Abstract
There is growing interest in understanding how diet affects the intestinal microbiota, including its possible associations with systemic diseases such as metabolic syndrome. Here we report a comprehensive and deep microbiota analysis of 14 obese males consuming fully controlled diets supplemented with resistant starch (RS) or non-starch polysaccharides (NSPs) and a weight-loss (WL) diet. We analyzed the composition, diversity and dynamics of the fecal microbiota on each dietary regime by phylogenetic microarray and quantitative PCR (qPCR) analysis. In addition, we analyzed fecal short chain fatty acids (SCFAs) as a proxy of colonic fermentation, and indices of insulin sensitivity from blood samples. The diet explained around 10% of the total variance in microbiota composition, which was substantially less than the inter-individual variance. Yet, each of the study diets induced clear and distinct changes in the microbiota. Multiple Ruminococcaceae phylotypes increased on the RS diet, whereas mostly Lachnospiraceae phylotypes increased on the NSP diet. Bifidobacteria decreased significantly on the WL diet. The RS diet decreased the diversity of the microbiota significantly. The total 16S ribosomal RNA gene signal estimated by qPCR correlated positively with the three major SCFAs, while the amount of propionate specifically correlated with the Bacteroidetes. The dietary responsiveness of the individual's microbiota varied substantially and associated inversely with its diversity, suggesting that individuals can be stratified into responders and non-responders based on the features of their intestinal microbiota.
Keywords: dietary intervention, intestinal microbiota, phylogenetic microarray, responder, short chain fatty acid, 16S rRNA
Introduction
The intestinal microbiota thrives in the interface of the host and environment, which are the two major components that control the development of diseases. The intestinal microbiota is currently acknowledged as a third factor in the etiology of many disorders, including obesity and associated metabolic conditions via numerous mechanisms (Flint, 2011; Burcelin et al., 2012; Everard and Cani, 2013). The microbiota consists of hundreds or thousands of bacterial species-level phylotypes that have a collective genome of several million genes (Qin et al., 2010; The Human Microbiome Project Consortium, 2012). Among the diverse predicted functions, those devoted to breakdown and bioconversion of otherwise indigestible dietary components, mainly plant-derived polysaccharides, have been most actively studied because of their high abundance and relevance for health (Flint et al., 2008, 2012a). Bacterial fermentation of dietary fiber produces butyrate and other short chain fatty acids (SCFAs) that have largely favorable physiological consequences for the host, ranging from tumor protection to glucose and lipid homeostasis (Macfarlane and Macfarlane, 2011; Russell et al., 2013).
The host and environmental factors, including life style, affect the intestinal microbiota to a largely unknown extent. Diet is considered one of the major modulators, although molecular analyses have only recently started to provide information about the extent and nature of the diet responsiveness of the gut ecosystem. These studies show that in laboratory mice, diet is a dominant source of variation in the microbiota composition (Zhang et al., 2009; Faith et al., 2011). The heterogeneous and highly personalized human microbiota shows a smaller dietary influence as the inter-individual variation decreases systematic dietary effects even under identical diet (Wu et al., 2011; David et al., 2014). Targeted analyses show that the amount and type of dietary carbohydrate affect the abundance of bacteria belonging to Clostridium clusters IV (Ruminococcaceae) and XIVa (Lachnospiraceae) along with bifidobacteria (Bouhnik et al., 2006; Duncan et al., 2007; Ramirez-Farias et al., 2009; Walker et al., 2011). Observational studies suggest that long-term dietary habits influence the intestinal microbiota composition, as comparisons of subjects who consume diets rich in fiber to subjects with a high intake of animal fat and protein have noted differences (for example, in the abundance of Prevotella spp. (De Filippo et al., 2010; Wu et al., 2011; Claesson et al., 2012; Yatsunenko et al., 2012)). Recent community-level microbiota analyses during dietary interventions have also shown marked effects on selected microbial taxa that are reversible and detectable within days (Martínez et al., 2010; Walker et al., 2011; David et al., 2014). Representation of certain Firmicutes increased in healthy Americans on diets enriched with whole grains (Martínez et al., 2012). Less pronounced effects were observed in Finnish subjects with high basal intake of whole grains (Lappi et al., 2013). Increased intake of fructans decreased Bacteroides spp. and in parallel increased the levels of bifidobacteria and Faecalibacterium prausnitzii (Dewulf et al., 2012). All these studies have noted strong individuality of the responses, the extent of which appears to depend on the initial microbiota composition (Korpela et al., 2014).
In this study, we have studied the intestinal microbiota of 14 subjects with metabolic syndrome who sequentially consumed four fully controlled diets, including fiber supplementations and weight-loss (WL) diet. Previously, quantitative PCR (qPCR) analysis of all subjects and sequencing of clone libraries for six of the individuals identified dominant, diet-responsive bacteria (Walker et al., 2011). The current microarray study was performed to deepen and widen the microbiota analysis from ca dozen dominant genera to cover over 1000 species-level phylotypes (Rajilić-Stojanović et al., 2009), and to investigate host response to the diets. To this end, we performed deep phylogenetic microarray and fecal SCFA analyses for all the subjects and diets, and in parallel analyzed the host's insulin metabolism based on blood samples. This set-up allowed us to perform both quantitative and qualitative analyses on the dietary influence on the human intestinal ecosystem, study the covariance between the microbiota composition, SCFA output and insulin sensitivity, and finally, differentiate between the dietary responders and non-responders.
Materials and methods
Subjects
Fourteen British males fulfilling the criteria for metabolic syndrome participated in the study. Mean age was 53 years (range 27–73), and mean body mass index was 39.4 kg m–2 (range 27.9–51.3). Details of the recruitment and characteristics of the participants have been published previously (Walker et al., 2011; Lobley et al., 2013). The study was approved by the North of Scotland Research Ethics Service and all volunteers provided informed, written consent.
Dietary interventions
The participants followed four different, fully controlled diets during the 10-week trial (Supplementary text; Supplementary Table S1 and Supplementary Figure S1). The details of the diets are described elsewhere (Walker et al., 2011; Lobley et al., 2013). Briefly, a run-in standard diet at weight maintenance (M) for 1 week was followed by two diets for 3 weeks each at weight maintenance in a randomized cross-over design. Both diets were similar in the overall macronutrient composition, but one was high in type 3 resistant starch (RS), whereas the other was high in non-starch polysaccharides (NSPs). For the last 3 weeks, all volunteers consumed a weight loss diet with high protein and medium carbohydrate levels.
Fecal samples and microbiota profiling
Fecal samples collected at the end of each dietary regime were used for in-depth microbiota analysis. The DNA extraction (Walker et al., 2011) and sample processing for HITChip microarray were performed as previously described (Rajilić-Stojanović et al., 2009). The HITChip microarray targets the V1 and V6 hypervariable regions of the 16S ribosomal RNA gene of >1000 bacterial species-level phylotypes detected in the human intestine. The data were normalized and preprocessed as explained in detail elsewhere (Rajilić-Stojanović et al., 2009; Jalanka-Tuovinen et al., 2011). Unless otherwise specified, probe signals summarized to 130 genus-like groups (⩾90% sequence similarity in the 16S ribosomal RNA gene), were used as a proxy of bacterial abundance, and are referred to as Species and relatives (shortened as ‘et rel.') according to the nearest cultured relative (Rajilić-Stojanović et al., 2009). The microarray data are available at the Dryad Digital Repository: http://doi.org/10.5061/dryad.7b37k (unique identifier provided upon acceptance).
The quantification of total bacteria and dominant bacterial groups with qPCR have been published previously (Walker et al., 2011). Primers for Ruminococcus spp. (Supplementary Table S2A) were designed and validated as detailed in Supplementary text and Supplementary Table S2B, and qPCR was carried out as described previously (Ramirez-Farias et al., 2009; Walker et al., 2011).
Analysis of acidic fermentation products in fecal samples
Fresh fecal samples were mixed and stored in 0.5 g amounts as described previously (Duncan et al., 2007). The SCFA content of the samples was determined by gas capillary chromatography following conversion to t-butylmethylsilyl derivatives (Richardson et al., 1989). The lower limit for reliable detection of the products is 0.2 mM.
Markers of insulin metabolism
Fasting blood samples collected at the end of each dietary regime were used for measurement of plasma glucose and insulin and determination of homeostasis model assessment of insulin resistance as explained elsewhere (Lobley et al., 2013).
Statistical analysis
The stability of the microbiota was estimated by calculating intra- and inter-individual Pearson's correlation coefficients (r) between the microbial profiles, and visualized with hierarchical clustering using complete linkage algorithm (Becker et al., 1988). An analysis of variance-like permutation test for redundancy analysis with subject as conditioning term was carried out to test whether the overall microbiota composition differed significantly between the diets using R package vegan (Oksanen et al., 2011). Dietary impact on the abundance of individual genus-like phylogenetic groups was assessed with linear mixed models using nlme package (Pinheiro et al., 2013) with contrasts estimated using R package multcomp (Bang et al., 2008). The effect of diet order and its interaction with diet were evaluated (to test for the carry-over effect) by including them as fixed effects in the linear mixed model. As these effects were nonsignificant, they were removed. Diet was modeled as fixed effect and subject as a random effect to account for the repeated-measures design of the experiment. Benjamini–Hochberg correction for multiple testing was applied to P-values from analysis of variance F-test statistic of diet effect. To identify which of the diets differed significantly, post-hoc analysis was carried out by pairwise comparisons of all diets, the dietary effects and their significances with (the default) single-step P-value adjustments for multiple comparisons in R package multcomp.
Diversity of the microbiota was quantified based on non-logarithmized HITChip oligo-level signals by inverse Simpson's index as implemented in the R package vegan. The amount of variability explained by diet and subject in different data sets was quantified with coefficient of determination (R2) using analysis of variance, and permutation test from the R package vegan. The responder status was evaluated based on within-subject Pearson correlations calculated between the bacterial profiles on the M diet and each of the three intervention diets.
SCFA analysis was carried out with linear mixed models as described for the HITChip data analysis. Also here, the cross-over effects were nonsignificant, resulting in a linear mixed model with subject as a random effect and diet as fixed effect. Associations between the microbiota, SCFA and clinical variables were assessed based on random effects regression with subject as a random effect. Additional information on the statistical methods can be found in the Supplementary text.
Results
Dietary impact on the structure, diversity and dynamics of the intestinal microbiota
Fecal and blood samples were analyzed from 14 obese male volunteers during an initial M diet, at the end of two diets rich in non-digestible carbohydrates, either RS or NSPs, and after a WL diet (Supplementary text and Supplementary Figure S1; Walker et al., 2011). We analyzed the dietary impact on the composition and ecology of the intestinal microbiota, SCFA output and insulin sensitivity, as well the inter-relations between the data sets.
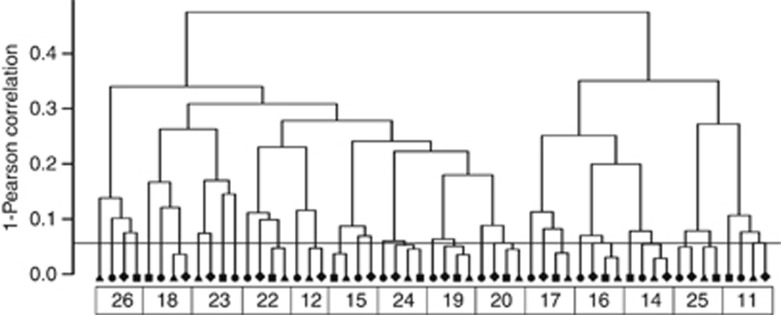
The microbiota composition was assessed using the HITChip phylogenetic microarray (Rajilić-Stojanović et al., 2009). Hierarchical clustering of the microbiota profiles representing four different diets unambiguously clustered by the donor (Figure 1). This subject-wise clustering demonstrates the individuality of the community structure and relatively limited effect on it. In most individuals (8/14), the sample taken during the RS diet was the most distinct from others, but the difference in intra-individual stabilities of the microbiota profiles did not reach significance in contrast to the inter-individual similarity (beta diversity) that was significantly lower on the M and RS diets (Supplementary Table S3).
Figure 1.
Hierarchical clustering of the bacterial fingerprints of fecal samples collected from 14 subjects during four different diets (M diet, square; RS diet, circle; NSP diet, triangle; WL diet, diamond). The subject-wise clustering is highlighted with boxes. Vertical line is drawn at Pearson correlation of 0.95, which represents a reference value for the temporal stability of the microbiota in subjects without dietary intervention (for details, see Discussion section).
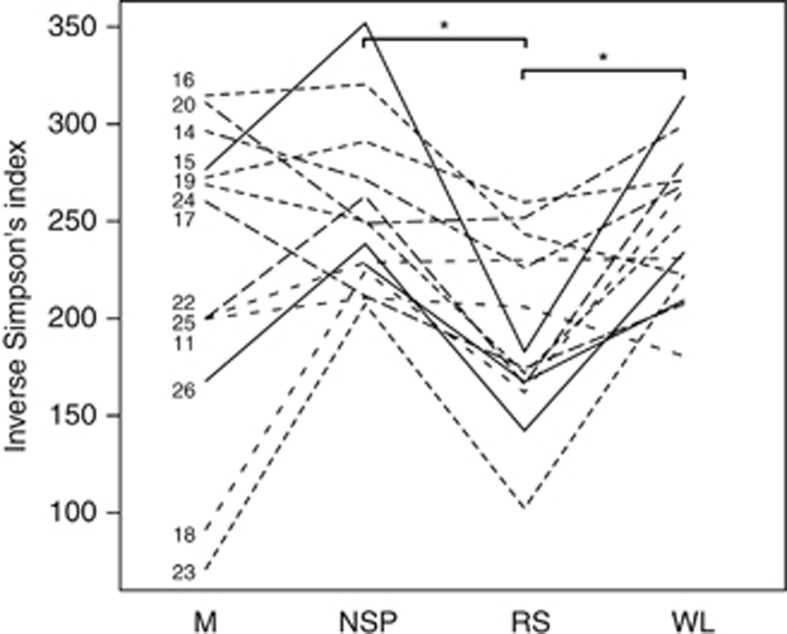
The alpha diversity, richness and evenness of the microbiota was evaluated, and found to be lowest after the RS diet (difference to NSP and WL diets P<0.05; M diet nonsignificant because of high variation; Supplementary Figure S2a). Although the subjects had highly variable initial microbiota diversities, each diet altered diversity in a relatively systematic way (Figure 2). The individual variation in the diversity was mainly ascribed to the microbiota richness that varied up to 2.5-fold on the M diet, ranging from 342 to 840 species-level phylotypes above the detection threshold (Supplementary Figure S2b). In addition, the evenness of the microbiota was the lowest after the RS diet, albeit nonsignificantly (Supplementary Figure S2c).
Figure 2.
Dynamics of the microbiota diversity per volunteer during dietary shifts. Diets with statistically significant (P<0.05) difference are indicated with asterisks.
The potential impact of study diets on the colonic bacterial biomass was estimated based on the recalculation of the qPCR data for total bacteria (Walker et al., 2011). The amount of bacteria g–1 feces was lowest during the RS and WL diets although high individual variation was observed for the RS diet (Supplementary Figure S2d).
Dietary impact on the abundance of individual bacterial taxa
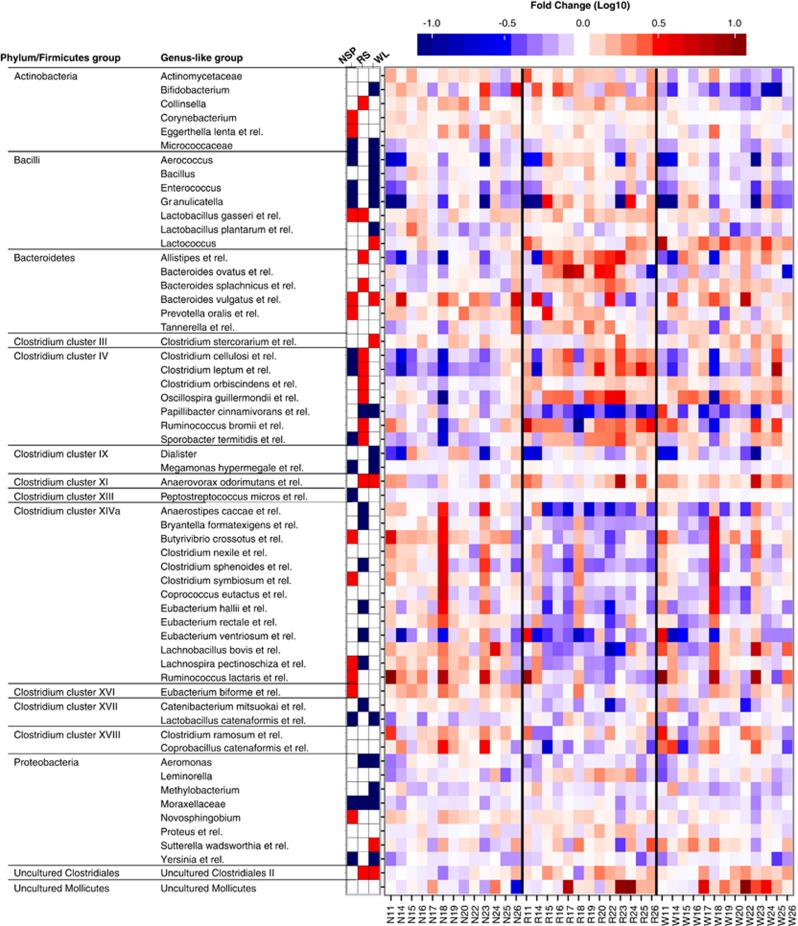
Permutation test of the microarray data indicated that the subjects' overall microbiota differed significantly between the diets (P<0.05). To identify the bacteria that were affected by the dietary switches, we compared the mean relative abundances of 130 genus-like phylogenetic groups pairwise between all the diets. Notable variation characterized the dietary responses across the individuals (Figure 3). The abundance of 59 (45%) bacterial groups changed significantly (P<0.05) in at least one pairwise comparison, 69% of the affected taxa showing >30% change (Supplementary Table S4). Among the 59 affected taxa, no phylum was significantly overrepresented. Most of the altered taxa belonged to predominant Clostridium clusters XIVa and IV; 68% and 58% of their genus-like groups reached statistical significance, respectively (45% for all Firmicutes). About one-third of the Bacteroidetes were significantly affected. Among the less abundant bacteria, Actinobacteria, Proteobacteria and Bacilli each contained a minimum of five significantly affected groups.
Figure 3.
Heatmaps of changes in bacterial abundance between the M diet and the three test diets (N=non-starch polysaccharides, R=resistant starch, W=weight loss). Each row represents genus-like phylogenetic groups of bacteria whose mean abundance differed significantly between at least one pairwise comparison of the diets. The leftmost three columns show the mean change of those taxa that reached statistical significance (P<0.05) compared with the M diet, red denoting increase and blue decrease. The large heatmap represents intra-individual changes. The logarithmic fold-change compared with the M diet is indicated by the colors ranging from dark blue (logFC –1 or lower) to dark red (logFC 1 and higher).
The RS diet stimulated bacteria related to Oscillospira guillermondii and R. bromii (as previously observed, Walker et al., 2011), but in addition Sporobacter termitis, Clostridium leptum and C. cellulosi within Clostridium cluster IV (Ruminococcaceae) (Figures 3 and 4). In contrast to other Ruminococcaceae, bacteria related to Papillibacter cinnamivorans were strongly reduced. Also certain Bacteroidetes were stimulated, Alistipes spp. the most.
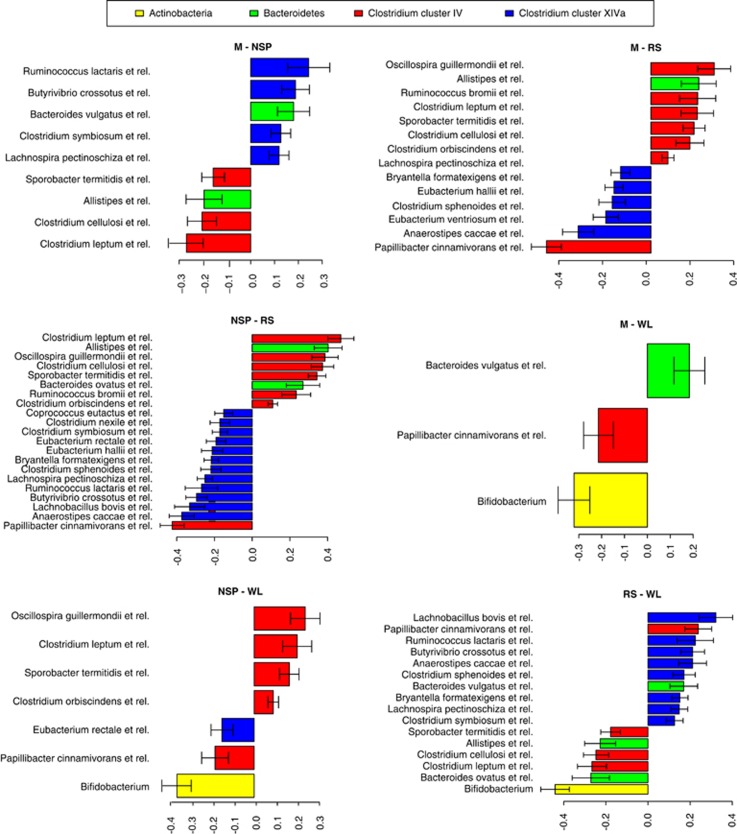
Figure 4.
Changes in abundant bacteria during dietary shifts as revealed by pairwise comparisons. The x axis shows logarithmic fold change of bacterial groups that differed significantly (P<0.05) and represented >0.5% of the total hybridization signal. The direction of bars indicates which of the two diets had higher abundance of the taxa listed on left side of each plot. The full list of significantly altered taxa is given in Supplementary Table S4.
Bacteria belonging to the Clostridium cluster XIVa (Lachnospiraceae) generally decreased during the RS diet (Figures 3 and 4). These included bacteria related to E. rectale (Figure 3), a finding that appeared to contradict the previous finding based on qPCR amplification of the Roseburia+E. rectale group from the same cohort (Walker et al., 2011). However, phylotype-level analysis of the microarray data showed that E. rectale and the uncultured bacterium D522 did increase on the RS diet, while the other related uncultured phylotypes and R. inulinivorans L1-83 decreased (Supplementary Figure S3), explaining the negative net effect of RS on this group.
Several members of the family Lachnospiraceae increased on the NSP diet compared with the M diet and especially to the RS diet (Figures 3 and 4). In addition, the genera Eggerthella, Collinsella and Corynebacterium within Actinobacteria, along with bacteria related to Bacteroides vulgatus and Prevotella oralis, were stimulated after the NSP diet. On the other hand, representatives of Ruminococcaceae such as C. leptum, C. cellulosi, Oscillospira spp. and Sprorobacter spp. that were increased after the RS diet, were notably reduced after the NSP diet.
During the WL diet, the abundance of bifidobacteria was significantly reduced compared with all other diets (Figure 4). On the other hand, there was a twofold increase in Lactococci compared with the M diet (P<0.05). Based on the study design, this likely reflected the high intake of fermented dairy products that was recently shown to lead to increase in food-derived bacteria in feces (David et al., 2014). The previously reported decrease in R. intestinalis group (Walker et al., 2011) was here borderline significant (P=0.06). Some low-abundant Bacilli and Proteobacteria were also decreased, except for bacteria related to Sutterella wadsworthia that were increased (Supplementary Table S4).
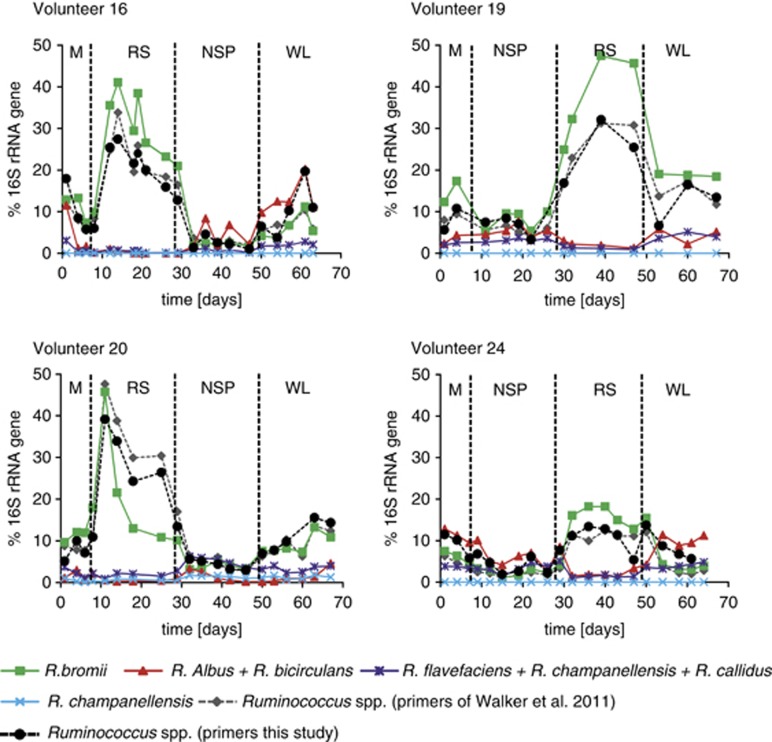
As a result of the central role inferred for Ruminococcus-related bacteria in the fermentation of RS (Ze et al., 2012), the abundance of members of this group was investigated further by using the qPCR approach. In addition to primer set targeting the ‘R-Ruminococcus' group (cluster IV Ruminococci; relatives of R. bromii, R. flavefaciens and R. albus) used previously, a new genus-level and four species-level primer pairs were used to amplify DNA from the fecal samples of four volunteers (Supplementary Table S2). The results (Figure 5) show that the increase in R. bromii largely accounted for the increase in the ‘R-Ruminococcus' group previously detected by Walker et al. (2011) during the RS diet. Interestingly, however, the representation of some other species belonging to this group, notably R. albus and R. bicirculans (Wegmann et al., 2013), showed the opposite trend, decreasing on the RS diet but increasing on the NSP and WL diets. As we concluded above from the microarray analysis for relatives of Roseburia and E. rectale, differences in substrate utilization exist at the species level that can result in very different responses to a dietary shift within a particular genus.
Figure 5.
Impact of diet on the abundance of Ruminococcus species in four volunteers, as assessed by qPCR. Primer pairs were designed that are specific for R. bromii, R. albus plus R. bicirculans, R. callidus plus R. flavefaciens plus R. champanellensis, and R. champanellensis together with R. champanellensis. Also shown are results obtained with two genus-level primer sets, the one used by Walker et al. (2011) and a newly designed set that gives better amplification of non-R.bromii species. Results are shown as % of the signal obtained with the universal 16S ribosomal RNA gene primer set. Primer sequences and amplification conditions are summarized in Supplementary Table S2. All of the available samples were analyzed from these four volunteers, providing a complete time course.
Impact of the dietary interventions on fecal SCFA concentrations and association with microbiota
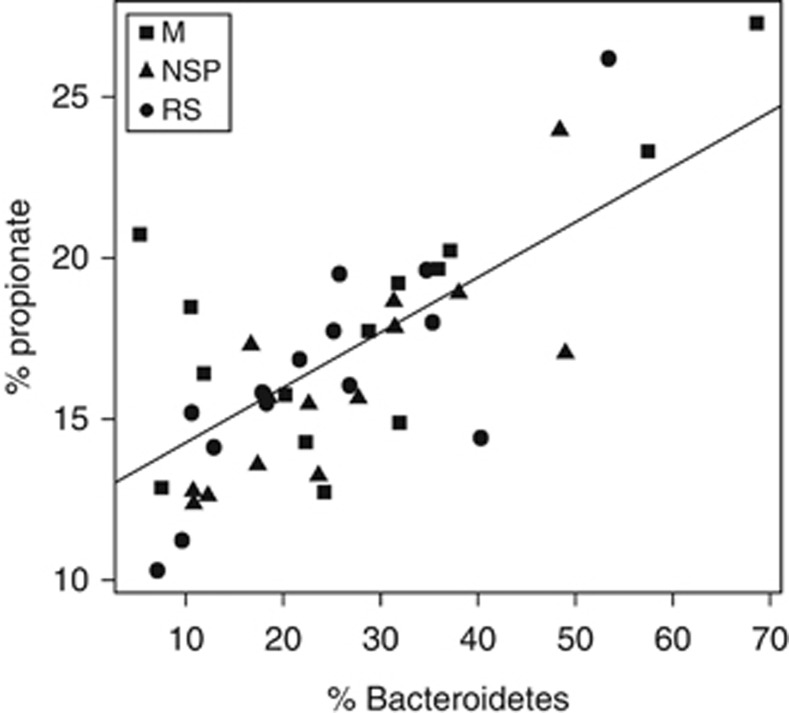
Fecal concentrations of the main SCFA acetate, propionate and butyrate, as well as succinate, were significantly lower on the RS and WL compared with the M and NSP diets, whereas most of the minor fermentation acids were increased (Supplementary Table S5A). The total 16S ribosomal RNA signal estimated by qPCR showed a significant positive correlation with total SCFA concentrations and with the concentrations of each of the three major SCFA for the three weight maintenance diets (Supplementary Table S5B). This indicates that, as may be expected, increased bacterial numbers coincided with increased rates of fermentation. Interestingly, propionate proportion among the total SCFA showed a significant positive correlation with the Bacteroidetes (Bacteroides and Prevotella-specific) qPCR signal (Figure 6) that was driven largely by inter-individual variation, since % propionate did not change with diet (Supplementary Table S5A). This is consistent with the Bacteroidetes being the largest contributor to propionate formation within the colonic microbiota (Flint et al., 2012b). Significant negative correlations with the major SCFA were seen for the cluster IV Ruminococcus group as detected by qPCR (Supplementary Table S5B). This appears to reflect the simultaneous impact of the RS diet in reducing total bacterial numbers and total SCFA, while promoting the Ruminococcus group.
Figure 6.
Relationship between proportion of propionate and of Bacteroides and Prevotella 16S ribosomal RNA sequences, as detected by qPCR. Data are means for all samples obtained from the last 2 weeks of each dietary period (Walker et al., 2011).
The proportions of branched chain SCFA (isobutyrate and isovalerate) increased on the two diets (RS and WL) that showed the lowest total SCFA and were negatively correlated with the total 16S ribosomal RNA gene signal (Supplementary Tables S5A and B). Since these SCFA are derived from amino-acid fermentation, this is likely explained by sustained protein fermentation under conditions of lower non-digestible carbohydrate fermentation in the distal colon (assumed to reflect mainly NSP fiber, with RS being fermented largely in the proximal colon) that support lower total bacterial numbers. A similar relationship applied to lactate, as also reported by Russell et al. (2011), but the reasons for this are unclear. Based on the microarray analysis, bacteria related to Anaerovorax odorimutans correlated positively with isobutyrate (q<0.05).
Quantification of the dietary impact on microbiota, SCFA and clinical variables
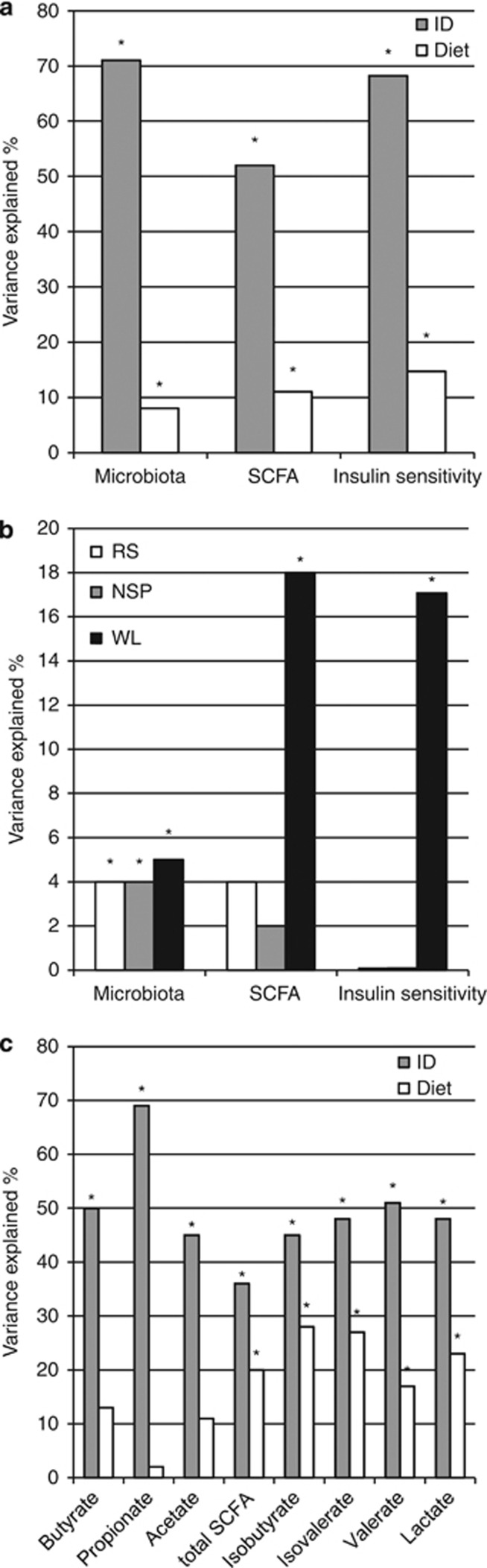
To quantify the effect of diet on the total variation in different data sets, we calculated the proportion of variance (r2) that was explained by diets and individuals. In all data sets, diet explained around 10% of the total variation, while the vast majority of the variation ascribed to the individual (Figure 7a). The ratio of individual and diet-driven variation was 2.6 times smaller for the SCFA than for the microbiota, indicating that diet had a stronger effect on the functional output of the microbiota than on its composition. Diet had a slightly stronger influence on insulin sensitivity than on the microbiota or SCFA (Figure 7a). The WL diet had a strikingly strong impact on SCFA (Figure 7b), as well as on insulin sensitivity as reported earlier (Lobley et al., 2013). For the microbiota, the variances explained by the three test diets were roughly equal (Figure 7b). In contrast to the major SCFA, the variance within the total SCFA, some minor SCFA as well as lactate were significantly explained not only by the individuality, but also by the diet (Figure 7c).
Figure 7.
Percentage of total variation in the data that is explained by diet and by individual (ID). (a) Proportion of variance explained in the microbiota (as analyzed with the HITChip), SCFAs and the markers of insulin sensitivity. (b) Variance explained by diet when looking at subsets of data collected during M diet and one of NSP, RS or WL diets. (c) Proportion of variance explained on individual metabolites. Data refer to relative proportion (%) of each metabolite. Graphs (a, b) are based on multivariate analysis of variance (ANOVA) using distance matrices allowing for effects of volunteer and diet, (c) is based on univariate ANOVA allowing for effects of volunteer and diet. Asterisks indicate variables where the explained proportion reached statistical significance (P<0.05).
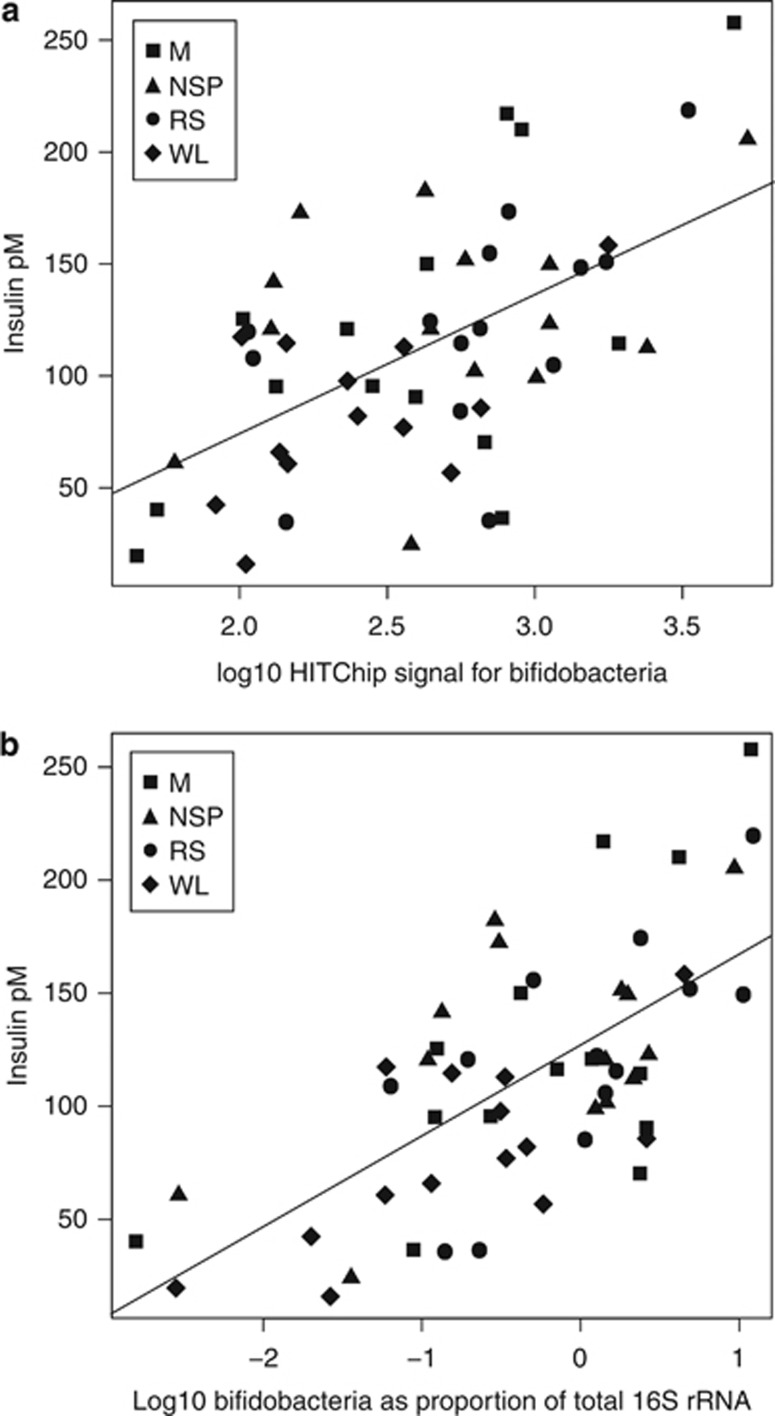
Finally, we addressed the potential associations between specific bacterial groups and insulin sensitivity. We included samples from all diets; analysis of baseline samples did not yield statistically significant correlations after correcting for multiple testing. A positive correlation between fecal bifidobacteria and plasma insulin was detected both with the microarray and qPCR (P<0.05; Figure 8; Supplementary Table S5C). The homeostasis model assessment score correlated positively with bifidobacteria and inversely with bacteria related to Bacteroides intestinalis (Supplementary Figure S4).
Figure 8.
Relationship between plasma insulin and the proportion of bifidobacteria in the fecal microbiota. (a) HITChip data, based on one sample per diet. (b) qPCR data, based on means of all available samples taken in the last 2 weeks of the NSP, RS and WL diets and in the single week of the M diet.
Individuality of the microbiota response: non-responders were characterized by high diversity
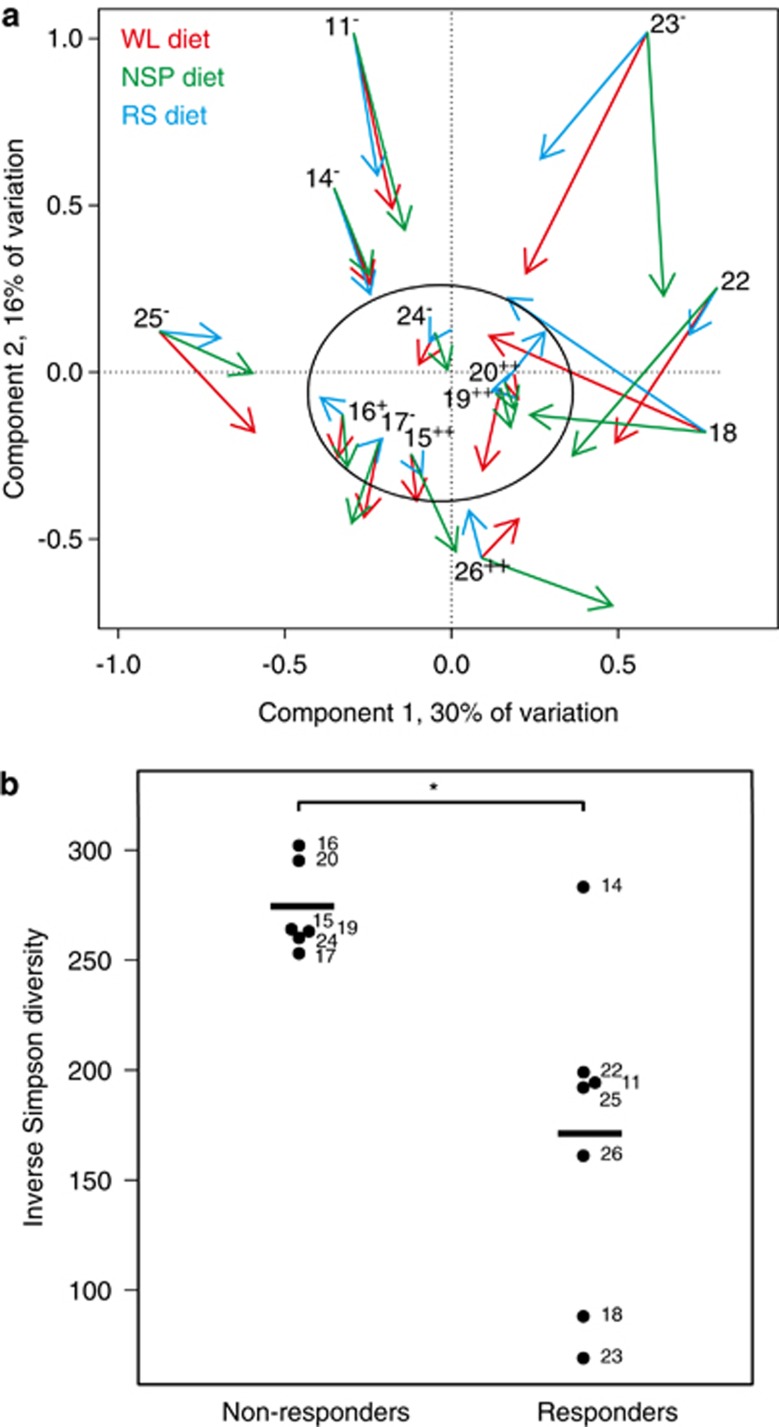
Owing to the high individuality of the subjects' microbiota and the dietary responses, we studied their potential interrelations. In a correlation-based similarity analysis, using the M diet as reference, the volunteers' microbiota tended to either respond to all dietary switches, or to remain stable throughout the trial. Hence, we divided the subjects into responders and non-responders based on the degree of microbiota stability and clustering in the principal coordinates analysis (Figure 9a). On the run-in diet, the six non-responders' microbiota was compositionally notably homogeneous (Figure 9a) and significantly higher in diversity compared with the seven responders (P<0.05; Figure 9b). The only exception was volunteer 14, a responder who showed high baseline diversity. The diversity was equally driven by the richness and evenness that correlated positively (r=0.69, P<0.05). The habitual food intake did not differ between the two groups (P=0.1), but interestingly, the prior intake levels of NSP correlated positively with the microbiota diversity during M diet in the responders, but not in the non-responders (Supplementary Figure S5). Individuals with high and low methanogenic archaeal populations (Walker et al., 2011) were found in both groups (Figure 9a). Volunteers 14 and 25, who were shown previously to exhibit incomplete RS fermentation and to have extremely low R. bromii populations, as replicated also in this study (Supplementary Figure S6), both grouped as responders.
Figure 9.
Dietary responsiveness of the individual's microbiota. (a) Biplot of the study subjects' baseline microbiota and its net response to diets indicated by arrow length. Subjects with stabile microbiota, that is, non-responders, are circled. Methanogen carriage status assessed by qPCR (based on an average of 20 samples per volunteer, Walker et al., 2011) is indicated as follows: – absence of detectable methanogens; + present in <50% of samples; ++ present in >50% of samples. (b) Diversity of the microbiota on the M (run-in) diet in the non-responders and responders. Numbers refer to specific study subjects. Asterisk indicates statistically significant difference (P<0.05).
Discussion
This study analyzed the intestinal microbiota in subjects consuming four different, fully controlled diets. Fecal samples collected at the end of each dietary regime were used for phylogenetic microarray analysis, enabling detection and relative quantification of over thousand intestinal phylotypes (Rajilić-Stojanović et al., 2009). In combination with the documentation of fecal SCFA and blood insulin levels, this enabled quantitative and qualitative assessment of the dietary impact on the ecology and metabolic output of the intestinal microbiota. The microarray analysis used here extends the previous qPCR and sequencing analyses (for details, see Introduction section; Walker et al., 2011). For abundant bacteria, the microarray results essentially recapitulated the prior data; the predominant species detected by sequencing (listed by Flint et al., 2012a) in clone libraries from six of the volunteers were highly represented also in the microarray profiles of all 14 subjects. Also previous reports have shown good correspondence for the proportions of the major phylogenetic groups between the HITChip microarray and other methods, including fluorescent in situ hybridization (Rajilić-Stojanović et al., 2009) and pyrosequencing (Claesson et al., 2009). However, the microarray analysis provided roughly 10-fold higher analytical depth compared with the previously performed sequencing; mean of 636 (s.d. 118) species-level phylotypes were detected per sample in comparison with the former 59 (s.d. 10) (Walker et al., 2011).
The cross-over study design, where each subject served his own control, coupled to a highly reproducible (Salonen et al., 2010) and in-depth community-level microbiota analysis allowed quantification of dietary impact on the human intestinal microbiota as well as on host metabolic markers. Approximately 10% of the total variance on the microbiota composition was ascribed to diet. This is substantially less than 60% reported for mice (Zhang et al., 2009; Faith et al., 2011). Such difference is not surprising regarding the notably smaller total variance of laboratory mice microbiota; the experimental diet should in practice be the sole source of variation as the animals are genetically and environmentally very similar. In that sense, the observed impact of diet upon the human microbiota is remarkable, given the high individual variation in genotype and life history, including medication and habitual diet.
The diet shifts did not markedly increase the temporal variance of the total microbiota, as the similarity of bacterial profiles, here sampled maximum of 9 weeks apart, paralleled to values observed as normal temporal variation using comparable time frame and same methodology (r=0.92 vs r=0.94; Jalanka-Tuovinen et al., 2011). However, as we have also shown previously (Walker et al., 2011), these diets induced major temporal changes in specific groups of bacteria that are diet responsive. In this work, the high analytical depth provided by the microarray enabled detection of bacterial groups that have relatively low abundance. An important outcome of the present work is identification of new groups of diet-responsive bacteria, in particular those Lachnospiracaeae that responded to the wheat bran (NSP) diet. Moreover, the microarray analysis revealed that while E. rectale responded positively to the RS diet, this does not apply to all members of the Roseburia/E. rectale cluster; thus E. rectale may have been largely responsible for the significant average increase detected previously (Walker et al., 2011). The use of species-level qPCR resulted in a similar conclusion for members of the Ruminococcaceae, highlighting the within-genus metabolic variability.
The current data show that the two chemically distinct non-digestible carbohydrates, NSP and RS, affect distinct bacteria, and have different impact on the community ecology of the human gut. Also the different forms of RS have been shown to bring about distinct compositional alterations (Martinez et al., 2010). Somewhat unexpectedly, the RS type 3 decreased both the total number as well as the diversity of the microbiota in this study. It should be noted that the bacteria were quantified per gram of feces; in the absence of data on the possible effect of the diets on the total fecal output, we cannot differentiate between relative and absolute decrease in fecal bacteria. Reduced diversity on the RS diet is assumed to reflect the promotion of a relatively small number of phylotypes, more specifically the abundant members of the Ruminococcaceae by this chemically homogeneous substrate. In contrast, the wheat bran added to the NSP diet is structurally highly complex, comprising cellulose, pectins and many different types of hemicellulose. Our findings provide insight into prebiotic mechanisms, suggesting that a simple, highly targeted prebiotic may decrease diversity as a result of dominance of abundant users, while promotion of a diverse microbiota probably requires the provision of multiple, or complex, substrates.
Bacterial fermentation products, the SCFA, are believed to provide a mechanistic link between physiological benefits associated to intake of dietary fiber. The interpretation of microbial fermentation in vivo is complex, as SCFA concentrations in feces represent the balance between production and absorption and will be influenced by many factors including the site of fermentation, transit rate and the moisture content of the digesta. The lower fecal total SCFA observed for the RS diet may, for example, reflect higher fermentation in the proximal colon and/or reduced fermentation of NSP fiber in the distal bowel (Govers et al., 1999). Nevertheless, we identified here a positive correlation between Bacteroidetes and propionate proportion, providing in vivo evidence for the association previously detected in vitro (Yang et al., 2013). Previous work revealed a clearcut relationship between the Roseburia+E. rectale group and fecal butyrate (Duncan et al., 2007; Russell et al., 2011). This relationship was also observed here when the WL diet was included in the analysis, but did not apply to the other three diets. An explanation for this is suggested by the phylogenetic microarray analysis, that is, that different phylotypes within the Roseburia+E. rectale grouping respond to changes in NSP and RS intake.
The positive correlation observed between the fecal bifidobacteria and plasma insulin and homeostasis model assessment score is in disagreement with the previously detected inverse relationship in mice (Cani et al., 2007) and humans (F.S. Teixeira et al., 2013). It should be noted that the detected positive association did not simply reflect a decrease in bifidobacteria on reduced carbohydrate WL diets that improved insulin status, as observed previously (Duncan et al., 2007; Lobley et al., 2013), but remained significant also when the WL data were omitted, highlighting the major contribution of inter-individual variation. Hence, data on the suggested protective effect of bifidobacteria in metabolic disease appear inconclusive.
Although the substantial individual differences in the dietary responsiveness are acknowledged as inherent property of the human intestinal microbiota (Martinez et al., 2010; Walker et al., 2011; Lampe et al., 2013), associating the responsiveness to the microbiota composition is an emerging concept (Korpela et al., 2014). In this cohort, high diversity associated to low dietary responsiveness of the microbiota, in line with the hypothesis that phylogenetic diversity promotes ecosystem stability. The basis of the non-responders' high diversity remains unclear, as it did not associate to available background information such as habitual diet. A ‘low gene count' (low microbiota diversity) group of 18 obese individuals identified recently by Cotillard et al. (2013) also showed a greater microbiota response to WL diets than did a ‘high gene count' comparator group (n=27). However, our recent study based on 78 obese individuals representing different backgrounds and dietary interventions identified the baseline abundance of certain Firmicutes, but not microbiota diversity, to predict the dietary responsiveness of individual's microbiota (Korpela et al., 2014). This suggests that diversity does not serve as a universal predictor for the microbiota responsiveness.
In conclusion, this analysis has identified numerous bacterial genera and phylotypes whose representation within the human colonic microbiota responds to specific dietary changes, although inter-individual variation accounted for the majority of the compositional variation, fermentation product profiles and host metabolic markers. Our data provide insight into the interconnections between diet, microbiota and host health that is the fundamental basis required for intentional modification of the microbiota toward health.
Acknowledgments
We thank the HITChip team from University of Wageningen for excellent technical assistance. This work was partly funded by the Finnish Funding Agency for Technology and Innovation (TEKES) grant 40274/06 (WMdV) and Academy of Finland grants (118602, 1141130, 137389, 141140 (WMdV) and 256950 (LL) and carried out in the context of the Finnish Centre of Excellence in Microbial Food Safety Research (CoE-MiFoSa). The Rowett Institute and Biomathematics and Statistics Scotland receive support from the Scottish Government Food, Land and People program. The human dietary study reported here was largely funded by a grant from the World Cancer Research Fund to HJF, GEL and AMJ.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Bang JW, Crockford DJ, Holmes E, Pazos F, Sternberg MJ, Muggleton SH et al. (2008). Integrative top-down system metabolic modeling in experimental disease states via data-driven Bayesian methods. J Proteome Res 7: 497–503. [DOI] [PubMed] [Google Scholar]
- Becker RA, Chambers JM, Wilks AR. (1988) The new S language: a programming environment for data analysis and graphics, Wadsworth and Brooks. Cole: Pacific Grove. [Google Scholar]
- Bouhnik Y, Raskine L, Simoneau G, Paineau D, Bornet F. (2006). The capacity of short-chain fructo-oligosaccharides to stimulate faecal bifidobacteria: a dose-response relationship study in healthy humans. Nutr J 5: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burcelin R, Garidou L, Pomié C. (2012). Immuno-microbiota cross and talk: the new paradigm of metabolic diseases. Sem Immunol 24: 67–74. [DOI] [PubMed] [Google Scholar]
- Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM et al. (2007). Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia 50: 2374–2383. [DOI] [PubMed] [Google Scholar]
- Claesson MJ, O'Sullivan O, Wang Q, Nikkilä J, Marchesi JR, Smidt H et al. (2009). Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 20: e6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S et al. (2012). Gut microbiota composition correlates with diet and health in the elderly. Nature 488: 178–184. [DOI] [PubMed] [Google Scholar]
- Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E et al. (2013). Dietary intervention impact on gut microbial gene richness. Nature 500: 585–588. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505: 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S et al. (2010). Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA 107: 14691–14696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewulf EM, Cani PD, Claus SP, Fuentes S, Puylaert PG, Neyrinck AM et al. (2012). Insight into the prebiotic concept: lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut 62: 1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. (2007). Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol 73: 1073–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Cani PD. (2013). Diabetes, obesity and gut microbiota. Best Prac Res Clin Gastroenterol 27: 73–83. [DOI] [PubMed] [Google Scholar]
- F.S. Teixeira T, Grześkowiak ŁM, Salminen S, Laitinen K, Bressan J, Gouveia Peluzio MdC. (2013). Faecal levels of Bifidobacterium and Clostridium coccoides but not plasma lipopolysaccharide are inversely related to insulin and HOMA index in women. Clin Nutr 32: 1017–1022. [DOI] [PubMed] [Google Scholar]
- Faith JJ, McNulty NP, Rey FE, Gordon JI. (2011). Predicting a human gut microbiota's response to diet in gnotobiotic mice. Science 333: 101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. (2008). Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol 6: 121–131. [DOI] [PubMed] [Google Scholar]
- Flint HJ. (2011). Obesity and the gut microbiota. J Clin Gastroenterol 45: S128–S132. [DOI] [PubMed] [Google Scholar]
- Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. (2012. a). Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3: 289–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Scott KP, Louis P, Duncan SH. (2012. b). The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol 9: 577–589. [DOI] [PubMed] [Google Scholar]
- Govers M, Gannon N, Dunshea F, Gibson P, Muir J. (1999). Wheat bran affects the site of fermentation of resistant starch and luminal indexes related to colon cancer risk: a study in pigs. Gut 45: 840–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalanka-Tuovinen J, Salonen A, Nikkilä J, Immonen O, Kekkonen R, Lahti L et al. (2011). Intestinal microbiota in healthy adults: temporal analysis reveals individual and common core and relation to intestinal symptoms. PLoS One 6: e23035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpela K, Flint H, Johnstone AM, Lappi J, Poutanen K, Dewulf E et al. (2014). Gut microbiota signatures predict host and microbiota responses to dietary interventions in obese individuals. PLoS One 9: e90702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe JW, Navarro SL, Hullar MAJ, Shojaie A. (2013). Inter-individual differences in response to dietary intervention: integrating omics platforms towards personalised dietary recommendation. Proc Nutr Soc 72: 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappi J, Salojärvi J, Kolehmainen M, Mykkänen H, Poutanen K, de Vos WM et al. (2013). Intake of whole-grain and fiber-rich rye bread versus refined wheat bread does not differentiate intestinal microbiota composition in finnish adults with metabolic syndrome. J Nutr 143: 648–655. [DOI] [PubMed] [Google Scholar]
- Lobley G, Holtrop G, Bremner D, Calder A, Milne E, Johnstone A. (2013). Impact of short term consumption of diets high in either non-starch polysaccharides or resistant starch in comparison with moderate weight loss on indices of insulin sensitivity in subjects with metabolic syndrome. Nutrients 5: 2144–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane GT, Macfarlane S. (2011). Fermentation in the human large intestine: its physiologic consequences and the potential contribution of prebiotics. J Clin Gastroenterol 45: S120–S127. [DOI] [PubMed] [Google Scholar]
- Martínez I, Kim J, Duffy PR, Schlegel VL, Walter J. (2010). Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS One 5: e15046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I, Lattimer JM, Hubach KL, Case JA, Yang J, Weber CG et al. (2012). Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J 7: 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen JFGB, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL et al. (2011). Package ‘vegan' version 2.0-2.
- Pinheiro JBD, DebRoy S, Sarkar D, the R Development Core Team (2013). nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1-110.
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajilić-Stojanović M, Heilig HG, Molenaar D, Kajander K, Surakka A, Smidt H et al. (2009). Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol 11: 1736–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. (2009). Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr 101: 541–550. [DOI] [PubMed] [Google Scholar]
- Richardson A, Calder A, Stewart C, Smith A. (1989). Simultaneous determination of volatile and non-volatile acidic fermentation products of anaerobes by capillary gas chromatography. Lett Appl Microbiol 9: 5–8. [Google Scholar]
- Russell WR, Gratz SW, Duncan SH, Holtrop G, Ince J, Scobbie L et al. (2011). High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am J Clin Nutr 93: 1062–1072. [DOI] [PubMed] [Google Scholar]
- Russell WR, Hoyles L, Flint HJ, Dumas M-E. (2013). Colonic bacterial metabolites and human health. Curr Opin Microbiol 16: 246–254. [DOI] [PubMed] [Google Scholar]
- Salonen A, Nikkilä J, Jalanka-Tuovinen J, Immonen O, Rajilić-Stojanović M, Kekkonen RA et al. (2010). Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbioll Methods 81: 127–134. [DOI] [PubMed] [Google Scholar]
- The Human Microbiome Project Consortium (2012). Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X et al. (2011). Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 5: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann U, Louis P, Goesmann A, Henrissat B, Duncan SH, Flint HJ. (2013). Complete genome of a new Firmicutes species belonging to the dominant human colonic microbiota ('Ruminococcus bicirculans') reveals two chromosomes and a selective capacity to utilize plant glucans. Environ Microbiol e-pub ahead of print 19 July 2013 doi:10.1111/1462-2920.12217. [DOI] [PubMed]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y-Y, Keilbaugh SA et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334: 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Martínez I, Walter J, Keshavarzian A, Rose DJ. (2013). In vitro characterization of the impact of selected dietary fibers on fecal microbiota composition and short chain fatty acid production. Anaerobe 23C: 74–81. [DOI] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M et al. (2012). Human gut microbiome viewed across age and geography. Nature 486: 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ze X, Duncan SH, Louis P, Flint HJ. (2012). Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J 6: 1535–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W et al. (2009). Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J 4: 232–241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.