Abstract
Nitrogen fixation, the biological reduction of dinitrogen gas (N2) to ammonium (NH4+), is quantitatively the most important external source of new nitrogen (N) to the open ocean. Classically, the ecological niche of oceanic N2 fixers (diazotrophs) is ascribed to tropical oligotrophic surface waters, often depleted in fixed N, with a diazotrophic community dominated by cyanobacteria. Although this applies for large areas of the ocean, biogeochemical models and phylogenetic studies suggest that the oceanic diazotrophic niche may be much broader than previously considered, resulting in major implications for the global N-budget. Here, we report on the composition, distribution and abundance of nifH, the functional gene marker for N2 fixation. Our results show the presence of eight clades of diazotrophs in the oxygen minimum zone (OMZ) off Peru. Although proteobacterial clades dominated overall, two clusters affiliated to spirochaeta and archaea were identified. N2 fixation was detected within OMZ waters and was stimulated by the addition of organic carbon sources supporting the view that non-phototrophic diazotrophs were actively fixing dinitrogen. The observed co-occurrence of key functional genes for N2 fixation, nitrification, anammox and denitrification suggests that a close spatial coupling of N-input and N-loss processes exists in the OMZ off Peru. The wide distribution of diazotrophs throughout the water column adds to the emerging view that the habitat of marine diazotrophs can be extended to low oxygen/high nitrate areas. Furthermore, our statistical analysis suggests that NO2− and PO43− are the major factors affecting diazotrophic distribution throughout the OMZ. In view of the predicted increase in ocean deoxygenation resulting from global warming, our findings indicate that the importance of OMZs as niches for N2 fixation may increase in the future.
Keywords: heterotrophic N2 fixation, nifH diversity, oxygen minimum zone off Peru
Introduction
High primary productivity in upwelling systems associated with eastern boundary currents results in enhanced oxygen (O2) consumption at depth related to the degradation of sinking organic matter (Duce et al., 2008).
The reduced O2 concentrations in subsurface waters favours N-loss processes, P release (Ingall and Jahnke, 1994; Deutsch et al., 2007) and may increase iron bioavailability, conditions that overall are favourable for N2 fixation (Deutsch et al., 2001; Deutsch et al., 2007). Similar to N2 fixation, other N-cycle processes such as anaerobic ammonium oxidation (anammox), denitrification and nitrification are sensitive to O2, thus low O2 concentrations make oxygen minimum zones (OMZs) hotspots for N-turnover processes (Capone, 2008; Codispoti, 2010; Kalvelage et al., 2013). Further, field observations have demonstrated that N2 fixation occurs concurrently with fixed N-loss processes (Halm et al., 2009; Fernandez et al., 2011; Gandhi et al., 2011), suggesting that OMZs, such as the large and persistent OMZ present in the eastern tropical Pacific Ocean off Peru, are an ideal environment for marine diazotrophy (Bopp et al., 2002).
N2 fixation rates in the ocean have largely been underestimated owing to a methodological bias (Grosskopf et al., 2012). Particularly, the contribution to fixed N budget from diazotrophs other than the classically known cyanobacteria, for example, the filamentous diazotroph Trichodesmium, may have been disproportionally underestimated owing to an uneven distribution of label in the classical 15N2 incubation experiments, which results in lower labeling efficiency of non-buoyant organisms (Grosskopf et al., 2012). This realization, together with the growing number of studies on the phylogenetic diversity of diazotrophs (Farnelid et al., 2011; Fernandez et al., 2011; Turk et al., 2011), demonstrates the need to investigate the diazotrophic community in environments that were previously not considered important for N2 fixation. Reports on the diversity of the nifH gene in OMZ waters suggested that the detected diazotrophic communities differ strongly from those present in oligotrophic oceanic waters (Zehr and Turner, 2001; Falcon et al., 2002; Langlois et al., 2005; Fernandez et al., 2011; Hamersley et al., 2011; Jayakumar et al., 2012; Farnelid et al., 2013). Along the Californian coast, the OMZ off Peru and Chile, in the Arabian Sea and in two basins of the Baltic Sea, diverse nifH gene sequences mostly related to heterotrophic proteobacteria have previously been identified (Fernandez et al., 2011; Hamersley et al., 2011; Jayakumar et al., 2012; Farnelid et al., 2013). The role and importance of those non-cyanobacterial diazotrophs for the oceanic fixed N budget is currently unclear due to the limited information on their biogeographical distribution, abundance and activity. To shed light on diazotrophy in OMZs and the potential role of non-phototrophic N2 fixers, we conducted a large-scale survey of functional gene abundances, O2 and nutrients, such as nitrite (NO2−), nitrate (NO3−), ammonia (NH4+), phosphate (PO43−), iron (Fe) concentrations and N2 fixation rates complemented with glucose enrichment and oxygen manipulation experiments throughout the OMZ off Peru in the eastern tropical South Pacific (ETSP).
Materials and methods
Hydrographic parameters and nutrients
Samples for salinity, O2 and macro-nutrient analysis (NO3−, NO2−, NH4+ and PO43−) were collected from a 24 Niskin bottle rosette equipped with a CTD (Conductivity, Temperature, Depth) sensor or from a pump-CTD. The pump-CTD used during M77/3 is a CTD, equipped with a pump that transfers water from depth directly up into the lab. CTD, O2, fluorescence, turbidity, and acoustic doppler counter profiler measurements can be combined with continuous water sampling over a water column of 350 m depth in high resolution (1 m maximum) along vertical profiles.
O2 concentrations were determined according to the Winkler method; salinity and nutrient concentrations were determined as previously described (Grasshoff et al., 1999). P* was calculated from phosphate and nitrate measurements according to Deutsch et al. (2007): P*=PO43−−NO3−/r16:1, where r16:1 is the ratio of nitrate to phosphate at Redfield conditions (Deutsch et al., 2007).
Trace metal sampling and analysis
Seawater samples were obtained using the modified Teflon-coated PVC General Oceanics (Miami, FL, USA) GoFlo (8 l) bottles on a trace metal clean hydrowire using established protocols (Bruland et al., 1979). After recovery, the bottles were transferred into a class 100 clean lab container and filtered immediately through a 0.2-μm membrane filter (Sartorius, Göttingen, Germany) under slight nitrogen overpressure (0.2–0.3 bar) into 1 liter acid-washed low-density polyethylene bottles. The seawater samples for trace metal analysis were acidified in a laminar flow hood with quartz-distilled concentrated hydrochloric acid (6 M QD-HCl) to pH 1.8 (17.8 μmol H+ l−1) and stored in the dark until analysis half a year later.
Dissolved Fe concentrations were determined via graphite furnace–atom absorption spectrometry (GF-AAS) (Zeemann Atomic Absorption Spectrometer 4100ZL, Perkin Elmer, Waltham, MA, USA). The samples were analysed by a method previously outlined by Danielson et al. (1978) and Bruland et al. (1979) and described in detail by Grasshoff et al. (1999). The accuracy of the analytical procedure was evaluated by measurement of the certified seawater standard NASS-5 (National Research Council of Canada) and the SAFe intercomparison standard. Our values for NASS 5 agreed within the stated values for NASS 5 and our SAFe data (SAFe S: 0.109±0.013 nmol kg−1 Fe; SAFe D2: 0.81±0.13 nmol kg−1 Fe) were close to the average consensus values for Fe (NASS 5: 3.70±0.63 nmol kg−1; SAFe S: 0.093±0.008 nmol kg−1; SAFe D2: 0.933±0.023 nmol kg−1). The precision for replicate analyses was between 3% at the concentrations found in this study. The analytical Fe blank was determined by re-extraction of samples and was found to be 0.041±0.024 nmol kg−1 (3σ detection limit=0.079 nmol kg−1).
Molecular genetic methods
Samples for the extraction of DNA/RNA were taken by filtering a volume of about 2 l (exact volumes and filtration times were determined and recorded for each sample) of seawater through 0.2-μm polyethersulfone membrane filters (Millipore, Billerica, MA, USA). The filters were immediately frozen and stored at −80 °C.
DNA and RNA were extracted using the Qiagen DNA/RNA All prep Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. DNA concentrations were determined fluorometrically using the PicoGreen (Invitrogen, Carlsbad, CA, USA) as described in the manufacturer's protocol using a Fluorometer (Fluoroscan Ascent Labsystems, Helsinki, Finnland). A standard dilution series (2.5–100 pg μl−1) was used for absolute quantification. Residual DNA was removed from the purified RNA by a DNase I treatment (Invitrogen). The RNA was checked for potential DNA contamination by 16S rDNA PCR amplification using the universal primer set 27F and 1492R (Lane et al., 1991) before reverse transcription and by a nested nifH PCR as described above. RNA concentrations were determined similar to DNA concentrations using the fluorescent dye RiboGreen (Invitrogen) and a RNA standard dilution series (2.5–80 pg μl−1).
The extracted RNA was reverse transcribed to cDNA using the Superscript III First Strand synthesis Kit (Invitrogen) following the manufacturer's protocol with primers nifH2 and nifH3 (Zani et al., 2000; Langlois et al., 2005).
NifH was amplified by a nested PCR with primers according to Zani et al. (2000) and Langlois et al. (2005), using 0.1 μl GoTaq polymerase (Promega, Madison, WI, USA) in a final volume of 25 μl. Based on nifH sequence information of clusters P1–P8, quantitative PCR (qPCR) primers and TaqMan MGB probes (6-carboxyfluorescein reporter) were designed with the Primer Express software package (Life Technologies, Darmstadt, Germany); oligonucleotide sequences and qPCR conditions are given in Table 1.
Table 1. Oligonucleotide sequences and quantitative PCR conditions.
| Cluster | Reference sequence | Forward primer 5′–3′ | Reverse primer 5′–3′ | Probe | Annealing temperature (°C) |
|---|---|---|---|---|---|
| P1 | M773_3_70_4 | GGACTACATTCGGACTAG | GTCGTAACCACGATCTAG | TCTTCAAAATCCCGCGTCCCG (antisense) | 60 |
| P2 | M773_3_150_7 | GGTGTTCTATGTGTTGAA | GTAGGAGTTACGAATTGG | TCGCCTAGCACATCATAGATCAC (antisense) | 50 |
| P3 | M773_56_100_1 | CACAGTTAGAGAGGTAGG | CAAGGTCGTCAGTAAAAG | AGCTCGACAAGGTAATGTTCACA (sense) | 54 |
| P4 | M773_22_30_1 | CTCGCACAGAAATCAGTG | GCATGTTAATGGAAGTGATG | ACGTCGAACTCGAAGACATCCG (sense) | 60 |
| P5 | M773_56_100_11 | GGAAGTCTTACTTGAAGG | CACCATTTTCCTCAAGAA | ATTGCTGTAATAACGCCTCTGC (antisense) | 60 |
| P6 | M773_56_85_3 | GCTCAATCTACAATTATGC | GCTGTAATAACTCCTCTAC | ACCACCTGACTCAGTACAATTAATGT (antisense) | 50 |
| P7 | M773_28_115_1 | GGTTCTGTTGAAGACATC | CGAAGTCTAAGTCTTCTTC | ATCGCTGTGATTACACCACGAC (antisense) | 52 |
| P8 | M774_800 m_11B14a | ACTCGTCTGATTCTTCAC | TTAATACATCTTCCAGTTCCA | AAAGCACAGAACACCATCATG (antisense) | 52 |
Primers and probes were checked against the NCBI database and an ARB nifH database using BLAST search to ensure that they were specific for the selected nifH cluster. Standards for the different nifH clusters were obtained by cloning the nifH amplicons using the Topo TA Cloning Kit (Invitrogen). Plasmid extraction and purification was performed using the Qiagen plasmid purification kit according to the manufacturer's protocol; plasmid concentrations were determined using a NanoDrop ND-1000 spectrophotometer (PeqLab, Erlangen, Germany) and diluted to 4 pg μl−1, which corresponds to 107 target sequence copies in a 5-μl volume. Serially diluted plasmid standards were used to calculate copy numbers in the qPCR assays. The primer and probe set we used to quantify UCYN-B in this study has been previously described (Langlois et al., 2008).
The specificity of the qPCR primer and probe sets was confirmed by testing for cross-reactivity and sensitivity against each available standard in our lab, diluted to 104 and 105 nifH copies. DNA amplification signals were detected only for the homologous phylotype. Additionally, the sensitivity of the primers and probes in a mixed DNA sample was tested in combinations of serially diluted standards mixed in ratios ranging from 107:101 to 101:107 copies of the specific and unspecific standard, which did not result in significant changes in the linear regressions, indicating that the presence of various amounts of closely related DNA sequences does not affect the quantification of a specific phylotype. qPCR mixtures contained 1 × TaqMan PCR buffer (Life Technologies), 100 nM TaqMan probe, 5 pmol μl−1 each of the forward and reverse primers, 400 ng μl−1 bovine serum albumin, 3 μl PCR water, and 5 μl of either standard or environmental sample. Addition of bovine serum albumin to the reactions was performed to avoid PCR inhibition without affecting standard curves or detection limits as previously described in Langlois et al. (2008). Environmental DNA samples and standards were run in duplicates. To avoid degradation, extracted DNA was frozen in aliquots and thawed only once for qPCR determination. Non-template controls were run in duplicate for each primer and probe set and were undetectable after 45 cycles, which translates into a theoretical detection limit of the qPCR assays of one nifH gene copy. The analytical detection limit depends on several other factors, for example, the amount of seawater filtered, the elution volume after extraction and the amount of sample loaded setting the detection limit for our samples to 60 copies l−1. Accordingly, we considered samples with CT values <36 (<102 copies l−1) detectable but not quantifiable, similar to previous studies (Langlois et al., 2008).
AmoA PCRs and qPCRs were performed as described in Loescher et al. (2012); nirS and hzo were amplified according to Lam et al. (2007) and Schmid et al. (2008), respectively.
Cloning of PCR amplicons was performed using the Topo TA Cloning Kit (Invitrogen) according to the manufacturer's instructions. Sanger sequencing was carried out by the Institute of Clinical Molecular Biology, Kiel, Germany. Nucleotide sequences were analysed using the ClustalW multiple alignment tool on a 321-bp fragment for nifH. Sequences were submitted to Genbank and were analysed using the ClustalW multiple alignment tool on a 321-bp fragment for nifH, sequence differences were set on a minimum of 5% and phylogenetic trees were made using the distance-based neighbor-joining analysis.
Statistical analysis
The impact of environmental parameters on the abundance and distribution of nifH clusters (copies l−1) as determined by qPCR was explored by redundancy analysis (RDA) based on 316 cases. Computations were performed in R v3.0.2 (R_Core_Team, 2013) by using the R package vegan (Oksanen et al., 2013). nifH cluster counts were subjected to Hellinger transformation in order to make them compatible with RDA (Legendre and Gallagher, 2001; Stratil et al., 2013).
The most parsimonious RDA model was determined via stepwise selection with vegan's function ordistep considering depth as well as concentrations of O2, NO2−, NO3− and PO43− as explanatory variables. Variable depth substitutes temperature and salinity to avoid collinearity and was used as a surrogate for variables changing with depth (for example, trace metals or viral infection pressure). Wherever possible, O2 concentrations determined according to the Winkler method were used. A correlation test between Winkler and CTD oxygen data showed that the two variables were strongly correlated (R(166)=0.98, R2=0.96, P<0.001). Correspondingly, a Wilcoxon signed rank test on existing data pairs did not indicate any systematic differences between Winkler and CTD oxygen data (two-sided test, n=168, P=0.41). Missing Winkler measurements were thus substituted by data from the CTD oxygen sensor. Significance of the selected model and of marginal effects (that is, of each parameter in the presence of all other parameters) was determined in permutation tests with 1000 random permutations.
15N2 seawater incubations
Seawater incubations were performed in triplicate at six stations in the OMZ off Peru (M77/3) according to an existing protocol (Montoya et al., 1996). In detail, seawater from the OMZ was sampled from a pump-CTD system and directly filled into 2 l glass bottles (Schott–Duran, Wertheim, Germany) from the bottom, allowing the displacement of about one bottle volume through the opening before closing the bottle bubble free with a Teflon covered butyl septum cap. Samples from the surface were taken by either the pump-CTD system or the conventional CTD and filled in 4.5 l polycarbonate bottles (Nalgene, Rochester, NY, USA) with Teflon-coated butyl rubber septum caps. Bottles were spiked with 2 ml (2-l flasks) or 4.5 ml (4.5-l flasks) of 98% 15N2 gas (Sigma-Aldrich, St Louis, MO, USA). This method has recently been found to underestimate the true N2 fixation rates (Mohr et al., 2010), therefore the rates measured in this study can be taken as minimum values.
For carbon fixation measurements, NaH13CO3 (Sigma-Aldrich) was dissolved in sterile deionized water (>18.2 MOhm cm−1, MilliQ, Millipore) (1 g/50 ml) and 0.5 ml (2-l bottles) or 1 ml (4.5-ml bottles) were added to the incubations with a syringe (∼3.5 atom % final). For glucose and oxygen addition experiments, 500 ml of seawater from the same location was aerated by vigorously bubbling with sterile filtered air from a compressor for 15 min. The amount of seawater leading to a final concentration of 10 μmol l−1 dissolved oxygen (depending on temperature conditions, for example, 39 ml l−1 at 12 °C) was added with a syringe to the bottom of the incubation bottles, while water from the top was replaced. Glucose (Sigma-Aldrich) was dissolved in MilliQ water (1.44 g l−1), and the concentrated solution was added through the septum with a syringe to yield a final concentration of 2 μM glucose. After amendments, bottles were inverted 100 times. Bottles from surface water were stored on deck in a seawater-cooled Plexiglas incubator covered with blue light foil (blue-lagoon, Lee filters, Andover, UK) that mimics the light environment of a 10-m depth. Samples from the OMZ were stored at 12 °C in the dark. After 24 h of incubation, 0.7–2.5 l of seawater were filtered onto precombusted (450 °C, 5 h) 47-mm diameter GF/F filters (Whatman, Maidstone, UK) under gentle vacuum (−200 mbar). The filtrations were stopped after 1 h, as high particle load of surface water often led to a clogging of the filters. Filters were oven dried (50 °C) for 24 h and stored over desiccant until analysis. Environmental samples of 2 l untreated seawater were filtered and prepared in the same way to serve as blank values. For isotope analysis, GF/F filters were acidified over fuming HCl overnight in a desiccator. Filters were then oven-dried for 2 h at 50 °C and pelletized in tin cups. Samples were analysed for particulate organic carbon and particulate organic nitrogen (PON) and isotopic composition using a CHN analyzer (Carlo Erba EA 1108, Milano, Italy) coupled to an isotope ratio monitoring mass spectrometer (Thermo Finnigan Delta S, Waltham, MA, USA). The content of PON of most samples exceeded 10 μg l−1. Lowest PON values were measured on the deep water offshore stations (∼3 μg l−1). Details of the statistical evaluation of the glucose and oxygen addition experiments are given in the Supplementary Information.
Results and discussion
Diversity and distribution of atypical nifH clusters in the OMZ off Peru
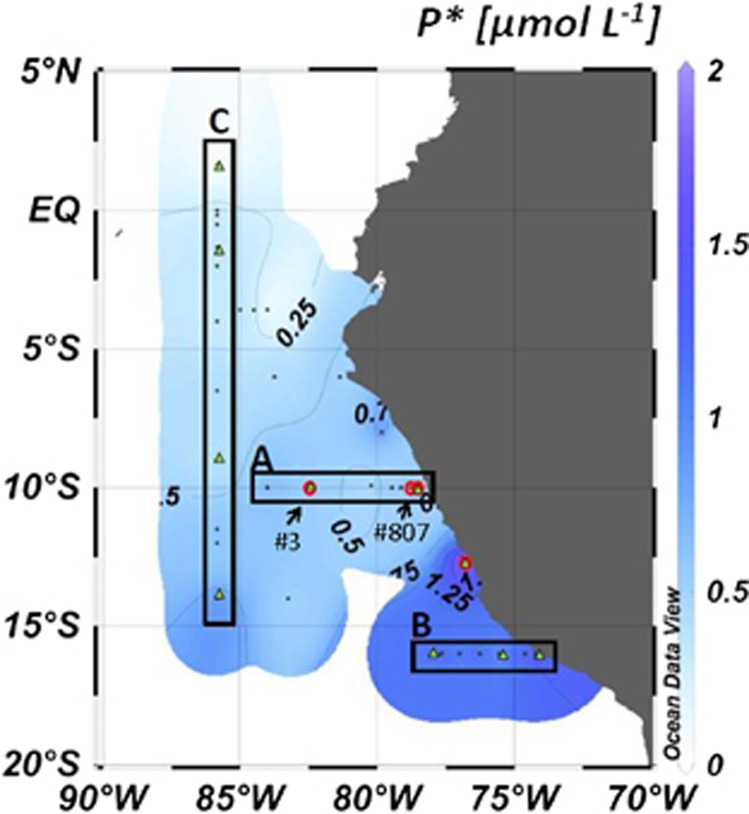
We conducted two consecutive expeditions with R/V Meteor in January (M77/3) and February 2009 (M77/4) to the OMZ off Peru. Samples from vertical depth profiles were collected along two transects from the shelf to the open ocean throughout the OMZ (Figure 1, transects A and B, M77/3) and a north–south transect in offshore waters (Figure 1, transect C, M77/4). On the shelf, the water column was anoxic between 20 m and 80 m (Supplementary Figure S1), while the oxycline deepened offshore reaching anoxic conditions at ∼100 m. Excess phosphorous (P* representing the excess of P relative to the standard N quota) previously implicated in the control of N2 fixation (Deutsch et al., 2007) was pervasive in the OMZ as well as in the surface layer (Figure 1, Supplementary Figure S1), with highest excess concentrations (up to 2 μM) on the shelf.
Figure 1.
Surface distribution of P* (μmol l−1) in the Peruvian OMZ, three transects (A, B and C) are marked with black boxes; red circles indicate the stations of N2 fixation experiments, green triangles indicate the stations, which were selected for clone library construction.
A total of∼600 nifH sequences were obtained from clone libraries (384 from DNA and 249 from cDNA libraries) constructed from 29 samples collected at 12 stations along transects A, B and C. Samples were chosen according to the detected O2 saturation to cover a spectrum reaching from full O2 saturation in surface waters, hypoxic saturations (15–75 μmol l−1) to the OMZ core ([O2]<15 μmol l−1) (Supplementary Figure S1).
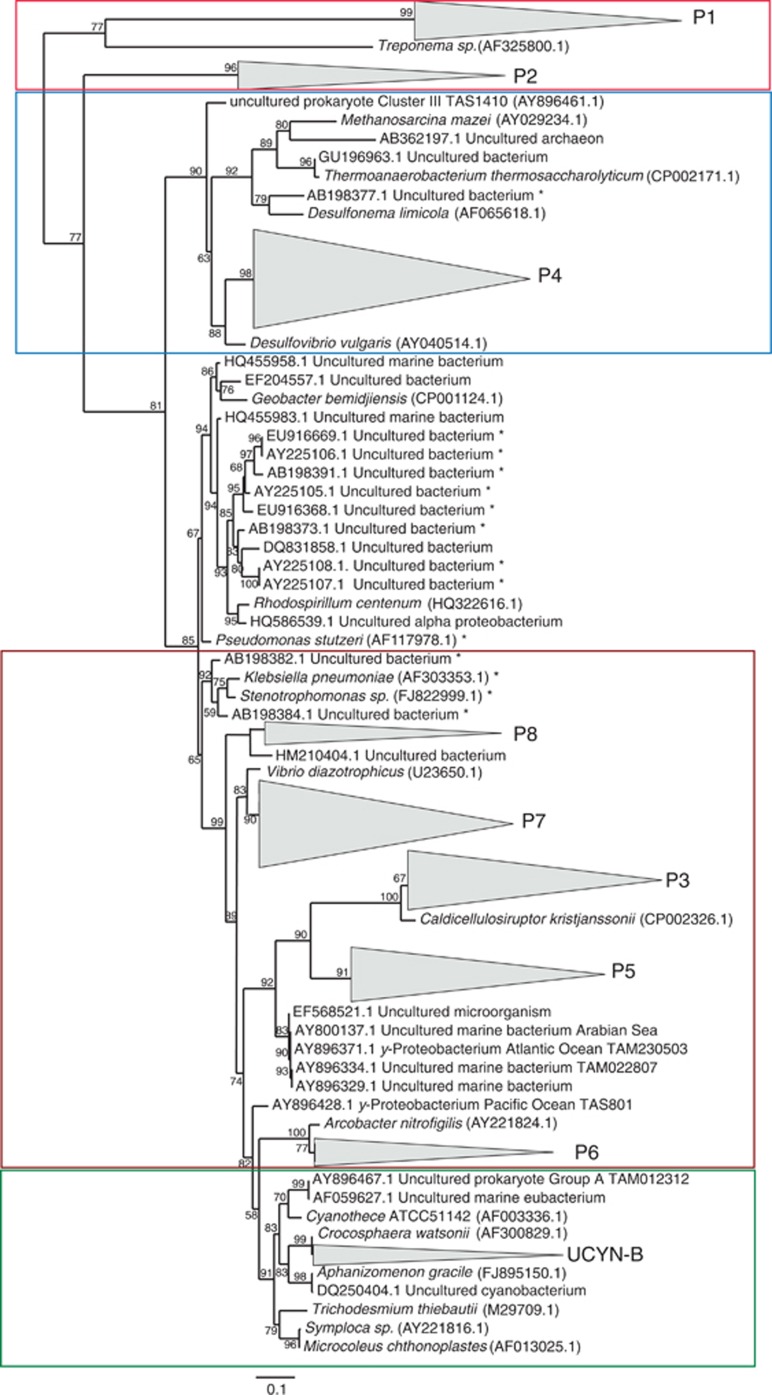
The nifH sequences grouped into nine clusters (further referred to as P1–P8 and UCYN-B; Figure 2). Those clusters were in large part (73%) recovered from OMZ waters ([O2]⩽15 μmol l−1 at ∼10-m depths onshore and at ∼50–100-m depths offshore (Kalvelage et al., 2013), see Supplementary Figure S1) in samples collected below the euphotic zone (Chl a was below the detection limit at ∼50-m depth onshore at 10°S and at ∼120-m depth offshore (Franz et al., 2012)). This indicates that the diazotroph community was most likely non-photoautotrophic. Combined with the phylogenetic analysis of nifH genes, this adds to the growing awareness of the importance of non-cyanobacterial diazotrophs for N2 fixation in the ocean (Fernandez et al., 2011; Hamersley et al., 2011; Jayakumar et al., 2012; Farnelid et al., 2013). The presence of those clusters points towards a potential to fix N2 in the OMZ off Peru.
Figure 2.
Phylogenetic tree based on the analysis of ∼600 nifH gene and transcript nucleotide sequences retrieved in this study. The detected clusters are indicated with grey triangles. Cyanobacterial sequences are highlighted in green, proteobacterial sequences are highlighted in brown, Cluster III sequences as defined by Zehr et al. (2003) are shown in blue and two distant clusters are highlighted red. Bootstrapped values (%)>50, out of 100 are shown on branches. The scale bar represents 10% estimated sequence divergence. Sequences marked with an asterisk indicate likely contaminated PCR products previously reported by Turk et al. (2011), the detected clusters are mostly distant from those sequences.
P1 and P2 nifH clades have never been reported before and were the most distant clades in our study compared with previously detected marine nifH genes. In addition to those two novel deep branching clusters, clusters P3–P7 were phylogenetically related to proteobacterial nifH genes amplified from hypoxic basins in the Californian Bight and the OMZ off Peru and Chile (97–99% similarity; see Supplementary Table S1 for sequence overlaps with previous studies). Sequences related to P4 and P8 clusters were previously detected in the tropical South Pacific Ocean (Fernandez et al., 2011; Halm et al., 2012) (Supplementary Table S1). The P4 sequences were mainly present in clone libraries derived from shelf waters (Figure 1, M77/3 transects A and B) while P8 sequences were recovered from clone libraries prepared from samples collected along a north–south transect at 85.83°W in open ocean waters during M77/4 (Figure 1, transect C from surface waters down to 350 m). Sequences from clusters P3, P7 and P8 have been detected before in the Indian Ocean (Farnelid et al., 2011).
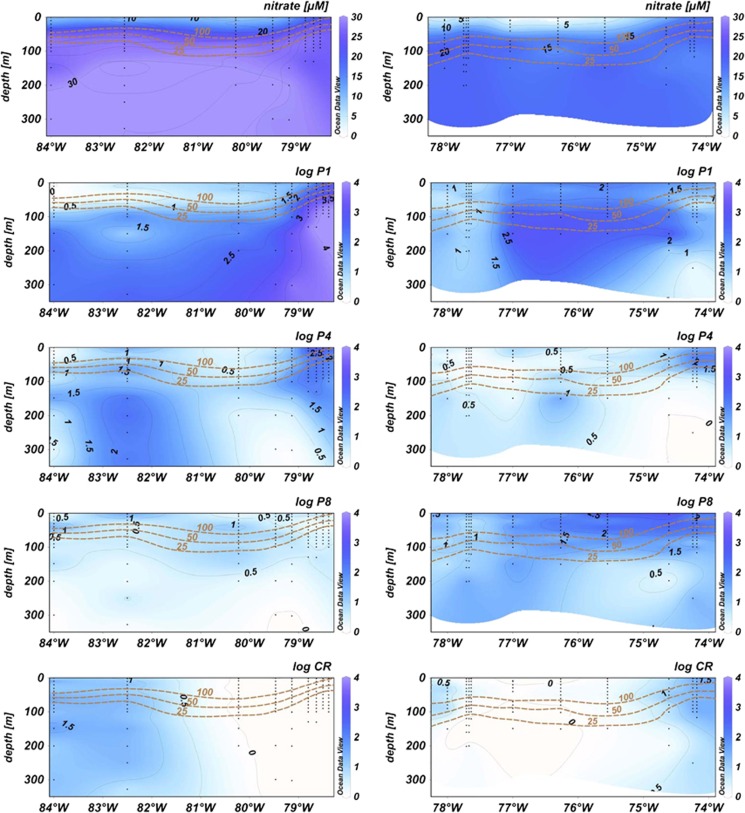
Cluster-specific qPCR-TaqMan assays targeting the nifH genes of the novel diazotrophs revealed that clusters P1 and P4 were dominating the diazotrophic community from the shelf to about 83°W at 10°S and to 77°W at 16°S (<105 nifH copies l−1, Figure 3), accounting for up to 93% of total nifH onshore at 10°S (from surface down to 100 m). The P1 cluster was generally associated with deeper waters (100–300 m) with low or non-detectable O2 concentrations and high PO43− (P1 abundances >103 copies l−1 were present at O2<5 μM and PO43−>2.4 μmol kg−1) concentrations, while other clusters (P2, P3, P4,) were present in surface to subsurface waters (Figure 3 and Supplementary Figure S1). Cluster P8 was present throughout the Peruvian OMZ (Figure 3, Supplementary Figure S2) dominating the nutrient-depleted open ocean region along the north–south offshore transect quantitatively (transect C, Figure 1, Supplementary Figure S2), with abundances reaching up to 106 nifH copies l−1. nifH gene sequences belonging to the filamentous non-heterocystous cyanobacterium Trichodesmium sp., the diatom endosymbiont Richelia sp. or Candidatus Atelocyanobacterium thalassa, which are all considered key diazotrophs in oligotrophic surface waters of the ocean (Church et al., 2005), were not recovered from our clone libraries; however, our sequencing approach allows only the detection of the predominant clusters. Crocosphaera-like nifH (Zehr et al., 1998) was the only cyanobacterial sequence recovered from some of our clone libraries (1 out of ∼384 sequences, 95.7% sequence identity to C. watsonii WH8501, GeneBank no.: AF300829)).
Figure 3.
Distribution of NO3− and the detected nifH clusters: NO3− (μM), nifH clusters P1, P4, P8 and Crocosphaera (log10 copies l−1), the oxycline (μM O2) is indicated with brown contour lines along (left) 10°S and (right) 16°S as marked A and B on the map (Figure 1).
The UCYN-B-like cluster was mainly present in abundances close to the detection limit offshore in waters with high NO3− concentrations, at 16°S. nifH genes affiliated to UCYN-B were detected on the shelf as well (1.8 × 102 copies l−1, Figure 3); however, it is unclear whether UCYN-B like diazotrophs had an active role in fixing N2 in the OMZ off Peru during our study.
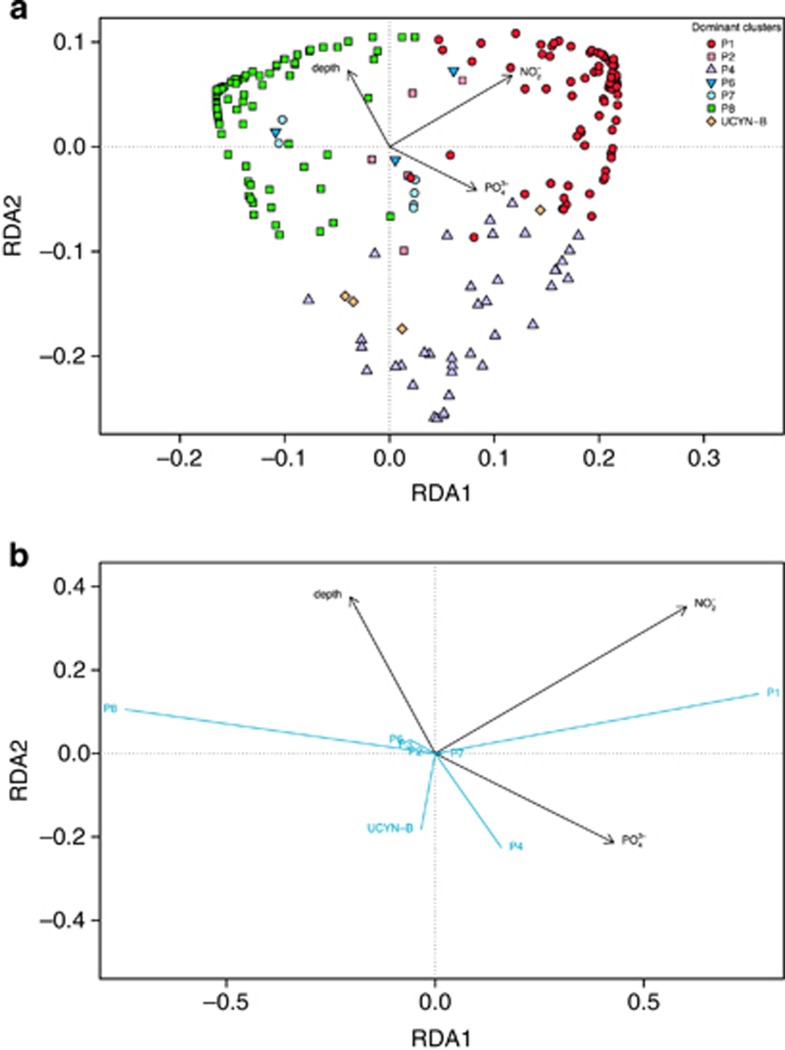
A RDA indicated that a model containing depth, NO2− and PO43− could explain roughly one-tenth of the total variance in the transformed nifH count data (Figure 4).
Figure 4.
Redundancy analysis (RDA) of Hellinger-transformed nifH cluster abundances. (a) The distance biplot (scaling 1) shows relations between samples (weighted sums of species scores) and environmental variables in the parsimonious model. (b) Correlation biplot (scaling 2) shows relations between nifH cluster vectors and environmental variables in the parsimonious model.
The dominant clusters P1, P4, P8 and, to a lesser extent, UCYN-B showed the most pronounced response to environmental variables in the parsimonious model (Figure 4, Supplementary Figure S3). Cluster P1 and P8 occupied complementary habitats, being positively and negatively correlated with NO2− and PO43− concentrations, respectively. Although P1 dominates the shelf OMZ, P8 is almost exclusively present offshore (transect C, Figure 1); this distribution may be explained by several factors, such as nutrient and/or trace metal quotas, or different requirements for organic material. P8 is identical with a cluster previously identified in the South Pacific Gyre (Gamma 4; Halm et al., 2012), suggesting that those organisms prefer more oligotrophic conditions. Cluster P4 was negatively correlated with depth and positively with PO43−, thus P4 occupies a very limited habitat in the OMZ, associated with the upper OMZ, where O2 is still available at low concentration while sinking particles potentially provide chemical energy to fuel their metabolism.
O2 and NO3− had no effect on the relation between nifH clusters and the other environmental variables (Supplementary Figure S4) and were dropped from the parsimonious model during the analyses. From the analysis of variance inflation factors, it became clear that the oxygen effect might be substituted by the effect of phosphate. The lack of an effect from NO3− might be explained by the findings of Grosskopf and LaRoche (2012) that assimilatory nitrate uptake loses its energetic advantage over N2 fixation in low O2 environments. Moreover, in our model, depth might substitute physical variables, such as temperature and salinity, as all of them mark different water masses collinearly. Thus, the observed depth-dependent nifH distribution patterns can potentially be linked to the presence of different water masses. Depth can further be thought of as a surrogate for secondary biotic and abiotic environmental variables. For example, the concentration of the trace metals such as Ni, Cu and Zn increase linearly with depth in our study area (Christian Schlosser, unpublished). The reason we did not incorporate trace metals into our statistical model is that we would have had to waive too many data points due to sparse sampling. It will be advantageous in future studies to directly measure these and other variables in addition to those we identified as relevant.
In situ N2 fixation
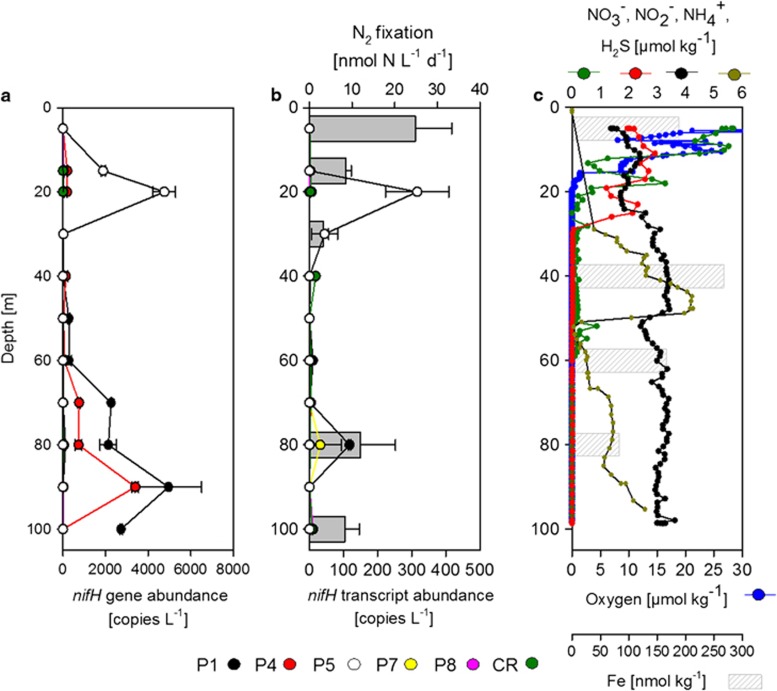
Measurements of the incorporation of 15N2 into particulate organic material (PON) revealed low but detectable N2 fixation rates throughout the OMZ, potentially favoured by P* (Figure 1, Supplementary Figure S1). In addition, dissolved iron was present in high concentrations in the waters on the shelf (10–60 nmol kg−1 in the OMZ core) and decreased towards open ocean locations down to 0.2–1.9 nmol kg−1 in the core of the OMZ in line with observations from earlier studies (Hong and Kester, 1986; Hutchins et al., 2002; Bruland et al., 2005). A broad maximum of N2 fixation extending into the OMZ (beginning at∼40 m) could be observed at 10°S from 79.134°W to 81.361°W with up to 0.4 nmol N l−1 d−1 at 200 m depth close to the coast, slightly decreasing towards offshore (Supplementary Figures S5 and S6). Further south on the Peruvian shelf (no.19, 12°21.88′S, 77°0.00′W), water column N2 fixation rates increased, showing higher activity in surface waters than at depth, consistent with low N:P ratios (resulting in high P*, Supplementary Figure S1) and high dissolved iron concentrations (up to 267 nmol kg−1, Figure 5), there. Highest N2 fixation rates of 24.8±8.4 nmol N l−1 d−1 were measured in surface waters and at lower rates in deeper waters, where nifH of UCYN-B, P1 and P7 was expressed (Figure 5). At this station, the water column below the mixed layer (∼20 m) was completely anoxic and hydrogen sulfide (H2S) was present from bottom to 26 m depth (Figure 5 (Schunck et al., 2013)). Below 30-m depth, the water was depleted in NO3− and NO2−, the key substrates for anammox and denitrification, suggesting a limitation of N-loss activity by substrate unavailability; however, both processes were active at that station (Kalvelage et al., 2013). Ammonia was present in concentrations of 2–4 μM below the oxycline, which is classically assumed to inhibit N2 fixation (Ullrich et al., 1990), but integrated water column N2 fixation exceeded 800 μmol N m−2 d−1, a rate comparable to those reported from major Trichodesmium blooms (Capone et al., 2005). The peak in P1 nifH expression present at 80 m along with a maximum in N2 fixation here indicates an involvement of this cluster in N2 fixation at depth. However, P1 sequences are the closest related to cluster IV (Zehr et al., 2003), which is classically not considered to be functional with regard to N2 fixation. Organisms associated with cluster IV, which actively fix N2, usually contain an additional cluster I or III nitrogenase, which was not detectable in our samples. Thus, we cannot fully exclude that P1 is associated with a non-N2-fixing microorganism.
Figure 5.
Vertical distribution of (a) the nifH gene abundance, (b) the measured N2 fixation and nifH gene expression (samples were specifically collected for RNA extraction), and (c) chemical parameters (O2, NO3−, NO2−, NH4+, H2S, Fe) at a coastal sulphidic station (M77/3, no.19, 12°21.88′S, 77°0.00′W).
The measured high N2 fixation rate (800 μmol N m−2 d−1) detected at this sulphidic station indicates that sulphidic events, which have previously been reported to occur in intense OMZs (Naqvi et al., 2000; Lavik et al., 2009) might sporadically significantly boost N2 fixation. This might be due to the supply of PO43− and Fe from the sediment to the water column assuming the sulfidic inputs are sediment derived. Further research in this direction is necessary to establish the potential relationship between these sulfidic events, which develop in nitrogen-depleted waters, and the input of new nitrogen through N2 fixation.
Heterotrophic diazotrophy in the OMZ off Peru
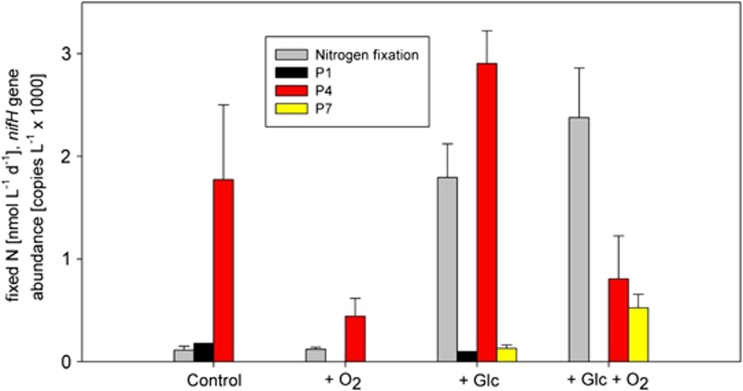
To investigate heterotrophic N2 fixation in the Peruvian OMZ, we performed glucose and oxygen manipulation experiments at one open ocean station (no.3, 10°S 81.3°W, Figure 1) and one coastal station (no.807, 10.001°S 78.38°W, Figure 1). In both experiments, N2 fixation was stimulated by the addition of glucose (2 μM) as well as by the simultaneous addition of glucose and oxygen (10 μM) pointing towards a contribution of heterotrophic diazotrophs to N2 fixation. However, only the glucose effect was significant (Supplementary Table S2). nifH analysis of end points of 24-h 15N2-incubation experiments at station 3 (95 m water depth) supplemented with glucose or O2 or a combination of both demonstrated the exclusive presence of P4 and P7 nifH genes in the different treatments. P1 was present in abundances close to the detection limit in the control and in the glucose treatment (Figure 6; Supplementary Figure S7 shows a similar experiment at the more coastal station 807 from 20-m water depth). For both P4 and P7, there was a conspicuous interaction of glucose with O2: in P4, combined glucose and O2 addition resulted in a marked increase of gene abundances compared with a (borderline) significant decrease with only one treatment factor applied (see Supplementary Material and Supplementary Table S2 for details of the statistical analysis). Combined glucose and O2 addition resulted in a clear decrease of P7 gene abundances compared with the glucose-only treatment. Gene abundances of cluster P1 did not show any systematic response to the experimental treatment (see Figure 6) and were therefore not further addressed statistically.
Figure 6.
The effect on the addition of glucose (2 μM) and oxygen (10 μM) on N2 fixation as well as on the nifH gene abundances of the detectable clusters P1, P4 and P7 (both as end point measurements in 24-h incubations, samples from no.3, 95 m) were determined. S.ds. of triplicate measurements are indicated, with the exception of P1 which was below the detection limit in two of three replicate samples. Rates are a minimum value of N2 fixation due to a probable underestimation by the applied method.
Highest N2 fixation rates in this experiment were measured in the combined glucose and oxygen addition experiment (in situ O2 concentrations ∼1.85 μM), which stimulated exclusively the growth of P7, whereas clusters P1 and P4 were negatively affected by the addition of oxygen, even in the presence of glucose. This suggests a potential switch to a dominance of the P7 cluster when O2 is transported into the OMZ (for example, by O2 intrusions or vertical mixing), thus demonstrating the capability of the diazotrophic community to react rapidly to changing O2 conditions. The increase in N2 fixation with the addition of glucose in both experiments (no.3, 95 m; and no.807, 20 m) points towards a heterotrophic mode of diazotrophy throughout the OMZ that might be limited by the availability of reduced carbon compounds rather than by nutrients.
Co-occurrence of N2 fixation and N-loss processes
Evidence for the co-occurrence of N-loss and N2 fixation mainly ascribed to Chlorobium-like diazotrophs has previously been documented for an anoxic lake (Halm et al., 2009) and is proposed to have a major role in marine N2 fixation as well (Deutsch et al., 2007). As mentioned above, Deutsch et al. (2007) predicted that the availability of excess phosphorous resulting from N-loss processes in the OMZ off Peru is favourable for N2 fixation. The dominance of heterotrophic diazotrophs along with active N2 fixation in waters below the oxycline implies the possibility for a close spatial coupling of N-loss and N2 fixation. Kalvelage et al. (2013) reported in their N-loss study from the same cruise that denitrification rates in the water column were not detectable except for two shelf stations (note that one of those was no.807 at 20 m with a denitrification rate of 5.42 nmol l−1 d−1, which has been discussed above with regard to N2 fixation rates) and the sulphidic station. In the presence of H2S, water column denitrification ranged from 126 nmol N l−1 d−1 to 2068 nmol N l−1 d−1, indicating a coupling with N2 fixation rates, which are significantly positively correlated to denitrification rates (n=5, r2=0.59) at that station. nifH gene abundances of cluster P1 coincided with the denitrification key functional gene nirS (coding for the cd1-containing nitrite reductase) on the shelf at 10°S (Supplementary Figures S2 and S8). Overall, Kalvelage et al. (2013) reported that anammox was the major N-loss process during the sampling period in that area and was detectable at all stations from the coast to 84°W, with highest rates (⩽225 nmol N l−1 d−1) over the central shelf (10°S–16°S) declining by two orders of magnitude towards the open ocean. Similarly, N2 fixation rates were higher on the shelf, declining by one to two orders of magnitude towards the open ocean (compare Figure 5, Supplementary Figures S5 and S6). This co-occurrence of N2 fixation and anammox is in line with the significant correlation of gene abundances of P1 and the hydrazine (N2H4) oxidoreductase anammox key gene hzo (n=113, r2=0.591) and a correlation of total nifH and hzo (n=113, r2= 0.544), both observed at 10°S. Both nifH gene abundance (see above) and anammox rates (Kalvelage et al., 2013) were significantly correlated to NO2− concentrations on the shelf. Further, nifH gene abundances of cluster P1 correlated significantly with the abundances of key functional genes of archaeal ammonia oxidation (archaeal amoA, coding for the ammonia monooxygenase, n=351, r=0.61) on the shelf pointing towards a close spatial coupling of all detected N turnover processes in this area.
Based on our findings, we hypothesize, that a close coupling of N-loss processes and N2 fixation exists in OMZs and that N2 fixation may be responsible for the progressive increase in N:P ratio from the inshore waters to the open ocean as previously suggested by the model study of Deutsch et al. (2007).
Conclusions
Our study demonstrates the presence of mainly heterotrophic diazotrophs in the OMZ off Peru. High primary production close to the coast might fuel heterotrophic diazotrophy in this area supported by the presence of elevated iron across the shelf region. In light of our results, this heterotrophic N2 fixation seems to be directly linked to anammox and other N-turnover processes, thus in principle confirming the model of Deutsch et al. (2007). However, in contrast to the suggestion of Deutsch et al. (2007) that the highest N2 fixation occurs in oligotrophic open ocean waters towards the South Pacific Gyre, our results support a much closer spatial coupling of N loss and N2 fixation, with the highest rates in shelf waters with high organic matter load and high iron availability. Contrary to the classical view, the diazotrophic distribution can statistically be explained by factors other than O2 and NO3−, namely NO2− and PO43−. The activity of diazotrophs in waters below the euphotic zone has currently not been considered in estimates of global N2 fixation rates neither from biogeochemical models (Codispoti, 2007) nor derived from geochemical tracer studies or direct measurements. Our findings may therefore facilitate a more realistic estimation of the marine nitrogen budget with regard to ocean de-oxygenation.
Furthermore, our findings contribute to the emerging view that the habitat of diazotrophs should be extended towards high fixed nitrogen environments in OMZs in which a unique diazotrophic community exists.
Acknowledgments
We thank the authorities of Peru for granting permission to work in their territorial waters. We acknowledge the support of the captain and crew of the R/V Meteor as well as the chief scientists Martin Frank and Lothar Stramma. Moreover, we thank I Grefe for sampling during M77/4 and O Baars and A Dammshäuser for trace metal sampling during M77/3; we also thank G Klockgether, K Stange, F Malien, V Leon and P Fritsche for oxygen and nutrient measurements, and also G Klockgether and P Streu for assisting with the mass spectrometry and trace metal analyses, respectively. We thank Hermann Bange, V Bertics and K Wuttig for helpful discussion. Financial support for this study was provided by the DFG Sonderforschungsbereich 754 (www.sfb754.de) and the Max Planck Society (MPG).
Author contributions
CRL, TG, HS and JLR collected samples and performed the experiments onboard. CRL and DG and FDD extracted the DNA. CRL and DG performed the qPCR of nifH genes from transect stations. FDD designed primers and probes for nifH qPCRs. CRL, TG, NP, FDD and DG cloned nifH gene fragments. TG, CRL and HS extracted DNA and RNA from experimental stations. TG and CRL performed nifH qPCRs of experimental stations. CRL performed amoA, hzo and nirS qPCRs of M77/3 stations. TG performed flow cytometry measurements and analysed the C and N fixation data. SCN performed the statistical analysis. CS and PLC performed the trace metal sampling and analysis. CRL wrote the manuscript with TG, HS, RAS and JLR. MMMK, GL, RAS, JLR, TG and CRL designed the experiments and analysed the data.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Bopp L, Le Quere C, Heimann M, Manning AC, Monfray P. (2002). Climate-induced oceanic oxygen fluxes: implications for the contemporary carbon budget. Global Biogeochem Cycles 16: 14. [Google Scholar]
- Bruland KW, Franks RP, Knauer GA, Martin JH. (1979). Sampling and analytical methods for the determination of copper, cadmium, zinc, and nickel at the nanogram per liter level in sea water. Anal Chim Acta 105: 233–245. [Google Scholar]
- Bruland KW, Rue EL, Smith GJ, DiTullio GR. (2005). Iron, macronutrients and diatom blooms in the Peru upwelling regime: brown and blue waters of Peru. Mar Chem 93: 81–103. [Google Scholar]
- Capone D. (2008). The marine nitrogen cycle. Microbe 3: 186–192. [Google Scholar]
- Capone DG, Burns JA, Montoya JP, Subramaniam A, Mahaffey C, Gunderson T et al. (2005). Nitrogen fixation by Trichodesmium spp.: an important source of new nitrogen to the tropical and subtropical North Atlantic Ocean. Global Biogeochem Cycles 19: 17. [Google Scholar]
- Church MJ, Jenkins BD, Karl DM, Zehr JP. (2005). Vertical distributions of nitrogen-fixing phylotypes at Stn ALOHA in the oligotrophic North Pacific Ocean. Aquat Microb Ecol 38: 3–14. [Google Scholar]
- Codispoti LA. (2007). An oceanic fixed nitrogen sink exceeding 400 Tg Na(-1) vs the concept of homeostasis in the fixed-nitrogen inventory. Biogeosciences 4: 233–253. [Google Scholar]
- Codispoti LA. (2010). Interesting times for marine N2O. Science 327: 1339–1340. [DOI] [PubMed] [Google Scholar]
- Danielsson L-G, Magnusson B, Westerlund S. (1978). An improved metal extraction procedure for the determination of trace metals in sea water by atomic absorption spectrometry with electrothermal atomization. Anal Chim Acta 98: 47–57. [Google Scholar]
- Deutsch C, Gruber N, Key RM, Sarmiento JL, Ganachaud A. (2001). Denitrification and N2 fixation in the Pacific Ocean. Global Biogeochem Cycles 15: 483–506. [Google Scholar]
- Deutsch C, Sarmiento JL, Sigman DM, Gruber N, Dunne JP. (2007). Spatial coupling of nitrogen inputs and losses in the ocean. Nature 445: 163–167. [DOI] [PubMed] [Google Scholar]
- Duce RA, LaRoche J, Altieri K, Arrigo KR, Baker AR, Capone DG et al. (2008). Impacts of atmospheric anthropogenic nitrogen on the open ocean. Science 320: 893–897. [DOI] [PubMed] [Google Scholar]
- Falcon LI, Cipriano F, Chistoserdov AY, Carpenter EJ. (2002). Diversity of diazotrophic unicellular cyanobacteria in the tropical North Atlantic Ocean. Appl Environ Microbiol 68: 5760–5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnelid H, Bentzon-Tilia M, Andersson AF, Bertilsson S, Jost G, Labrenz M et al. (2013). Active nitrogen-fixing heterotrophic bacteria at and below the chemocline of the central Baltic Sea. ISME J 7: 1413–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnelid H, Andersson AF, Bertilsson S, Al-Soud WA, Hansen LH, Sørensen S et al. (2011). Nitrogenase gene amplicons from global marine surface waters are dominated by genes of non-cyanobacteria. PLoS One 6: e19223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez C, Farias L, Ulloa O. (2011). Nitrogen fixation in denitrified marine waters. PLoS One 6: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz J, Krahmann G, Lavik G, Grasse P, Dittmar T, Riebesell U. (2012). Dynamics and stoichiometry of nutrients and phytoplankton in waters influenced by the oxygen minimum zone in the eastern tropical Pacific. Deep-Sea Res Pt I 62: 20–31. [Google Scholar]
- Gandhi N, Singh A, Prakash S, Ramesh R, Raman M, Sheshshayee MS et al. (2011). First direct measurements of N2 fixation during a Trichodesmium bloom in the eastern Arabian Sea. Global Biogeochem Cycles 25: 10. [Google Scholar]
- Grasshoff G, Kremling K, Erhardt M. (1999) Methods of Seawater Analysis. Wiley VCH: Weinheim, Germany. [Google Scholar]
- Grosskopf T, LaRoche J. (2012). Direct and indirect costs of dinitrogen fixation in Crocosphaera watsonii WH8501 and possible implications for the nitrogen cycle. Front Aquat Microbiol 3: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskopf T, Mohr W, Baustian T, Schunck H, Gill D, Kuypers MMM et al. (2012). Doubling of marine dinitrogen-fixation rates based on direct measurements. Nature 488: 361–364. [DOI] [PubMed] [Google Scholar]
- Halm H, Lam P, Ferdelman TG, Lavik G, Dittmar T, LaRoche J et al. (2012). Heterotrophic organisms dominate nitrogen fixation in the South Pacific Gyre. ISME J 6: 1238–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halm H, Musat N, Lam P, Langlois R, Musat F, Peduzzi S et al. (2009). Co-occurrence of denitrification and nitrogen fixation in a meromictic lake, Lake Cadagno (Switzerland). Environ Microbiol 11: 1945–1958. [DOI] [PubMed] [Google Scholar]
- Hamersley MR, Turk KA, Leinweber A, Gruber N, Zehr JP, Gunderson T et al. (2011). Nitrogen fixation within the water column associated with two hypoxic basins in the Southern California Bight. Aquat Microb Ecol 63: 193. [Google Scholar]
- Hong H, Kester DR. (1986). Redox state of iron in offshore waters of Peru. Limnol Oceanogr 31: 512–524. [Google Scholar]
- Hutchins DA, Hare CE, Weaver RS, Zhang Y, Firme GF, DiTullio GR et al. (2002). Phytoplankton iron limitation in the Humboldt Current and Peru Upwelling. Limnol Oceanogr 47: 997–1011. [Google Scholar]
- Ingall E, Jahnke R. (1994). Evidence for enhanced phosphorus regeneration from marine sediments overlain by oxygen depleted waters. Geochim Cosmochim Acta 58: 2571–2575. [Google Scholar]
- Jayakumar A, Al-Rshaidat MMD, Ward BB, Mulholland MR. (2012). Diversity, distribution, and expression of diazotroph nifH genes in oxygen-deficient waters of the Arabian Sea. FEMS Microbiol Ecol 82: 597–606. [DOI] [PubMed] [Google Scholar]
- Kalvelage T, Lavik G, Lam P, Contreras S, Artega L, Löscher CR et al. (2013). Nitrogen cycling driven by organic matter export in the South Pacific oxygen minimum zone. Nat Geosci 6: 228–234. [Google Scholar]
- Lam P, Jensen MM, Lavik G, McGinnis DF, Muller B, Schubert CJ et al. (2007). Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc Natl Acad Sci USA 104: 7104–7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane BG, Bernier F, Dratewka-Kos E, Shafai R, Kennedy TD, Pyne C et al. (1991). Homologies between members of the germin gene family in hexaploid wheat and similarities between these wheat germins and certain Physarum spherulins. J Biol Chem 266: 10461–10469. [PubMed] [Google Scholar]
- Langlois RJ, LaRoche J, Raab PA. (2005). Diazotrophic diversity and distribution in the tropical and subtropical Atlantic ocean. Appl Environ Microbiol 71: 7910–7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois RJ, Hummer D, LaRoche J. (2008). Abundances and distributions of the dominant nifH phylotypes in the Northern Atlantic Ocean. Appl Environ Microbiol 74: 1922–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavik G, Stuhrmann T, Bruchert V, Van der Plas A, Mohrholz V, Lam P et al. (2009). Detoxification of sulphidic African shelf waters by blooming chemolithotrophs. Nature 457: 581–U586. [DOI] [PubMed] [Google Scholar]
- Legendre P, Gallagher ED. (2001). Ecologically meaningful transformations for ordination of species data. Oecologia 129: 271–280. [DOI] [PubMed] [Google Scholar]
- Loescher CR, Kock A, Koenneke M, LaRoche J, Bange HW, Schmitz RA. (2012). Production of oceanic nitrous oxide by ammonia-oxidizing archaea. Biogeosci Discuss 9: 2095–2122. [Google Scholar]
- Mohr W, Grosskopf T, Wallace DWR, LaRoche J. (2010). Methodological underestimation of oceanic nitrogen fixation rates. PLoS One 5: e12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya JP, Voss M, Kahler P, Capone DG. (1996). A simple, high-precision, high-sensitivity tracer assay for N2 fixation. Appl Environ Microbiol 62: 986–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi SWA, Jayakumar DA, Narveka PV, Naik H, Sarma VVSS, D'Souza W et al. (2000). Increased marine production of N2O due to intensifying anoxia on the Indian continental shelf. Nature 408: 346–349. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB et al. (2013). vegan: Community ecology package. R package version 2.0-10. http://CRAN.R-project.org/package=vegan.
- R_Core_Team. (2013). R: A language and environment for statistical computing. In: http://www.R-project.org/.
- Schmid MC, Hooper AB, Klotz MG, Woebken D, Lam P, Kuypers MMM et al. (2008). Environmental detection of octahaem cytochrome c hydroxylamine/hydrazine oxidoreductase genes of aerobic and anaerobic ammonium-oxidizing bacteria. Environ Microbiol 10: 3140–3149. [DOI] [PubMed] [Google Scholar]
- Schunck H, Lavik G, Desai DK, Großkopf T, Kalvelage T, Löscher CR et al. (2013). Giant hydrogen sulfide plume in the oxygen minimum zone off peru supports chemolithoautotrophy. PLoS One 8: e68661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratil SB, Neulinger SC, Knecht H, Friedrichs AK, Wahl M. (2013). Temperature-driven shifts in the epibiotic bacterial community composition of the brown macroalga Fucus vesiculosus. Microbiologyopen 2: 338–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk KA, Rees AP, Zehr JP, Pereira N, Swift P, Shelley R et al. (2011). Nitrogen fixation and nitrogenase (nifH) expression in tropical waters of the eastern North Atlantic. ISME J 5: 1201–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich W, Rigano C, Fuggi A, Aparicio P, Paneque A, Cejudo FJ et al. (1990). Nitrogen metabolism in heterotrophic bacteria. Simultaneous ammonia inhibition of nitrogen fixation and nitrate uptake and divalentation regulation of nitrate uptake in Azotobacter chroococcum. In Inorganic Nitrogen in Plants and Microorganisms. Springer: Berlin, Heidelberg, Germany, pp 93–98. [Google Scholar]
- Zani S, Mellon MT, Collier JL, Zehr JP. (2000). Expression of nifH genes in natural microbial assemblages in Lake George, New York, detected by reverse transcriptase PCR. Appl Environ Microbiol 66: 3119–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr JP, Turner PJ. (2001). Nitrogen fixation: nitrogenase genes and gene expression. In: Methods in Microbiology vol 30. Academic Press Inc: San Diego, CA, USA, pp 271–286. [Google Scholar]
- Zehr JP, Mellon MT, Zani S. (1998). New nitrogen-fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes (vol 64, pg 3444, 1998). Appl Environ Microbiol 64: 5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr JP, Jenkins BD, Short SM, Steward GF. (2003). Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ Microbiol 5: 539–554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.