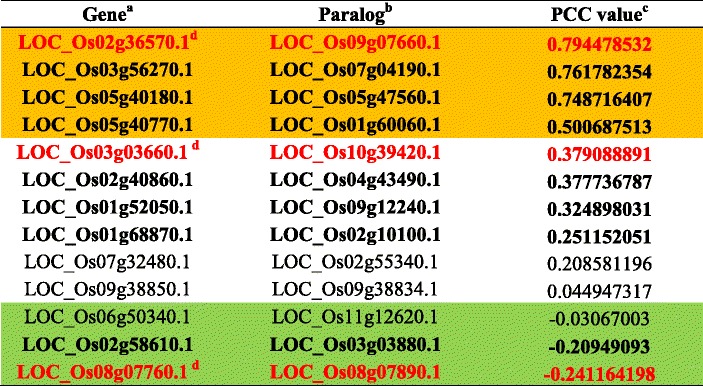
aIndicates the rice kinase genes which have been functionally characterized and identified from OGRO database (http://qtaro.abr.affrc.go.jp/ogro/table)
bIndicates paralogs of rice kinase genes in gene column which are identified from RKD 2.0 databases
cIndicates Pierson correlation coefficient (PCC) value between paralog pairs in gene and paralog columns
dIndicates rice kinase genes (marked with red letters) used in Figure 6. Brown box has 0.5 > PCC value and is estimated to have redundant roles among paralogs; green one with negative PCC value and uncolored area with 0 < PCC value < 0.5 are estimated to have predominant roles among paralogs
Fig. 5.
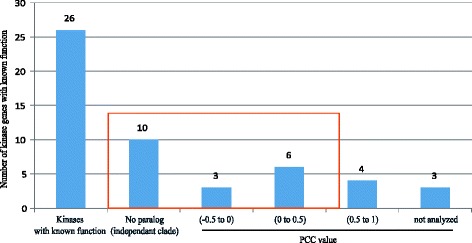
Estimations of functional dominancy for rice kinases, based on meta-expression data. Y-axis, number of kinases with known functions; X-axis, different groups of kinases with known functions, based on range of PCC values or existence of paralogs in subfamilies
Fig. 6.
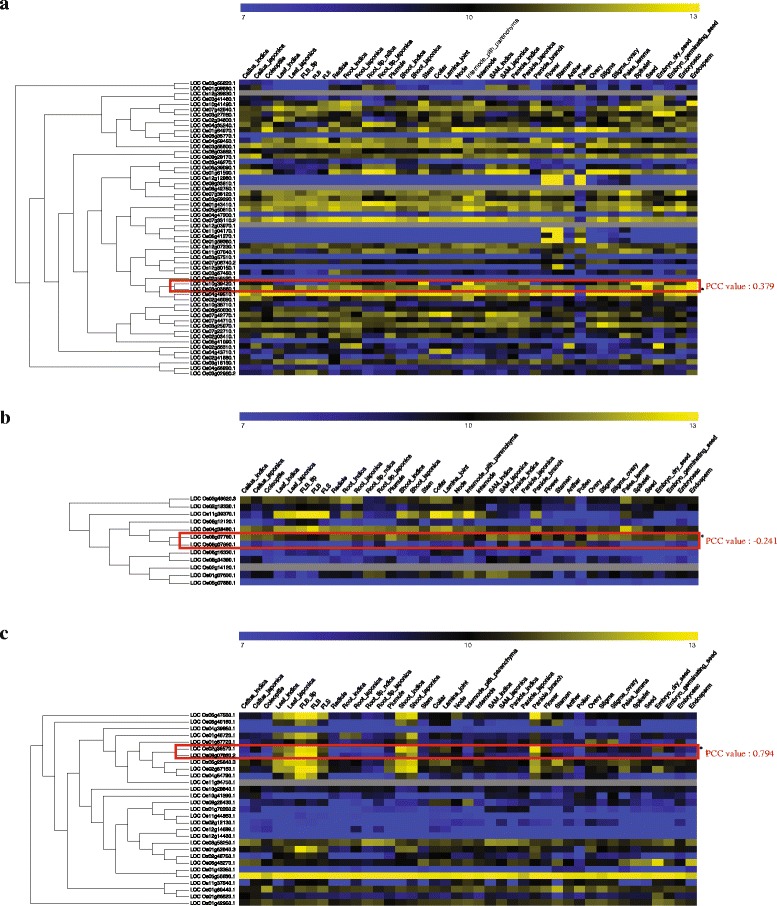
Phylogenomics data for three kinase subfamilies that include members with known functions. Integrating anatomical expression data within context of phylogenic tree suggested functional dominancy of OsCDPK1 (LOC_Os03g03660) with PCC value <0.379 (a), OsBAK1 (LOC_Os08g07760) with PC value of −0.241 (b), and functional redundancy of OsABC1-2 (LOC_Os02g36570) with PCC value of 0.794 = (c). Red boxes indicate pairs of kinases used for PCC analysis, gray color indicates the genes without probes in affymetrix array platform, and asterisks (*) indicate the selected kinases
Conclusions
Of the 1508 kinases featured in RKD 2.0, functions for all except 61 kinases require further examination. This new database will enable researchers to select target kinases more effectively for additional characterization. The predominant expression found among paralogous kinases might be a valuable indicator of functional dominance. Based on loss-of-function analyses and application of PCC values between a kinase and its paralog, we were able to explain the dominancy of 19 kinases with PCC values above 0.5 as well as those that form independent clades in a phylogenetic tree. Primary candidates for loss-of-function studies can be chosen by identifying kinase genes with dominant expression and then comparing them with closely linked family members. These meta-expression data also serve as a source of information about the location and/or conditions that can be applied in subsequent assessments as well as situations in which true functional redundancy may exist. Additional datasets, such as predicted protein–protein interaction networks, further enhance database utility by indicating direct targets regulated by kinases. With the use of gene-indexed mutants that cover at least half of the rice genome, accumulation of diverse omics data will further enhance the functionality of such phylogenomics databases.
Methods
Collection of microarray data
Anatomy: Expression data for the anatomical tissues/organs were retrieved from ROAD (Cao et al. 2012).
Abiotic stress: The dataset for abiotic stresses comprised multiple samples from NCBI GEO under the series GSE6901, GSE16108, GSE21651, GSE24048, GSE25176, GSE26280, GSE33204, GSE37940, and GSE38023. Experiment IDs in the Array express dataset (Array Express; https://www.ebi.ac.uk/arrayexpress/. Accessed 2 December 2015) included E-MEXP-2267and E-MEXP-2401. Altogether, 145 individual experiments were parsed for this analysis.
Biotic stress: The GEO series for the biotic stress database involved GSE7256, GSE30941, GSE41798, GSE18361, GSE36272, GSE29967, and GSE11025. In all, 103 individual samples were analyzed under these seven series.
Hormone response: For analyzing hormone-related meta-expression, we relied upon the Agilent 44 K array data GSE39429, as generated by Sato et al. (2013).
Collection of RNA-Seq data
RNA-Seq anatomical expression dataset consisting of 27 tissues/organs were retrieved from source data developed by Wang et al. (2015). In addition, spatio-temporal transcriptome data of rice root in response to phosphate starvation and recovery were used (Secco et al. 2013).
PCC analysis
An anatomy dataset was used for estimating PCC values among neighboring genes within a subclade. For each gene, the intensity level of the RGAP gene model probes was fetched and formatted in pairs using in-house scripts. For every pair of genes that showed expression across a range of experiments, a PCC value was calculated using the Microsoft Excel function.
Heatmap analysis
To examine the expression patterns of kinase genes in various rice tissues/organs as well as in response to abiotic/biotic stresses or hormone treatments, we used a meta-expression analysis based on 1150 Affymetrix array data plus GEO series GSE39429, which utilized the Agilent 44 K microarray platform (GEO platform GPL6854). We then uploaded the log2 normalized intensity data (tab-delimited text format) into Multi Experiment Viewer (MEV; http://www.tm4.org/, accessed on October 20, 2015) and created the desired heatmaps (Fig. 1; Additional file 1: Table S1). In addition, we developed a meta-expression database for biotic stress responses to MG, MO, RSV, Xoo, and BPH (Fig. 2; Additional file 2: Table S2), a meta-expression database for responses to drought, salt, cold, heat, and submergence (Fig. 3; Additional file 3: Table S3), and a meta-expression database for responses to six hormones (ABA, GA, IAA, BL, JA, and cytokinin/trans-Zeatin) (Fig. 4; Additional file 5: Table S5). Expression data for selected kinases were presented by heatmaps. For other methods used for construction of RKD 2.0, RKD original version retains the details (Dardick et al. 2007).
Abbreviations
NCBI GEO, National Center for Biotechnology Information Gene Expression Omnibus; non-TE, non-transposable element; PCC, Pearson correlation coefficient; RGAP, rice genome annotation project database; RKD, rice kinase database; ROAD, rice oligonucleotide array database
Acknowledgements
This work was carried out with the support of “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01182602)”, Rural Development Administration, Republic of Korea, and was part of the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the U. S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U. S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes.
Additional files
Rice kinases with patterns of anatomical expression. (XLSX 25 kb)
Rice kinases with abiotic stress responses. (XLSX 19 kb)
Rice kinases with biotic stress responses. (XLSX 28 kb)
Rice kinases differentially expressed under root heavy metal treatment. (XLSX 62 kb)
Rice kinases with hormone responses. (XLSX 25 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Author contributions
JKH, PCR and CD conceived and designed the experiments. AKNC and YYH performed the experiment. JKH, PC, WJH, RS and MS analyzed the data, AKNC, YYH. PCR and JKH prepared the manuscript. All authors read and approved the final manuscript.
Contributor Information
Pamela C Ronald, Email: pcronald@ucdavis.edu.
Ki-Hong Jung, Email: khjung2010@khu.ac.kr.
References
- Alexandrov N, Tai S, Wang W, Mansueto L, Palis K, Fuentes RR, Ulat VJ, Chebotarov D, Zhang G, Li Z, Mauleon R, Hamilton RS, McNally KL. SNP-seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res. 2015;43:D1023–D1027. doi: 10.1093/nar/gku1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Holko M, Ayanbule O, Yefanov A, Soboleva A. NCBI GEO: archive for functional genomics data sets--10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao PJ, Bartley LE, Jung KH, Ronald PC. Construction of a rice glycosyltransferase phylogenomic database and identification of rice-diverged glycosyltransferases. Mol Plant. 2008;1:858–877. doi: 10.1093/mp/ssn052. [DOI] [PubMed] [Google Scholar]
- Cao P, Jung KH, Choi D, Hwang D, Zhu J, Ronald PC. The rice oligonucleotide array database: an atlas of rice gene expression. Rice (N Y) 2012;5:17. doi: 10.1186/1939-8433-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran AKN, Jung KH. Resources for systems biology in rice. J Plant Biol. 2014;57:80–92. doi: 10.1007/s12374-014-0903-6. [DOI] [Google Scholar]
- Dardick C, Chen J, Richter T, Ouyang S, Ronald P. The rice kinase database. A phylogenomic database for the rice kinome. Plant Physiol. 2007;143:579–586. doi: 10.1104/pp.106.087270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IRGSP The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Itoh T, Tanaka T, Barrero RA, Yamasaki C, Fujii Y, Hilton PB, Antonio BA, Aono H, Apweiler R, Bruskiewich R, Bureau T, Burr F, Costa de Oliveira A, Fuks G, Habara T, Haberer G, Han B, Harada E, Hiraki AT, Hirochika H, Hoen D, Hokari H, Hosokawa S, Hsing YI, Ikawa H, Ikeo K, Imanishi T, Ito Y, Jaiswal P, Kanno M, Kawahara Y, Kawamura T, Kawashima H, Khurana JP, Kikuchi S, Komatsu S, Koyanagi KO, Kubooka H, Lieberherr D, Lin YC, Lonsdale D, Matsumoto T, Matsuya A, McCombie WR, Messing J, Miyao A, Mulder N, Nagamura Y, Nam J, Namiki N, Numa H, Nurimoto S, O’Donovan C, Ohyanagi H, Okido T, Oota S, Osato N, Palmer LE, Quetier F, Raghuvanshi S, Saichi N, Sakai H, Sakai Y, Sakata K, Sakurai T, Sato F, Sato Y, Schoof H, Seki M, Shibata M, Shimizu Y, Shinozaki K, Shinso Y, Singh NK, Smith-White B, Takeda J, Tanino M, Tatusova T, Thongjuea S, Todokoro F, Tsugane M, Tyagi AK, Vanavichit A, Wang A, Wing RA, Yamaguchi K, Yamamoto M, Yamamoto N, Yu Y, Zhang H, Zhao Q, Higo K, Burr B, Gojobori T, Sasaki T. Curated genome annotation of Oryza sativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana. Genome Res. 2007;17:175–183. doi: 10.1101/gr.5509507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Lee J, Dardick C, Seo YS, Cao P, Canlas P, Phetsom J, Xu X, Ouyang S, An K, Cho YJ, Lee GC, Lee Y, An G, Ronald PC. Identification and functional analysis of light-responsive unique genes and gene family members in rice. PLoS Genet. 2008;4:e1000164. doi: 10.1371/journal.pgen.1000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Cao P, Seo YS, Dardick C, Ronald PC. The rice kinase phylogenomics database: a guide for systematic analysis of the rice kinase super-family. Trends Plant Sci. 2010;15:595–599. doi: 10.1016/j.tplants.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Jung KH, Kim SR, Giong HK, Nguyen MX, Koh HJ, An G. Genome-wide identification and functional analysis of genes expressed ubiquitously in rice. Mol Plant. 2015;8:276–289. doi: 10.1016/j.molp.2014.10.013. [DOI] [PubMed] [Google Scholar]
- Kim JA, Cho K, Singh R, Jung YH, Jeong SH, Kim SH, Lee JE, Cho YS, Agrawal GK, Rakwal R, Tamogami S, Kersten B, Jeon JS, An G, Jwa NS. Rice OsACDR1 (Oryza sativa accelerated cell death and resistance 1) is a potential positive regulator of fungal disease resistance. Mol Cells. 2009;28:431–439. doi: 10.1007/s10059-009-0161-5. [DOI] [PubMed] [Google Scholar]
- Lin H, Ouyang S, Egan A, Nobuta K, Haas BJ, Zhu W, Gu X, Silva JC, Meyers BC, Buell CR. Characterization of paralogous protein families in rice. BMC Plant Biol. 2008;8:18. doi: 10.1186/1471-2229-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, Orvis J, Haas B, Wortman J, Buell CR. The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K. Over-expression of a single Ca2 + −dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 2000;23:319–327. doi: 10.1046/j.1365-313x.2000.00787.x. [DOI] [PubMed] [Google Scholar]
- Sato Y, Takehisa H, Kamatsuki K, Minami H, Namiki N, Ikawa H, Ohyanagi H, Sugimoto K, Antonio BA, Nagamura Y. RiceXPro version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 2013;41:D1206–D1213. doi: 10.1093/nar/gks1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D, Jabnoune M, Walker H, Shou H, Wu P, Poirier Y, Whelan J. Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell. 2013;25:4285–4304. doi: 10.1105/tpc.113.117325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, Fauquet C, Ronald P. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Wang J, Qi M, Liu J, Zhang Y. CARMO: a comprehensive annotation platform for functional exploration of rice multi-omics data. Plant J. 2015;83:359–374. doi: 10.1111/tpj.12894. [DOI] [PubMed] [Google Scholar]
- Yamamoto E, Yonemaru J-I, Yamamoto T, Yano M. OGRO: the overview of functionally characterized genes in rice online database. Rice. 2012;5:1–10. doi: 10.1186/1939-8433-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Laule O, Schmitz J, Hruz T, Bleuler S, Gruissem W. Genevestigator transcriptome meta-analysis and biomarker search using rice and barley gene expression databases. Mol Plant. 2008;1:851–857. doi: 10.1093/mp/ssn048. [DOI] [PubMed] [Google Scholar]
- Zou Y, Liu X, Wang Q, Chen Y, Liu C, Qiu Y, Zhang W. OsRPK1, a novel leucine-rich repeat receptor-like kinase, negatively regulates polar auxin transport and root development in rice. Biochim Biophys Acta. 2014;1840:1676–1685. doi: 10.1016/j.bbagen.2014.01.003. [DOI] [PubMed] [Google Scholar]


