Abstract
Fucosyltransferase 2 (FUT2) is an enzyme that is responsible for the synthesis of the H antigen in body fluids and on the intestinal mucosa. The H antigen is an oligosaccharide moiety that acts as both an attachment site and carbon source for intestinal bacteria. Non-secretors, who are homozygous for the loss-of-function alleles of FUT2 gene (sese), have increased susceptibility to Crohn's disease (CD). To characterize the effect of FUT2 polymorphism on the mucosal ecosystem, we profiled the microbiome, meta-proteome and meta-metabolome of 75 endoscopic lavage samples from the cecum and sigmoid of 39 healthy subjects (12 SeSe, 18 Sese and 9 sese). Imputed metagenomic analysis revealed perturbations of energy metabolism in the microbiome of non-secretor and heterozygote individuals, notably the enrichment of carbohydrate and lipid metabolism, cofactor and vitamin metabolism and glycan biosynthesis and metabolism-related pathways, and the depletion of amino-acid biosynthesis and metabolism. Similar changes were observed in mice bearing the FUT2−/− genotype. Metabolomic analysis of human specimens revealed concordant as well as novel changes in the levels of several metabolites. Human metaproteomic analysis indicated that these functional changes were accompanied by sub-clinical levels of inflammation in the local intestinal mucosa. Therefore, the colonic microbiota of non-secretors is altered at both the compositional and functional levels, affecting the host mucosal state and potentially explaining the association of FUT2 genotype and CD susceptibility.
Keywords: glycan foraging, intestinal microbiome, microbial ecology, multi'omic analysis
Introduction
The human intestinal microbiome contributes vital biological functions to healthy hosts, including maintenance of immune homeostasis, modulation of intestinal development and enhanced metabolic capabilities (Li et al., 2008; Turnbaugh et al., 2009a; Lee and Mazmanian, 2010; Qin et al., 2010; Human Microbiome Project C, 2012). Dysbiosis, which refers to perturbations of the normally stable intestinal microbiota, has been associated with the development and progression of many conditions, including inflammatory bowel diseases (IBDs) (Frank et al., 2007; Willing et al., 2010; Lepage et al., 2011; Morgan et al., 2012), type 2 diabetes (Qin et al., 2012) and obesity (Turnbaugh et al., 2009a). The reasons for such associations are not yet clear and may reflect either causal or secondary processes due to the impact on microbial composition and function of inter-individual variability and the contributions of environment and host genetics (Spor et al., 2011). The contributions of such factors on human microbial composition are beginning to emerge through dietary and environmental studies (Wu et al., 2011; Claesson et al., 2012; Morgan et al., 2012; Smith et al., 2013), as well as twin studies (Zoetendal et al., 2001; Stewart et al., 2005; Turnbaugh et al., 2009a; Smith et al., 2013). However, much work is still necessary to fully understand the extent of host genetic influence on the composition and function of the gut microbiome and the mechanisms linking these genetic traits with microbial function and disease biology.
A recent genome-wide association study published by our group identified Fucosyltransferase 2 (FUT2) gene as a Crohn's disease (CD) risk locus (McGovern et al., 2010), a finding that has been validated in a meta-analysis of CD and ulcerative colitis genome-wide association scans (Jostins et al., 2012). However, the molecular mechanism of the association between non-secretor status and CD remains unknown. Mucin 2 (muc2), the predominantly secreted mucin in the colon, has an important barrier role in intercepting and excluding bacteria from the mucosal cell surface, thereby reducing host susceptibility to colitis (Van der Sluis et al., 2006; Johansson et al., 2008; McGuckin et al., 2011). Research from our group has shown that aberrant glycosylation of muc2 core proteins causes spontaneous colitis in mice (Fu et al., 2011). Both the core 1- and core 3-derived O-glycans of mucin core proteins are terminally fucosylated, which serve as interceptive binding structures for bacteria (Linden et al., 2008). Moreover, a subset of human intestinal microbiota produce glycosidases capable of hydrolyzing α-1,2-fucosyl linkages present in various mucin-type glycoproteins, as well as mucus glycan structures that are not capped by fucose (Katayama et al., 2004). A mass spectrometry-based analysis of insoluble colonic mucin of both Fut2-null and wild-type mice (Hurd et al., 2005) identified 17 different oligosaccharides with up to eight sugar residues of which 11 were neutral, five sulfated and one sialylated. The most abundant structures were composed of core 2 (Galβ1-3(GlcNAc β1-6)GalNAc-) glycan sequence with some based on core 1 (Galβ1-3GalNAc-) glycan structures. The primary difference in oligosaccharides was the presence of terminal fucose residues forming the blood group H-type epitope in most of the oligosaccharides in wild-type mice. In contrast, all the peaks for oligosaccharides carrying blood group H-type epitopes were absent in the Fut2-null mice. Therefore, FUT2 deficiency may alter the composition of intestinal microbiota by affecting either microbial adhesion and/or utilization of host-derived glycans, potentially leading to dysbiosis.
The phylogenetic composition in non-secretor individuals have been characterized in two studies recently (Rausch et al., 2011; Wacklin et al., 2011), showing that the FUT2 genotype was associated both with deviations of overall community ecology and with altered abundances of specific microbes. However, these descriptions did not address the degree to which these alterations were functional nor their potential mechanisms of action in IBD risk. Both questions are of particular interest, because microbial composition can exhibit large inter-individual variations compared with function-based analyses even in healthy individuals (Qin et al., 2010). This may also be one of the reasons for existing between-study discrepancies (Rausch et al., 2011; Wacklin et al., 2011), other than the difference between measurements of the luminal/fecal microbiota and those at the mucosal surface (Eckburg et al., 2005). As bacterial colonization largely occurs in the outer mucous layers (Johansson et al., 2008) where the residual glycans that fuel bacterial growth are degraded, lavage sampling of the mucosal surface compartment coupled with functional and metabolic profiling is arguably more biologically relevant to host–microbial glycan metabolism.
We present here a comprehensive description of the mucosal luminal interface of healthy individuals distinguished by secretor status, capturing multiple aspects of the microbial ecosystem including microbiome composition, imputed function, metabolome and proteome. 16S rRNA gene sequencing can be used to characterize the composition and diversity of the microbiota, and with recent advances, it allows us to impute functionality of the microbiome. Deeper insight into microbial functionality can be provided by combining 16S rRNA gene sequencing with proteomic and metabolomic data (Erickson et al., 2012). We detailed the phylogenetic and functional profiles of the mucosal microbiome associated with FUT2 polymorphism, indicating a strong effect of host genetics on the re-programing of energy metabolisms into dysbiotic setting. The combination of multi'omic analysis also provided us with unprecedented understanding of the dynamics of host–microbial interaction.
Materials and methods
Subject cohort and lavage sample collection
A subject cohort of 39 individuals (Supplementary Table S1) was recruited from patients presenting for screening colonoscopy at Cedars-Sinai Medical Center. Following institutional review board approval, subjects were consented and then included in the study if colonoscopy did not reveal any mucosal abnormalities. Enrolled subjects were prepared for colonoscopy by taking Golytely the day before the procedure. The mucosal lavage samples representing the mucosal luminal interface were collected from cecum and sigmoid colon as described previously (Li et al., 2011).
Animals
All animal protocols were in accordance with Administrative Panel on Laboratory Animal Care, the Stanford Institutional Animal Care and Use Committee. Conventionally housed Fut2−/− mice (B6.129 × 1-Fut2tm1Sdo/J; backcrossed 12 generations with C57BL/6J) were re-derived as germ free and maintained in gnotobiotic isolators. Eight-week-old non-littermate germ-free Fut2-deficient Fut2−/− (n=10), wild-type Fut2+/+ (n=10; C57BL/6J) and heterozygous Fut2+/− (n=8) mice were colonized with feces obtained from a healthy human donor (secretor) by oral gavage of 200 μl of human fecal sample (male, age 38 years, American diet). The sample was prepared by mixing stored frozen human fecal sample with filter-sterilized pre-reduced phosphate-buffered saline. Mice were singly housed and maintained in gnotobiotic isolators on a strict 12-h light cycle for the experiment. Mice were fed a standard autoclaved mouse diet (Purina LabDiet 5K67, LabDiet, St Louis, MO, USA). Fecal samples were collected 4 weeks after humanization for 16S rRNA gene sequencing using the 454 titanium platform.
Genotyping
The single-nucleotide polymorphism rs601338 (G>A) defines secretor status in Europeans and Africans (Ferrer-Admetlla et al., 2009). We used rs516246, which is in strong linkage disequilibrium with rs601338, to infer secretor status. Estimate of linkage between rs516246 and rs601338 is 100%. These two single-nucleotide polymorphisms tag each other perfectly, as they are in perfect LD with one another, with R2 of 1.0 and D′ of 1.0, according to Hapmap3 release 2, CEU population. The individuals with the homozygote A/A genotype are defined as non-secretors (Supplementary Materials and Methods). In this cohort, 97% (38/39) of the subjects are Caucasian. One subject is African American, who is heterozygous G/A genotype for rs516246 and therefore categorized as Sese. Mouse genomic DNA was prepared from ear tissue obtained by ear punch. PCR amplification using three primers (F: 5′-CCTGCCATGCTTTCTTTCCTG-3′, R: 5′-ATTCCTTCTCTGACAGGGTTTGG-3′ (WT), 5′-TGGGTAACGCCAGGGTTTTC-3′ (KO)) yielded either a 191-bp band (Fut2−/−) or 154-bp band (Fut2+/+) or both (Fut2+/−).
16S rRNA gene sequencing and microbial composition analysis
Genomic DNA was extracted as previously described (McHardy et al., 2013). The V4 region of 16S ribosomal RNA genes were amplified and sequenced on an Illumina HiSeq 2000 (Illumina, Inc., San Diego, CA, USA) as previously described (McHardy et al., 2013). HiSeq reads were processed using QIIME v1.5.0 (Caporaso et al., 2010) with parameters of: minimum Q-score considered high quality: 20, maximum number of consecutive low-quality base calls allowed before truncating: 3, and maximum number of N characters allowed: 0. All filtered reads had a length of 101 bp. The number of reads per sample ranged from 326 481 to 1 021 473, with a mean of 646 140 and totaling 48 460 491. Sequence sub-sampling was performed for each sample at the depth of 300 000 reads per sample. This normalized data set was used for all the following analysis, including alpha-diversity analysis, beta-diversity analysis and imputed metagenomic analysis. For mouse fecal pellets, after DNA isolation (MoBio fecal DNA kit, MO BIO Laboratories, Inc., Carlsbad, CA, USA), 626 bp amplicons spanning 16S variable regions 3′–5′ (V3–V5) were generated using barcoded forward primer (338F, 906R) (Kashyap et al., 2013). Samples were sent for pyrosequencing to Duke ISGP (Durham, NC, USA) using the Roche 454 titanium platform (454 Life Sciences, Branford, CT, USA). Operational taxonomic units (OTUs) were picked against the 4 February 2011 version of the Greengenes database (http://greengenes.lbl.gov/cgi-bin/nph-index.cgi) (DeSantis et al., 2006), pre-filtered at 97% identity. For quality control, all the singletons were removed. After reference-based OTU picking, 97.5% of the total reads were successfully mapped to the reference Greengenes database. These steps were performed using QIIME v1.5.0 (Caporaso et al., 2010). Alpha rarefaction was performed using the number of observed species, Chao1 and phylogenetic diversity. The comparison of alpha diversity between the two groups at certain sampling depths was performed using a two-sided Student's t-test. Beta diversity of 16S rRNA gene and imputed metagenomic data sets were estimated by computing unweighted UniFrac and Bray–Curtis distances between samples, respectively, using QIIME. Ordination of the resulting distance matrix was performed using principal coordinate analysis (PCoA). Pair-wise comparisons between SeSe, Sese and sese individuals were conducted using the Kruskal–Wallis test to identify differentially abundant bacterial phylotypes at phylum and 97% OTU levels. Multiple hypothesis tests were adjusted to produce a final Benjamini and Hochberg false discovery rate (FDR; Benjamini and Hochberg, 1995), and significant association was considered below a FDR q-value threshold of 0.25.
Identification of functional microbial communities (FMCs)
We used 97% OTUs with a minimum of 30 reads in the data set that were present in>50% of samples. We first defined a co-occurrence similarity measure, which was used to define the network. Assume that the vector xi specifies the abundance of the i-th OTUs across the samples, the pair-wise Sparse Correlations for Compositional data (SparCC) ρij was inferred from the abundance profile of each OTU xi and xj as the measurement of co-occurrence relationship (Friedman and Alm, 2012). A signed weighted adjacency matrix (network) was defined by raising ρij to a power aij=(0.5+0.5ρij)β, with β=4 (Zhang and Horvath, 2005). Modules were defined as branches of a hierarchical clustering tree based on the topological overlap measure. The modules were detected after applying the dynamic tree cut method (Langfelder et al., 2008). These network modules (clusters) were interpreted as functional microbial communities (FMCs). The classical multidimensional scaling of the topological overlap distance matrix was plotted using the ‘cmdscale' function in R (v.2.15.1). To summarize the profiles of co-occurrence modules, we calculated the eigenOTU, which is defined as the first right-singular vector of the standardized module abundance matrix. The computation is implemented in the R function module Eigengenes (Langfelder and Horvath, 2008). As a weighted average abundance profile, it provides a mathematically optimal way of summarizing the co-occurrence patterns of all OTUs belonging to each module (Tong et al., 2013). To identify modules (FMCs) that were correlated with clinical traits, we used correlation tests to relate each eigenOTU to the clinical traits. These steps were performed using the WGCNA package (version 1.13) in R (version 2.13.1) (Langfelder and Horvath, 2008). R tutorials explaining the analysis steps can be found on the webpage: http://www.genetics.ucla.edu/labs/horvath/CoexpressionNetwork/Rpackages/ WGCNA/Tutorials/.
Imputation of microbial gene content and metagenomes
This study takes advantage of PICRUSt, a program that infers the metagenome of a sample from its phylogenetic composition and was recently validated against conventional deep-sequencing metagenomics (Langille et al., 2013). The OTU table was used as the input file for metagenome imputation of individual human and mouse samples. For the metagenomic profiling of FMCs, the OTU table of the six FMCs was generated with one count for each 97% OTU in a given FMC. The gene content of 2590 KEGG (Kyoto Encyclopedia of Genes and Genomes) reference genomes was used to infer the approximate gene content of the detected phylotypes using PICRUSt (v0.1) (http://picrust.sourceforge.net/) (Langille et al., 2013). The program output the inferred metagenome represented by KEGG Orthology (KO) for each FMC. Taking the PICRUSt KO gene abundance inferences as inputs, the metabolic pathways were re-constructed using HUMAnN (v0.98) (Abubucker et al., 2012). We restricted our analysis to the KEGG pathways that were present in at least 90% of the samples. Pair-wise Kruskal–Wallis tests between SeSe, Sese and sese individuals were performed to identify imputed KEGG pathways with differential relative abundance. Multiple hypothesis tests were adjusted to produce a final Benjamini and Hochberg FDR (Benjamini and Hochberg, 1995), and significant association was considered below a FDR q-value threshold of 0.05.
Mass spectrometry analysis
For metabolomic analysis, each human lavage samples was subjected to solid-phase extraction to eliminate a polymeric contaminant believed to originate from the lubricant used during colonoscopy preparation. The eluate was dried and reconstituted in 2% acetonitrile in water before mass spectrometry analysis. A 5-μl aliquot of extracted metabolites from each sample was injected onto a reverse-phase 50 × 2.1 mm ACQUITY 1.7-μm C18 column (Waters Corp., Milford, MA, USA) using an ACQUITY UPLC system (Waters Corp.). A Waters Q-TOF Premier was operated in negative-ion (ESI−) or positive-ion (ESI+) electrospray ionization mode with a capillary voltage of 3200 V and a sampling cone voltage of 20 V in negative mode and 35 V in positive mode. Data were acquired in centroid mode with a mass window of 50 to 850 m/z and processed using MassLynx software (Waters Corp.) (Supplementary Materials and Methods). To profile the meta-proteome of lavage samples, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) was performed using the soluble fraction of samples as previously described in Li et al. (2011). The abundances of metabolomic and proteomic peaks were compared using analysis of variance (ANOVA) to identify features associated with FUT2 genotype. Multiple hypothesis tests were adjusted to produce a final Benjamini and Hochberg FDR (Benjamini and Hochberg, 1995), and significant association was considered below a FDR q-value threshold of 0.25. The relatively high FDR was used to avoid excessively strict filtering of metabolomic and proteomic features.
Results
Whole-community microbiome ecology differs according to secretor status
To study the host–microbial interaction at the mucosal luminal interface, 75 lavage samples were collected from the cecum and sigmoid colons of 39 healthy subjects (Supplementary Table S1). We assessed differences in overall microbial ecology between secretors (both homozygous SeSe and heterozygous Sese for the functional allele) and non-secretors (sese).
We first examined the microbial composition in these samples, to affirm that the present cohort matched previously reported differences between secretors and non-secretors in microbial composition (Rausch et al., 2011; Wacklin et al., 2011). The microbiota from these samples was profiled by multiplex sequencing, and a total of 47 171 132 reads (628 948±130 744 s.d. reads per sample) were generated after quality control. A total of 4074 OTUs were then identified by grouping reads at a 97% sequence similarity threshold. Compared with SeSe individuals, both Sese and sese individuals exhibited lower alpha-diversity based on the number of observed species (t test, P=0.012 and 0.085, respectively), although the difference between SeSe and sese individuals was not statistically significant (Supplementary Figure S1a). We also measured other diversity indexes, including Chao1 and phylogenetic diversity. Compared with SeSe individuals, Sese individuals exhibited significantly lower alpha-diversity as indicated by Chao1 and phylogenetic diversity indexes at the depth of 300 000 reads per sample (t test, P=0.019 and 0.02, respectively). The same trend was observed in sese individuals, although not statistically significant (t test, P=0.10 for Chao1 and 0.18 for phylogenetic diversity) (Supplementary Figures S1b and c).
The beta-diversity measured by unweighted UniFrac distance matrix was calculated for each sample to evaluate the similarity between microbial communities. PCoA demonstrated that the phylogenetic compositions of SeSe microbiomes were significantly different from those of Sese (Adonis test, P=0.016) but not of sese individuals (Adonis test, P=0.092) (Supplementary Figure S2a). The significant difference in phylotype abundances reported previously, namely, the increase in Bacteroidetes among non-secretors, was confirmed at the phylum level (Rausch et al., 2011) (Supplementary Figure S2b).
To analyze at lower taxonomic levels, we filtered out low-abundant 97% OTUs based on the criteria of (1) minimum total observation count of 30 across all samples and (2) being observed in at least 60% of the samples, reducing the number of OTUs from 4074 to 419. Among these OTUs, 19 (4.5%) of them were depleted in Sese and sese compared with SeSe individuals (Kruskal–Wallis, FDR q<0.25) (Supplementary Data set S1). In summary, the FUT2 polymorphism was significantly associated with selected phylotypes of colonic microbiota in Sese and sese individuals, and the alterations in Sese individuals resulted in a significant shift of microbial composition compared with SeSe. These data revealed the gardening effect of FUT2 polymorphism on phylogenetic composition of the colonic microbiota.
Non-secretor-associated functional changes revealed by imputed metagenomes
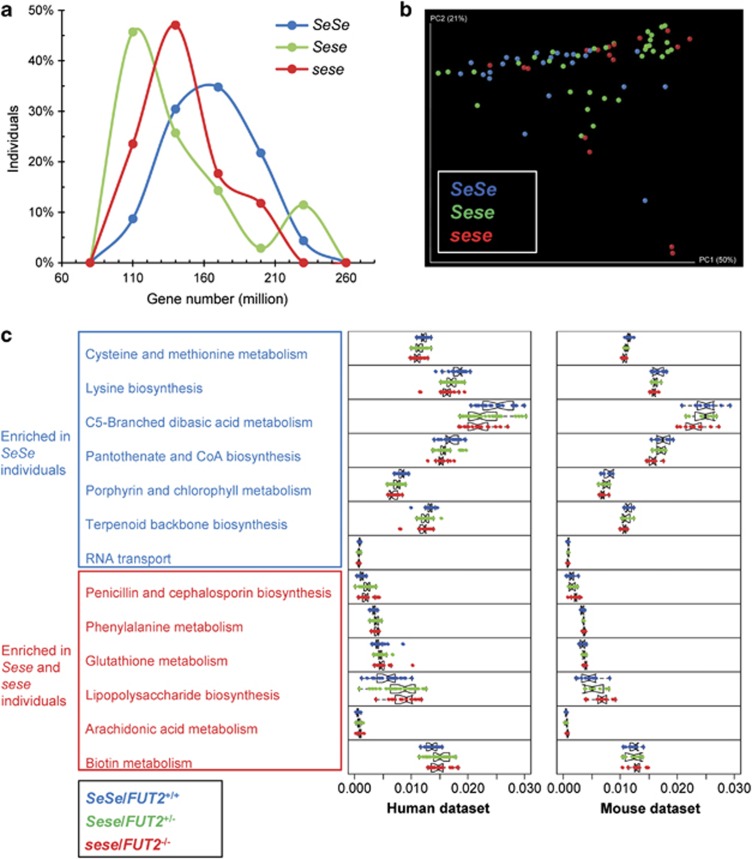
We hypothesized that these compositional changes result in selectively augmented or deficient functional capabilities that may be relevant to CD susceptibility. To test this idea, we inferred the metabolic capacities of mucosal microbiota associated with secretor status, using a recently developed bioinformatic pipeline centering on the PICRUSt (Langille et al., 2013) and HUMAnN tools (Abubucker et al., 2012). In the ‘gene content inference' step, the gene contents and 16S rRNA gene copy number of the detected phylotype were predicted based on its evolutionary similarity with the 1119 KEGG reference genomes. In the subsequent ‘metagenome inference' step, the resulting gene content predictions for all microbial taxa with the relative abundance of 16S rRNA genes in each samples are combined and corrected for expected 16S rRNA gene copy number, to generate the expected abundances of gene families in the entire community represented by KOs. The prediction accuracy of PICRUSt has been validated using human and mammalian gut microbiome with paired 16S rRNA gene and metagenome sequencing data (Langille et al., 2013). The relative abundances of KEGG pathways in each sample were then reconstructed by mapping KOs to these pathways using HUMAnN. At our routine sampling depth, both Sese and sese individuals harbored 15% fewer microbial genes on average than SeSe individuals (Figure 1a), which is consistent with the significant lower compositional diversity observed in Sese individuals compared with SeSe individuals.
Figure 1.
Imputed metagenomes reveal the significant enrichment of KEGG pathways in secretors and non-secretor individuals. (a) Distribution of bacterial genes in SeSe, Sese and sese individuals. The proportions of individuals having a given number of genes are shown. (b) Communities clustered using PCoA of the Bray–Curtis distance matrix. Each colored point corresponds to a sample. The clustering of SeSe was significant compared with both Sese and sese individuals (Adonis test, P=0.004 and 0.004, respectively). (c) Relative abundance of KEGG metabolic pathways in microbiome samples was colored by secretor status. Only the 13 pathways showing concordant alterations in both human and murine data sets were plotted.
The similarity of functional states of the microbiomes from secretors and non-secretors was evaluated by the composition of imputed metagenomes. PCoA using Bray–Curtis distance demonstrated separation of the samples from SeSe, Sese and sese individuals along PC1. The clustering of SeSe was significant compared with both Sese and sese individuals (Adonis test, P=0.004 and 0.004, respectively), suggesting that variations at the metagenomic level were more profound than those at the compositional level and that FUT2 exhibited haploinsufficiency in programing the metagenomic functions (Figure 1b). In support of the distinction between the phylogenetic and imputed metagenomic data sets, Procrustes analysis of the Bray–Curtis PCoA plots derived from 16S rRNA gene and imputed metagenome data sets showed that the clustering of samples across data sets was not significant (P=0.510; Supplementary Figure S3). Among the 154 imputed metabolic pathways, 23 (14.9%) were differentially abundant between SeSe and Sese individuals. The alterations in sese individuals were greater, as shown by the changes of abundances of 43 (27.9%) of the imputed pathways compared with SeSe individuals (Kruskal–Wallis, FDR q<0.05) (Supplementary Data set S2, Supplementary Figures S4 and S5).
The FUT2-associated changes were more robust at the imputed metagenomic versus the phylotypic level. As compared with SeSe individuals, a diverse consortium of metabolic functions represented by 27 KEGG pathways were depleted in sese individuals, including amino-acid metabolism pathways, cofactors and vitamins metabolism pathways and genetic information processing pathways (Supplementary Figure S6). A broad-based decrease in amino-acid biosynthesis was observed, including lower abundances of lysine (KO00300), valine, leucine and isoleucine (KO00290), and phenylalanine, tyrosine and tryptophan (KO00400) biosynthesis pathways. Accompanying the depletion, 16 microbial pathways were enriched in these individuals, highlighted by aspects of energy metabolism, including carbohydrate and lipid metabolism, cofactors and vitamins metabolism and glycan biosynthesis and metabolism (Supplementary Figure S7). These data suggest that FUT2 gene polymorphism acted in a haploinsufficient manner to perturb metabolic pathways such as amino-acid biosynthesis encoded by the gut microbiome at the mucosal interface.
To validate these findings, we performed the same analysis on a 16S rRNA gene data set of the fecal samples collected from humanized FUT2−/− mice (germ-free FUT2−/− mice colonized with human feces from a healthy secretor) (Kashyap et al., 2013). Among the 146 metabolic pathways reconstructed, 47 (32.2%) of them were differentially abundant between the FUT2+/+ and FUT2−/− mice (Supplementary Data set S3), comparable to our findings with the human samples. After cross comparing the two data sets, we identified 13 pathways that were consistently enriched or depleted with FUT2 haploinsufficiency (Kruskal–Wallis, FDR q<0.05) (Figure 1c). Specifically, the carbohydrate and lipid metabolism and glycan biosynthesis-related pathways were over-represented in Sese/sese individuals and FUT2+/−/FUT2−/− mice, whereas the relative abundances of five amino acid and vitamin metabolism-related pathways were enriched in SeSe individuals and FUT2+/+ mice (Figure 1c). These findings suggest that FUT2 genotype had a similar impact on imputed microbial metagenomic functions in humans and mice, notably in reduced amino-acid synthesis capabilities.
FMCs associated with non-secretor status
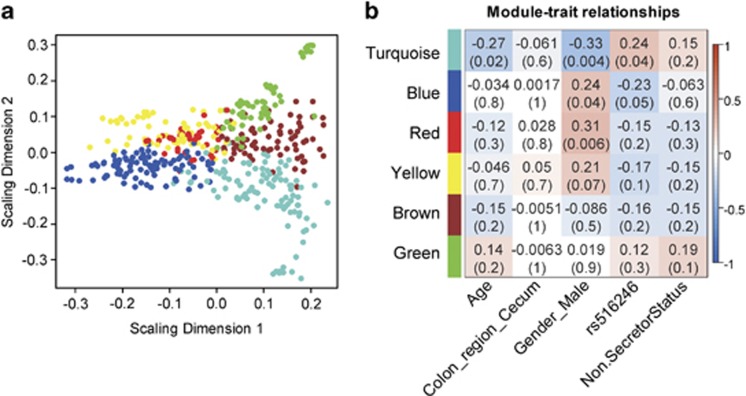
It is well understood that even healthy individuals differ remarkably in their fecal and mucosal surface gut microbial composition, especially at the genus and species level. Mucosal microbiota can be further assessed for functional relatedness based on their co-occurrence patterns (Faust et al., 2012; Koren et al., 2013). To determine whether such ecological structures can be observed in this data set, we developed a methodology to infer microbial co-occurrence networks (see Materials and methods). Nodes (OTUs) of these networks were grouped based on their topological overlaps using hierarchical clustering. Using this approach, six modules, ranging from 39 to 97 OTUs, were identified (Supplementary Figure S8). These modules of OTUs represent putatively key ecological features, which we term as functional microbial communities (FMCs). Multi-dimensional scaling was used to depict module structure and network connections (Figure 2a). Phylogenetically related OTUs were clustered into the same FMC, presumably because preferences for ecological niche are more likely to be shared between more closely related microbes. However, each FMC also included phylogenetically distinct OTUs from different phyla, suggesting that in addition to phylogenetic relatedness the formation of FMC depended upon additional ecological affinities, which could range from syntrophic dependences to convergent functionality between distinct phylotypes (Supplementary Data set S4).
Figure 2.
FMCs associated with non-secretor status. (a) Classical multi-dimensional scaling plot in which OTUs in each FMC represented by colored dots tend to form distinct clusters. (b) FMC-trait correlations and P values. Each cell reports the Pearson correlation coefficient (and P-value) derived from correlating FMC eigenvectors (rows) to traits (columns). For the association with non-secretor status, the SeSe and Sese individuals were grouped together as secretor. For the association with FUT2 genotype (rs516246), additive genetic model was used. The table was color-coded by correlation according to the color legend.
The ‘abundance' of each FMC can be quantified by defining the OTU abundance profiles of each FMC (its module eigenvector; defined in Material and methods). One can thus correlate the abundance of the FMCs with the metadata, including host genotype, disease phenotype, age and so on. When using an additive genetic model, the abundances of turquoise and blue FMCs significantly associated with the copy number of the FUT2 loss-of-function allele reciprocally (P=0.04 and 0.05, respectively, with rs516246 by Pearson correlation) (Figure 2b). This observation was concordant with results at the individual OTU level: when examining the membership of the FMCs, we found that 12 of the 19 OTUs enriched in SeSe individuals were assigned to the blue FMC. Gender was another subject phenotype that significantly associated with microbial composition: the turquoise (P=0.004, Pearson correlation) FMC was enriched in females, whereas the blue and red FMCs (P=0.04 and 0.006, respectively, Pearson correlation) were more abundant in males. The gender effect on intestinal microbiome has also been reported previously in humans and murine model (Mueller et al., 2006; Markle et al., 2013). Because of the difference of male/female ratio in SeSe, Sese and sese individuals (Supplementary Table S1), the gender effect could potentially contribute to the association of turquoise and blue FMCs with FUT2 genotype.
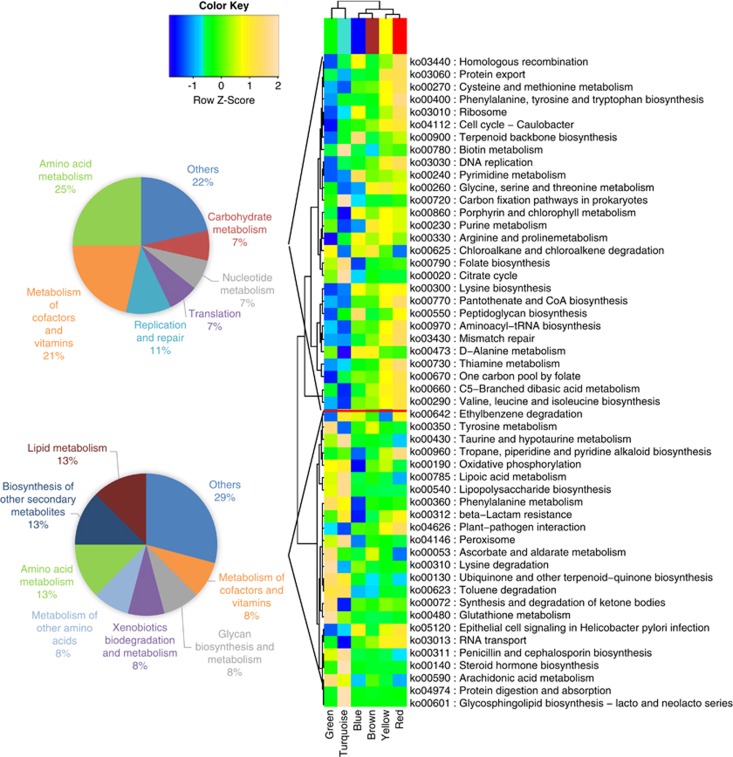
To determine whether these co-occurring microbial communities represented distinct functional units at the mucosal surface, we profiled the metabolic capabilities of FMCs using the approximate gene contents imputed previously. After aggregating the individual inferred genomes according to module membership, the relative abundances of metabolic pathways in each FMC were re-constructed. The functional profiles of FMCs were highly variable (Figure 3). The pathways associated with FUT2 clustered into two groups that were over-represented in the FUT2 loss-of-function allele-associated turquoise FMC and functional allele-associated blue FMC, respectively. Moreover, the pathways enriched in SeSe individuals tended to cluster into amino-acid metabolism class and cofactors and vitamins metabolism class, whereas the pathways enriched in Sese and sese individuals highlighted amino-acid metabolism, lipid metabolism and biosynthesis of secondary metabolites (Figure 3). These metabolically specialized microbial communities were therefore responsible for the imputed metagenomic alterations associated with FUT2 polymorphism.
Figure 3.
Variations of KEGG metabolic pathways in the functional microbial communities. The heatmap shows the functional profiles of FMCs (columns) based on the relative abundance of FUT2-associated metabolic pathways (rows) after z-score transformation. The color bar on top shows module membership. The dendrograms show the hierarchical clustering of columns and rows, respectively, using Euclidean distance. The two pie-charts show the number of pathways in each functional class for the cluster associated with turquoise and blue FMC, respectively.
Non-secretor-associated metagenomic changes reflected by metabolomic and proteomic profiling
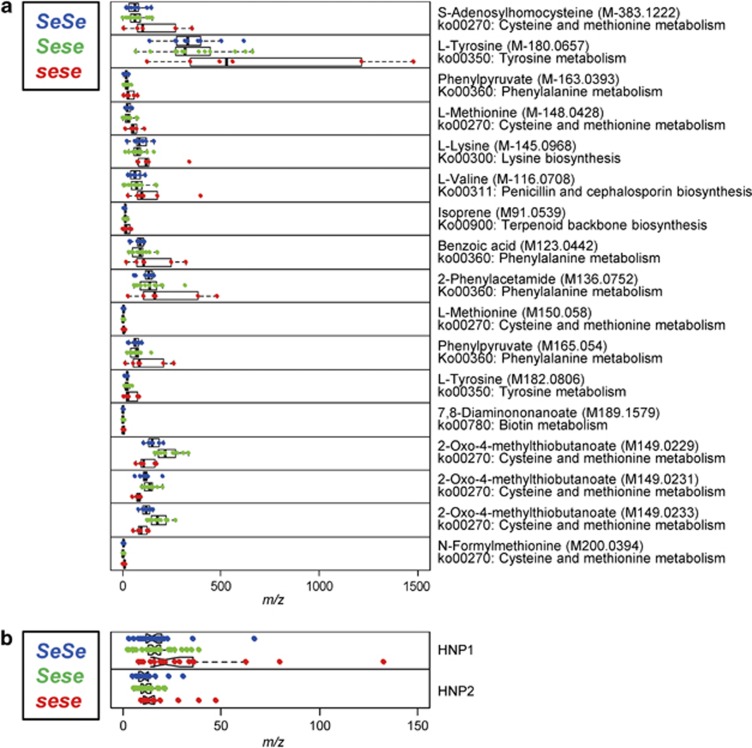
To determine whether the imputed metagenomic alterations associated with FUT2 polymorphism correlate with changes in metabolic activities of mucosal microbiota, we profiled the soluble metabolites of the same lavage samples using Q-TOF MS. The analysis generated a rich metabolomic data set consisting of 649 and 576 spectral features in the cecum and sigmoid regions, respectively. Putative IDs were assigned to 372 ions by comparing their m/z values to those available in online databanks using a predefined mass error window of 20 p.p.m. The putative IDs were then used to map out the ions to various metabolomic pathways in the KEGG data set. In accord with our previous study,∼50% of all metabolites were located at the terminal end of metabolic pathways, suggesting enrichment for end-products (McHardy et al., 2013). In the cecum, 48 metabolites were mapped to the 13 KEGG pathways associated with non-secretor individuals in the human and murine data sets and were present in>90% of the samples. Of these, 13 (27.1%) were differentially abundant among secretors (SeSe and Sese) and non-secretors (sese) (ANOVA, FDR q<0.25). In the sigmoid, 30 metabolites were mapped to the non-secretor-associated pathways; 4 (13.3%) were differentially abundant (ANOVA, FDR q<0.25; Figure 4a and Supplementary Data set S5). A less stringent q-value, up to 0.25, was used to avoid missing significant associations, as shown in recent comparable study designs (Wu et al., 2011; Morgan et al., 2012). When using more stringent q-value threshold (FDR q-value<0.1), these metabolites did not show significant association, which is comparable with the results from study with similar design (Wu et al., 2011). Thus, differences in imputed microbial metagenome content corresponded to abundances of metabolic end-products directly detected in the same samples.
Figure 4.
Meta-metabolomic and meta-proteomic features that differentiate secretors and non-secretors. Relative abundance of meta-metabolomic (a) and meta-proteomic (b) features in lavage samples is colored by secretor status.
To determine how the host reacted to the changes of the functional state of the microbiota, we also profiled the proteomic features of the same samples using MALDI-TOF MS. We focused our analysis on peaks of human origin. Of the 453 peaks included, the abundances of 16 (3.5%) were significantly different among secretors and non-secretors (ANOVA, FDR q<0.25). Although only 17 (3.8%) molecular features could be identified, we found that the expression levels of human neutrophil peptides 1 and 2 (HNP-1 and -2) were significantly higher in non-secretors, consistent with a sub-clinical inflammatory state at the mucosal surface (Figure 4b). This difference could be driven by one particular sese sample that had high expression of HNP-1 and -2 relative to the other samples (Figure 4b). To exclude such possibility, we repeated the analysis excluding this sample, and found that the levels of HNP1 and HNP2 were still significantly higher in non-secretors based on nominal P-values (ANOVA, nominal P=0.007 and 0.047, respectively), although FDR q-values were>0.25 (FDR q=0.27 and 0.49, respectively). Our clinical records did not indicate that this individual had gastrointestinal symptoms or chronic inflammation in the intestine at the time of sampling. These data suggest that the non-secretor state of the mucosa, via alteration of the mucosal surface ecosystem, changes the inflammatory state of the human intestinal mucosa.
Discussion
We combined 16S rRNA gene sequencing, metagenome imputation, meta-metabolomic and meta-proteomic profiling to delineate the integrated landscape of the mucosal surface ecosystem. The phylogenetic diversity and composition of intestinal mucosal microbiota in non-secretor individuals were significantly different from that of secretors. Compared with SeSe, the metabolic functions encoded and expressed by the gut microbiome in non-secretors were enriched for carbohydrate and lipid metabolism, cofactors and vitamins metabolism and glycan biosynthesis and metabolism and depleted for seven pathways related to amino-acid metabolism. These alterations in humans were highly consistent with analogous changes in the murine genetic counterpart. Changes in the imputed metagenomes were reflected by concordant metabolite pools as determined by meta-metabolomic assays, providing validation for certain imputed metagenomic changes as functionally consequential. Moreover, these microbial functional changes were accompanied by sub-clinical intestinal inflammation. FUT2 therefore appears to have a role in shaping the functional state of the mucosal surface by affecting not only microbial composition but also the resulting functional state of the microbiota at the human intestinal mucosal surface.
It was surprising that difference with the SeSe group were in several cases (for example, alpha-diversity) greater in the Sese rather than in the sese group. This raises two issues. One is the extent of glycan difference produced by haploinsufficiency. As there was a strong haploinsufficiency phenotype in all the facets of this study, we surmise that a substantial glycan change is produced by haploinsufficiency. However, to our knowledge, the glycan profile in heterozygous individuals has not been well described in the literature. The other issue is why the Sese group had a larger and more significant difference than the sese group. One possible explanation is that the inter-individual variation of gut microbial phylogenetic composition is inherently large (Human Microbiome Project C, 2012; Yatsunenko et al., 2012). In this context, it is notable that the sizes of the test groups were modest (9 for sese, 18 for Sese). It is possible that the particularly small size of the sese group made it prone to outliers and potentially underpowered for establishing mean phenotypes and robust statistical comparisons (for example, the 95% confidence interval for alpha-diversity was larger for the sese (4.4) than for the Sese (1.8) group).
The imputed metagenome represents an accurate but approximate inference of the reference microbial genomes currently available. Potential bias may result from unmappable 16S rRNA gene reads and lack of sufficient reference genomes. After the reference-based OTU picking, 97.5% of the total reads were successfully mapped to the reference Greengenes database. The 2.5% unmappable 16S rRNA gene reads might cause the loss of metagenomic content that can be captured by shot-gun sequencing. Also, only the 2590 microbial genomes that had identifiers in the Greengenes reference tree were used as the reference to predict unknown genomes. Despite these bias and limitations, PICRUSt predictions usually reach high agreement with metagenomically measured gene content (Spearman r=0.8–0.9) (Langille et al., 2013). In this study, the findings of imputed metagenomic analysis in human subjects were validated by metabolomic and proteomic data as well as by comparison to an independent 16S rRNA gene data set from humanized FUT2−/− mice. In the future, adding meta-transcriptomic data will further enrich our understanding of microbial functional capability at any given time.
Inconsistencies of FUT2-associated imputed metagenomic changes were also observed between human and murine data sets. In the humanized mouse gut microbiota, there are changes of microbial compositions as compared with the donor (Turnbaugh et al., 2009b). The fecal sample used for humanization was from only one healthy donor, which might cause inherent bias due to the limited sample size. Also, taxonomic biases between the two different 16S rRNA gene data sets may exist due to different PCR primer sequences, amplicon lengths and sequencing technologies (Claesson et al., 2010).
One of the key drivers of gut microbiota composition and function is the type and quantity of complex carbohydrates, which are typically derived from either diet or host mucus (Wu et al., 2011; Koropatkin et al., 2012). These polysaccharides serve as a primary metabolic input for the abundant carbohydrate-fermenting bacteria within the microbiota (Martens et al., 2011). However, different microbes within the gut are differentially endowed with abilities to use specific types of glycans (Sonnenburg et al., 2010), and so differences in carbohydrate availability, such as the presence or absence of fucose in mucosal glycans, translate into selective and regulatory events that result in discrete alterations in the microbiota's functional properties (Kashyap et al., 2013). Individual members of the microbiota can alter gene expression to accommodate the absence of fucose, while other members may be lost, and others recruited (Kashyap et al., 2013). Additionally a change in carbohydrate utilization by relatively few members can cascade into ecosystem-wide alterations, given the interconnectedness of metabolic functions within the microbiome. Fucose processing by a gut-resident symbiont allows expansion of pathogenic species, such as Salmonella typhimurium. Similarly, increased sialic acid release by certain bacterial species (Bacteroides thetaiotaomicron) allows expansion of Clostridium difficile (Ng et al., 2013). Differences in host glycan fucosylation result in distinct microbial ecosystems at the mucosal interface, the compositions and metabolic activities of which are likely precursors and predictors of ensuing disease phenotypes. This concept has been validated by the upstream role of microbial fucose processing for the expansion of the enteric pathogen S. typhimurium (Ng et al., 2013).
Among the phylotypes that are depleted in Sese and sese compared with SeSe individuals (Supplementary Data set S1), Roseburia and Faecalibacterium are both short-chain fatty acid-producing bacteria (Scheppach, 1994; Wong et al., 2006) and reported to be anti-inflammatory (Sokol et al., 2008; Atarashi et al., 2011). Moreover, depletion of Firmicutes and expansion of Proteobacteria members are also characteristic of changes associated with the IBD microbiome (Frank et al., 2007; Sokol and Seksik, 2010). Both Sese and sese individuals harbored 15% fewer microbial genes on average than SeSe individuals (Figure 1a). The lower metagenome diversity is an unfavorable feature for the host, which has also been observed in IBD (Qin et al., 2010) and obesity (Le Chatelier et al., 2013). Among the pathways that were consistently depleted or enriched in Sese and sese individuals, the increase in glutathione metabolism and decrease in amino-acid biosynthesis (particularly lysine) pathways have been reported as a feature of the metagenome in IBD patients (Morgan et al., 2012). These data indicated that the non-secretor-associated changes of microbial composition and imputed metagenomic functions are also characteristics of other chronic inflammatory conditions and therefore are unfavorable for the host.
FUT2−/− mice have a marked alteration in gastric mucosa glycosylation, characterized by diminished expression of alpha(1,2)fucosylated structures (Magalhaes et al., 2009). As gut microbes have developed the ability to degrade host-derived glycans (Katayama et al., 2004; Sonnenburg et al., 2005), the deprivation of terminal fucosylation may affect the metabolic activity of the gut microbiota and thus its fermentation products potentially available to the host. Recent work reported that enterohemorrhagic Escherichia coli encodes a two-component system, termed FusKR, which responds to fucose and controls metabolic gene expression (Pacheco et al., 2012). The imputed metagenomic changes in non-secretors highlighted the depletion of indispensable amino-acid biosynthesis. This group of metagenomic functions complements that encoded by the host genome (Metges, 2000; Gill et al., 2006; Qin et al., 2010). In the IBD metagenome, amino-acid biosynthesis and carbohydrate metabolism are reduced in favor of nutrient uptake (Morgan et al., 2012). Such changes might reflect compensation by the microbiota for the lower availability of carbon sources. Amino-acid starvation can lead to host stress response and the induction of autophagy of intestinal epithelial cells (Tattoli et al., 2012), which may increase the risk of IBD. Although the current imputed metagenomic analysis is limited to the KEGG pathway level, further insights could be gained by extending the analysis to individual KEGG module or enzyme. It would be helpful to further identify the individual genes or reactions that are differentially abundant in these pathways, which would serve as potential candidates for therapeutic manipulation.
Low richness of gut microbiota is a well-known feature of patients with IBD (Manichanh et al., 2006; Lepage et al., 2011), and other chronic conditions such as obesity (Turnbaugh et al., 2009a), and of elderly patients with inflammation (Claesson et al., 2012). A recent study defined two groups of individuals who differed by the number of gut microbial gene as low gene count (LGC) and high gene count (HGC) (Le Chatelier et al., 2013). LGC individuals exhibited an imbalance of pro- and anti-inflammatory bacterial species and evidence of low-grade inflammation. We have shown that both Sese and sese individuals harbored 15% fewer microbial genes on average than SeSe individuals and therefore exhibited low metagenomic richness. Similarly, in Sese and sese individuals, genera associated with LGC individuals, including Faecalibacterium and Coprococcus, were also more dominant. Moreover, the sub-clinical inflammatory state at the mucosal surface was reflected by the higher expression levels of HNP-1 and -2 in non-secretors.
The data presented here supports the hypothesis that the FUT2 loss-of-function allele increased the risk of CD by shaping the functional states of mucosal microbiota. Meta-analysis of genome-wide association studies has increased the number of confirmed IBD (both CD and ulcerative colitis) susceptibility loci to 167 (Jostins et al., 2012), indicating that IBD is biologically heterogeneous. The analysis presented in this study focused on individuals without clinical symptoms. It will be important to extend the same analysis to patients with CD to determine to what extent the changes we present are recapitulated in the disease setting. In addition to FUT2, other risk genes have also been shown to affect the gut microbial composition, such as NOD2 (Petnicki-Ocwieja et al., 2009; Frank et al., 2011) and several defensin genes (Ivanov et al., 2008; Salzman et al., 2010). It is currently unclear whether the microbiota associated with genes of similar functions has the same compositional and functional signatures. The stratification of gut microbiome by host genetics is a crucial step for elucidating the pathogenic mechanism of IBD as well as the design of personalized therapeutic interventions.
To achieve unprecedented understanding of the ecological structures and biomolecular activities of the gut microbiome, it is necessary to extend the analysis to multiple levels of biological organization—genome content, gene expression, protein expression and metabolism (Raes and Bork, 2008; Nicholson et al., 2012). In this study, we used multiple 'omic approaches to disentangle the complex host–microbial metabolic interplay. Meta-proteomic analysis in this case focused on the host side, but metabolomics could be extended to incorporate richer microbial analysis in the future. Although only a limited number of integrative ‘omics profiles of the gut microbiota currently exist (Erickson et al., 2012; Perez-Cobas et al., 2012), multi'omic studies have shown great potential in providing a holistic picture of the metabolic status of the gut microbiota and the host response to functional changes.
Acknowledgments
This work was supported by NIH DK046763 (to JB, DPBM), DK08569 (to JB), AI078885 (to JB, DK062413 (to DPBM), NCRR and NCATS UL1TR000124 (to JB), the Crohn's and Colitis Foundation of America 3153 (to JB), the Burroughs Wellcome Fund Inter-school Program in Metabolic Diseases Fellowship (to MT), Cedars-Sinai F. Widjaja Family Foundation Inflammatory Bowel and Immunobiology Research Institute Research Funds (to DPBM) and the Helmsley Foundation (DPBM). We thank Georgetown University's Proteomics and Metabolomics shared resource partially supported by the NIH/NCI Grant P30-CA051008.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL et al. (2012). Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol 8: e1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y et al. (2011). Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331: 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995). Controlling the false discovery rate—a practical and powerful approach to multiple testing. J Roy Stat Soc Ser B Meth 57: 289–300. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S et al. (2012). Gut microbiota composition correlates with diet and health in the elderly. Nature 488: 178–184. [DOI] [PubMed] [Google Scholar]
- Claesson MJ, Wang Q, O'Sullivan O, Greene-Diniz R, Cole JR, Ross RP et al. (2010). Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res 38: e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M et al. (2005). Diversity of the human intestinal microbial flora. Science 308: 1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson AR, Cantarel BL, Lamendella R, Darzi Y, Mongodin EF, Pan C et al. (2012). Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn's disease. PLoS One 7: e49138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J et al. (2012). Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol 8: e1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Admetlla A, Sikora M, Laayouni H, Esteve A, Roubinet F, Blancher A et al. (2009). A natural history of FUT2 polymorphism in humans. Mol Biol Evol 26: 1993–2003. [DOI] [PubMed] [Google Scholar]
- Frank DN St, Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 104: 13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, Robertson CE, Hamm CM, Kpadeh Z, Zhang T, Chen H et al. (2011). Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis 17: 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Alm EJ. (2012). Inferring correlation networks from genomic survey data. PLoS Comput Biol 8: e1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Wei B, Wen T, Johansson ME, Liu X, Bradford E et al. (2011). Loss of intestinal core 1-derived O-glycans causes spontaneous colitis in mice. J Clin Invest 121: 1657–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS et al. (2006). Metagenomic analysis of the human distal gut microbiome. Science 312: 1355–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project C (2012). Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd EA, Holmen JM, Hansson GC, Domino SE. (2005). Gastrointestinal mucins of Fut2-null mice lack terminal fucosylation without affecting colonization by Candida albicans. Glycobiology 15: 1002–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB et al. (2008). Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4: 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. (2008). The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA 105: 15064–15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY et al. (2012). Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491: 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap PC, Marcobal A, Ursell LK, Smits SA, Sonnenburg ED, Costello EK et al. (2013). Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci USA 110: 17059–17064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T, Sakuma A, Kimura T, Makimura Y, Hiratake J, Sakata K et al. (2004). Molecular cloning and characterization of Bifidobacterium bifidum 1,2-alpha-L-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95). J Bacteriol 186: 4885–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R et al. (2013). A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput Biol 9: e1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Cameron EA, Martens EC. (2012). How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 10: 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Zhang B, Horvath S. (2008). Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 24: 719–720. [DOI] [PubMed] [Google Scholar]
- Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31: 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500: 541–546. [DOI] [PubMed] [Google Scholar]
- Lee YK, Mazmanian SK. (2010). Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 330: 1768–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage P, Hasler R, Spehlmann ME, Rehman A, Zvirbliene A, Begun A et al. (2011). Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 141: 227–236. [DOI] [PubMed] [Google Scholar]
- Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H et al. (2008). Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci USA 105: 2117–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, LeBlanc J, Truong A, Vuthoori R, Chen SS, Lustgarten JL et al. (2011). A metaproteomic approach to study human-microbial ecosystems at the mucosal luminal interface. PLoS One 6: e26542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. (2008). Mucins in the mucosal barrier to infection. Mucosal Immunol 1: 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes A, Gomes J, Ismail MN, Haslam SM, Mendes N, Osorio H et al. (2009). Fut2-null mice display an altered glycosylation profile and impaired BabA-mediated Helicobacter pylori adhesion to gastric mucosa. Glycobiology 19: 1525–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L et al. (2006). Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 55: 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U et al. (2013). Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339: 1084–1088. [DOI] [PubMed] [Google Scholar]
- Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP et al. (2011). Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol 9: e1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern DP, Jones MR, Taylor KD, Marciante K, Yan X, Dubinsky M et al. (2010). Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn's disease. Hum Mol Genet 19: 3468–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuckin MA, Linden SK, Sutton P, Florin TH. (2011). Mucin dynamics and enteric pathogens. Nat Rev Microbiol 9: 265–278. [DOI] [PubMed] [Google Scholar]
- McHardy I, Goudarzi M, Tong M, Ruegger P, Schwager E, Weger J et al. (2013). Integrative analysis of the microbiome and metabolome of the human intestinal mucosal surface reveals exquisite inter-relationships. Microbiome 1: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metges CC. (2000). Contribution of microbial amino acids to amino acid homeostasis of the host. J Nutr 130: 1857S–1864S. [DOI] [PubMed] [Google Scholar]
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV et al. (2012). Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 13: R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Saunier K, Hanisch C, Norin E, Alm L, Midtvedt T et al. (2006). Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol 72: 1027–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S et al. (2013). Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502: 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W et al. (2012). Host-gut microbiota metabolic interactions. Science 336: 1262–1267. [DOI] [PubMed] [Google Scholar]
- Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG et al. (2012). Fucose sensing regulates bacterial intestinal colonization. Nature 492: 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Cobas AE, Gosalbes MJ, Friedrichs A, Knecht H, Artacho A, Eismann K et al. (2012). Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut 62: 1591–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, Tlaskalova-Hogenova H et al. (2009). Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci USA 106: 15813–15818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490: 55–60. [DOI] [PubMed] [Google Scholar]
- Raes J, Bork P. (2008). Molecular eco-systems biology: towards an understanding of community function. Nat Rev Microbiol 6: 693–699. [DOI] [PubMed] [Google Scholar]
- Rausch P, Rehman A, Kunzel S, Hasler R, Ott SJ, Schreiber S et al. (2011). Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci USA 108: 19030–19035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E et al. (2010). Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol 11: 76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheppach W. (1994). Effects of short chain fatty acids on gut morphology and function. Gut 35: S35–S38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J et al. (2013). Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339: 548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ et al. (2008). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA 105: 16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Seksik P. (2010). The intestinal microbiota in inflammatory bowel diseases: time to connect with the host. Curr Opin Gastroenterol 26: 327–331. [DOI] [PubMed] [Google Scholar]
- Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN et al. (2010). Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141: 1241–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J et al. (2005). Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307: 1955–1959. [DOI] [PubMed] [Google Scholar]
- Spor A, Koren O, Ley R. (2011). Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 9: 279–290. [DOI] [PubMed] [Google Scholar]
- Stewart JA, Chadwick VS, Murray A. (2005). Investigations into the influence of host genetics on the predominant eubacteria in the faecal microflora of children. J Med Microbiol 54: 1239–1242. [DOI] [PubMed] [Google Scholar]
- Tattoli I, Sorbara MT, Vuckovic D, Ling A, Soares F, Carneiro LA et al. (2012). Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe 11: 563–575. [DOI] [PubMed] [Google Scholar]
- Tong M, Li X, Wegener Parfrey L, Roth B, Ippoliti A, Wei B et al. (2013). A modular organization of the human intestinal mucosal microbiota and its association with inflammatory bowel disease. PLoS One 8: e80702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE et al. (2009. a). A core gut microbiome in obese and lean twins. Nature 457: 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. (2009. b). The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1: 6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Sluis M, De Koning BA, De Bruijn AC, Velcich A, Meijerink JP, Van Goudoever JB et al. (2006). Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 131: 117–129. [DOI] [PubMed] [Google Scholar]
- Wacklin P, Makivuokko H, Alakulppi N, Nikkila J, Tenkanen H, Rabina J et al. (2011). Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS One 6: e20113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing BP, Gill N, Finlay BB. (2010). The role of the immune system in regulating the microbiota. Gut Microbes 1: 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. (2006). Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol 40: 235–243. [DOI] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334: 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M et al. (2012). Human gut microbiome viewed across age and geography. Nature 486: 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. (2005). A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4: Article17. [DOI] [PubMed] [Google Scholar]
- Zoetendal EG, Akkermans ADL, Akkermans-van Vliet WM, de Visser AGM, de Vos WM. (2001). The host genotype affects the bacterial community in the human gastronintestinal tract. Microb Ecol Health Dis 13: 129–134. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.