Abstract
Polymicrobial biofilms are of large medical importance, but relatively little is known about the role of interspecies interactions for their physiology and virulence. Here, we studied two human pathogens co-occuring in the oral cavity, the opportunistic fungus Candida albicans and the caries-promoting bacterium Streptococcus mutans. Dual-species biofilms reached higher biomass and cell numbers than mono-species biofilms, and the production of extracellular polymeric substances (EPSs) by S. mutans was strongly suppressed, which was confirmed by scanning electron microscopy, gas chromatography–mass spectrometry and transcriptome analysis. To detect interkingdom communication, C. albicans was co-cultivated with a strain of S. mutans carrying a transcriptional fusion between a green fluorescent protein-encoding gene and the promoter for sigX, the alternative sigma factor of S. mutans, which is induced by quorum sensing signals. Strong induction of sigX was observed in dual-species biofilms, but not in single-species biofilms. Conditioned media from mixed biofilms but not from C. albicans or S. mutans cultivated alone activated sigX in the reporter strain. Deletion of comS encoding the synthesis of the sigX-inducing peptide precursor abolished this activity, whereas deletion of comC encoding the competence-stimulating peptide precursor had no effect. Transcriptome analysis of S. mutans confirmed induction of comS, sigX, bacteriocins and the downstream late competence genes, including fratricins, in dual-species biofilms. We show here for the first time the stimulation of the complete quorum sensing system of S. mutans by a species from another kingdom, namely the fungus C. albicans, resulting in fundamentally changed virulence properties of the caries pathogen.
Keywords: biofilm, Candida, Streptococcus, EPS, quorum sensing
Introduction
It has been estimated that 80% of human infections result from pathogenic biofilms (Harriott and Noverr, 2011). Clinically, biofilm infections represent an overwhelming problem, as the microorganisms embedded in the extracellular matrix are resistant to antibiotics as well as to the host defence. Polymicrobial pathogenic biofilms are not only found in the oral cavity, but also in the respiratory tracts, skin, the reproductive and urinary tract, in patients with chronic lung diseases and on in-dwelling mechanical devices (Peleg et al., 2010).
Candida albicans is the most prevalent opportunistic human pathogenic fungus (Kim and Sudbery, 2011) and can cause infections of mucosal membranes (candidiasis) and the blood stream (candidemia). It is able to form biofilms on mucosal membranes as well as on implants (Cuellar-Cruz et al., 2012). Biofilm formation and virulence of C. albicans are connected with the transition from the yeast to the hyphae morphotype, which represents a crucial step towards pathogenesis. Yeast cells colonise predominantly surfaces, whereas the hyphal form of C. albicans is invasive (Sudbery, 2011; Gow et al., 2012). Hyphae provide structural integrity to biofilms (Finkel and Mitchell, 2011; Banerjee et al., 2013). C. albicans has been found in periodontal pockets in both the chronic and aggressive forms of periodontitis (Urzua et al., 2008).
Streptococcus mutans is another opportunistic pathogen inhabiting the human oral cavity (Ajdic et al., 2002). S. mutans belongs to the phylum Firmicutes and is a Gram positive, facultative anaerobic bacterium that can ferment a large spectrum of dietary sugars. The excreted organic acids result in a strong localized pH drop that can cause lesions of the dental enamel and thus initiate caries development. S. mutans is a common inhabitant of the oral cavity and has long been recognized as one of the causes of dental caries, a highly prevalent biofilm-dependent polymicrobial oral infectious disease (Kutsch and Young, 2011; Takahashi and Nyvad, 2011; Burne et al., 2012). Biofilm formation in S. mutans is readily induced by dietary sugars that are transformed to extracellular polysaccharides by dedicated enzymes, which are therefore important targets of anti-caries strategies (Bowen and Koo, 2011).
C. albicans and S. mutans are found together in early childhood caries (Marchant et al., 2001; de Carvalho et al., 2006; Raja et al., 2010) and on bracket materials (Rammohan et al., 2012). Early childhood caries is a very aggressive form of caries. It has been shown that C, albicans is present in 96% of caries-positive children (age group 6–12 years) but only in 24% of caries-free children (Raja et al., 2010). Denture plaque was shown to contain both S. mutans and C. albicans in 25.5% of healthy individuals (Ribeiro, 2012). However, data showing actual cell–cell contact between those two organisms in vivo is currently lacking.
A tight co-aggregation between C. albicans and streptococci has been observed (Jenkinson et al., 1990; Metwalli et al., 2013) and could be owing to specific adhesins similar to those found in S. gordonii (Silverman et al., 2010) or to a glucan layer formed on the Candida cells by the glucosyltransferase B (GtfB) exoenzyme (Gregoire et al., 2011). Interaction between C. albicans and streptococci can contribute to enhanced biofilm formation. Oral streptococci produce cell wall-anchored proteins facilitating binding to C. albicans (Bamford et al., 2009). The S. gordonii cell wall-associated polypeptide SspB3 interacts directly with the C. albicans hyphae-specific agglutinin-like sequence 3 (Nobbs et al., 2010). A synergistic partnership between S. oralis, S. sanguinis and C. albicans was observed, where the fungus promoted biofilm formation by the streptococci on abiotic surfaces and on mucosa (Diaz et al., 2012). Thus not only S. mutans, but also C. albicans might have crucial roles for the cariogenic biofilm succession (Metwalli et al., 2013). This is supported by a recent study showing that an increased load of Candida cells correlated with decreased diversity of the saliva microbiome and a shift of the microbial community towards streptococci (Kraneveld et al., 2012).
Quorum sensing signalling has been shown to be involved in interactions between C. albicans and bacteria. Hyphae development in C. albicans can be inhibited by 3-oxo-C12-homoserine lactone from Pseudomonas aeruginosa (Hogan et al., 2004), the S. mutans pheromone CSP (competence-stimulating peptide) (Jarosz et al., 2009) or by the diffusible signal factor trans-2-decenoic acid (Vilchez et al., 2010). Conversely, the signalling molecule farnesol from C. albicans inhibits the swarming motility of P. aeruginosa (McAlester et al., 2008) and induces the Pseudomonas quinolone signal synthesis (Cugini et al., 2010). No report exists of the induction of the quorum sensing signalling circuit of an oral pathogen by a eukaryotic fungus.
The main virulence traits of S. mutans—acidogenicity, aciduricity, biofilm formation and mutacin production—as well as its ability to incorporate foreign DNA into its genome (genetic competence) are controlled by quorum sensing. S. mutans secretes the so-called AI-2, for which no signalling function could so far be identified in this organism (Sztajer et al., 2008). It also secretes two peptide pheromones, the CSP (Li et al., 2001) and XIP (alternative sigma factor sigX-inducing peptide) (Mashburn-Warren et al., 2010; Khan et al., 2012) that induce genetic competence through two different signalling pathways converging on SigX, the only alternative sigma factor of S. mutans, which is therefore a master regulator of quorum sensing (Federle and Morrison, 2012).
The overwhelming majority of quorum sensing studies have been performed in mono-species systems. However, the main quorum sensing-controlled traits of S. mutans, that is, mutacin synthesis and genetic competence, have their largest impact in multi-species systems because the DNA that is taken up is different from the DNA released from the competent cell, and especially in biofilms, which provide high local concentrations of chemical cues as well as direct cell–cell contact. For example, S. mitis has been shown to acquire antibiotic resistance genes by transformation in a multi-species biofilm (Hannan et al., 2010). As S. mutans and C. albicans have been found together in oral infections and biofilm growth is essential for the virulence of both of them, here we studied morphology, physiology, transcriptomics and genetics of their dual-species biofilms in vitro under conditions of robust hyphal growth of C. albicans and strong biofilm formation of S. mutans. We were particularly interested in the role of cell–cell communication in shaping the physiology of dual-species biofilms, and for this purpose used a reporter strain for the promoter of the alternative sigma factor SigX (Lemme et al., 2011). The data show that both microorganisms profit from growth in dual-species biofilms, and that C. albicans has a profound influence on the physiology and quorum sensing-mediated traits of S. mutans.
Materials and methods
Strains and culture conditions
Microorganisms used were S. mutans UA159 wild type (ATCC 700610, Manassas, VA, USA), S. mutans UA159 SMPsigXgreen fluorescent protein (GFP) (Lemme et al., 2011) and C. albicans (DSM 11225, Braunschweig, Germany). Pre-cultures of S. mutans strains were grown in THBY medium (Becton, Dickinson and Company, Sparks, MD, USA), if necessary (for the reporter strain and deletion mutants) with erythromycin (3 μg ml−1) at 37 °C aerobically with 5% CO2 without shaking. C. albicans pre-cultures were grown in yeast nitrogen base (YNB) synthetic medium (Difco Laboratories, Detroit, MI, USA) supplemented with maltose (1 g l−1) and glucose (2 g l−1) at 37 °C aerobically with 5% CO2 without shaking for 16 h. The medium that supported growth of S. mutans and C. albicans in biofilms (YNBB) contained YNB (6.7 g l−1, Difco Laboratories), 75 mM Na2HPO4–NaH2PO4 (pH 7.3), N-acetylglucosamine (2.5 mM, Sigma-Aldrich, Taufkirchen, Germany), casamino acids (2 g l−1, Becton, Dickinson and Company) and sucrose (5 g l−1). The pH during biofilm growth was in the range of pH 7.0–6.6 (S. mutans single biofilm), pH 7.0–6.4 (dual-species biofilm) and pH 7.0–6.95 (C. albicans single biofilm).
Biofilm formation
Pre-cultures of S. mutans and C. albicans were inoculated from single colonies or frozen glycerol stocks, respectively, grown for 16 h, harvested by centrifugation (5000 r.p.m., 20 min, 4 °C) and suspended in YNBB medium to an OD600 of 0.1. Equal volumes of each strain (500 μl) were inoculated into the wells of 24-well microtitre plates. Suspensions (1 ml) of one strain only (S. mutans or C. albicans) were prepared for single-strain inoculums. The microtitre plates (Thermo Fisher Scientific, Rochester, NY, USA) used for biofilm formation were coated with artificial saliva for 1 h at 37 °C to mimick the conditions in the oral cavity (Wong and Sissons, 2001). Natural saliva has a crucial role for the colonization of dental enamel (Scannapieco, 1994). Following the removal of artificial saliva, the inoculum was pipetted into the coated well (1 ml for 24-well microtitre plates and 0.2 ml for 96-well microtitre plates). The plates were incubated aerobically (5% CO2) at 37 °C. Biofilms were allowed to develop for 4, 6, 8, 10, 12 and 24 h. Growth was monitored by crystal violet staining, quantitative PCR and microscopical analysis.
Induction of the alternative sigma factor SigX
The S. mutans reporter strain SMPsigXGFP carrying a plasmid with the promoter of sigX fused to GFP was cultivated together with C. albicans in dual-species biofilms. Single-species biofilms were prepared in parallel. Sterile medium was used as a control. After removal of the supernatant, the GFP fluorescence intensity was recorded with a Wallac 1420 Multilabel counter (PerkinElmer, Waltham, MA, USA). Fluorescence microscopic analysis of biofilms was carried out using an Olympus BX60 microscope (Olympus, Seelze, Germany) equipped with a colour view II camera and a × 100 immersion oil objective. The filter U-MWIBA3 (excitation 460–495 nm, emission 510–550 nm and dichromatic filter 505 nm) was used.
Two pheromones of S. mutans (CSP and XIP) and farnesol of C. albicans were tested for induction of sigX in biofilms of the reporter strain SMPsigXGFP. The pheromones were added at concentrations between 0.1 and 100 μM, and the biofilm was incubated for 4, 6, 8, 10 and 24 h. Afterwards the planktonic phase of the culture was withdrawn and the fluorescence intensity was recorded with a Wallac 1420 Multilabel counter (PerkinElmer). Fresh YNBB medium was used as a control. In addition, we have tested induction of sigX by CSP, XIP and farnesol added to the mature biofilm (8 h). After 2 and 4 h of treatment the fluorescence intensity was recorded.
Field emission scanning electron microscopy
Biofilm samples were fixed with a fixation solution containing 2% glutaraldehyde and 5% formaldehyde in HEPES buffer (100 mM HEPES, 10 mM MgCl2, 10 mM CaCl2, 90 mM sucrose, pH 7.0) and stored at 4 °C. After washing the samples three times with TE buffer (10 mM TRIS, 2 mM EDTA, pH 7.0), they were dehydrated with a graded series of acetone (10%, 30%, 50%, 70%, 90% and 100%) on ice. The 100% acetone step was repeated at room temperature. Samples were then subjected to critical point drying with CO2 and sputter coated with gold-palladium. The analyses were carried out using a Zeiss Merlin field emission scanning electron microscope (Zeiss, Oberkochen, Germany) at an acceleration voltage of 5 kV using the Everhart-Thornley SE-detector and the inlens SE-detector in a 25:75 ratio. Contrast and brightness were adjusted in Adobe Photoshop CS3.
Staining with concanavalin A
The procedure was adapted from Kolodkin-Gal et al. (2012). Biofilms were rinsed with 200 μl of 1 × phosphate-buffered saline buffer and stained with 50 μl Concanavalin A labelled with AlexaFluor 488 (100 μg ml−1) for 40 min at room temparature in the dark. Subsequently the staining solution was withdrawn, the biofilm was washed with 50 μl of 1 × phosphate-buffered saline and then counterstained with 10 μM DAPI (4′,6-diamidino-2-phenylindole) for 15 min at room temperature in the dark. After removal of DAPI the biofilm was analyzed under the fluorescence microscope (Olympus BX 60, Shinjuku, Japan) using the filter U-MWIBA3 for AlexaFluor 488 and U-MINUA2 for DAPI respectively.
Uptake of labelled DNA
The amplicon of the 16S RNA gene of S. mutans was used as a source of DNA for the competence test and was obtained as described above. Purified DNA amplicon (2 μg) was labelled with 4 μl of Cy3 (Kreatech Biotechnology, Amsterdam, the Netherlands) for 15 min at 85 °C. The labelled DNA was purified on the column of the ULS labelling kit (Kreatech Biotechnology) according to the manufacturer's instructions. The degree of labelling was determined by measuring absorbance at 260 and 550 nm. Material with a degree of labelling of 1.5 was used. DNA uptake was tested as described with modifications (Lemme et al., 2011). Biofilms of the S. mutans reporter strain SMPsigXGFP alone and together with C. albicans were cultivated as decribed for 10 h. The supernatants were removed and biofilm of three wells was suspended in 100 μl of fresh YNBB medium without sucrose. To disrupt cell chains, the biofilm was sonified for 10 cycles of 5 s with 10 s pause at 10% power (Bandelin Electronic, Berlin, Germany) on ice. The Cy3-labelled DNA (final concentration 5 μg ml−1) was added to the biofilm cells and incubated in the dark for 30 min at 37 °C. Excess DNA was digested with DNAse I and the biofilms were analysed by fluorescence microscopy. The red channel was used to visualize Cy3, and the green channel to observe GFP.
Methods for preparation of conditions media, construction of knockout mutants, quantitative PCR, extraction of DNA and RNA, microarray analysis and gas chromatography–mass spectrometry analysis are described in Supplementary Methods S1. Primers and a comparison between qPCR and microarray data are provided in Supplementary Tables S1–S3.
Results
Cultivation medium
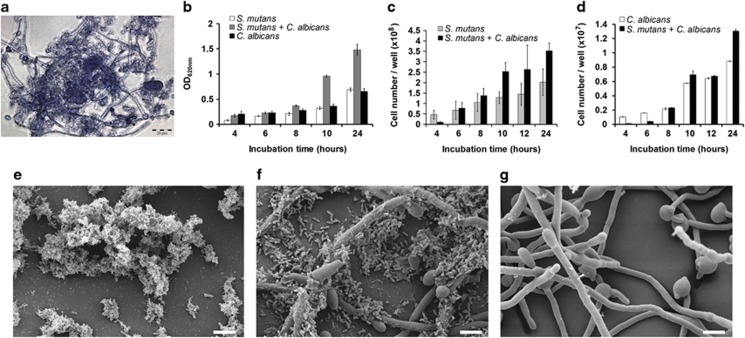
We used a chemically defined synthetic medium commonly used for cultivating C. albicans as a basis. Sucrose was provided as a carbon source, as it is fermented by both species, and in S. mutans induces strong biofilm formation. S. mutans is auxotrophic for several amino acids (Ajdic et al., 2002), therefore we added casamino acids. GlcNAc was provided to induce hyphae formation in C. albicans (Sudbery, 2011) and the medium was buffered to exclude different pH effects in single and dual-species biofilms. In this medium, designated YNBB, both microorganisms formed biofilms and C. albicans grew predominantly in the hyphal form. Phase contrast microscopy (Figure 1a) showed that C. albicans and S. mutans were in direct cell–cell contact, with many S. mutans cells adhering to the hyphae of C. albicans.
Figure 1.
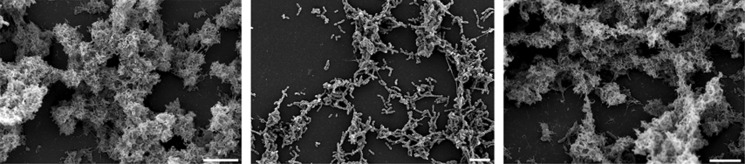
Growth and morphology of S. mutans and C. albicans in single- and dual-species biofilms. (a) Phase contrast micrograph of a dual-species biofilm. C albicans mainly grows in the hyphal form; some cells growing in the yeast form are also visible. S. mutans attaches to the hyphae of C. albicans. (b) Biofilm mass determined by crystal violet staining (mean and s.d. from three independent experiments). (c) Cell numbers of S. mutans (c) and C. albicans (d) determined by quantitative PCR of the 16S rRNA gene and the 18S rRNA gene, respectively. Mean and s.d. of three independent experiments with two technical replicates each are shown. (e and f) Scanning electron micrographs of 10-h biofilms of S. mutans (e), dual-species biofilm with C. albicans (f) and C. albicans (g). Scale bar (e–g) 4 μm.
Growth and morphology of dual-species biofilms of S. mutans and C. albicans
Biofilm biomass increased continuously for both species throughout the 24 h of cultivation, both in mono-culture and in co-culture (Figure 1b). At 10 and 24 h the biomass of dual-species biofilms was almost twice that of single-species biofilms. Quantitative PCR of the 16S rRNA and 18S rRNA gene revealed that cell numbers of both species increased throughout the experiment, in accordance with the biomass data. Cell numbers in dual-species biofilms were significantly higher than in mono-species biofilms, both for S. mutans and C. albicans (Figures 1c and d). The data show that co-cultivation of S. mutans and C. albicans in dual-species biofilms resulted in better growth of each species suggesting that metabolic interactions may have taken place, which provided additional nutrients.
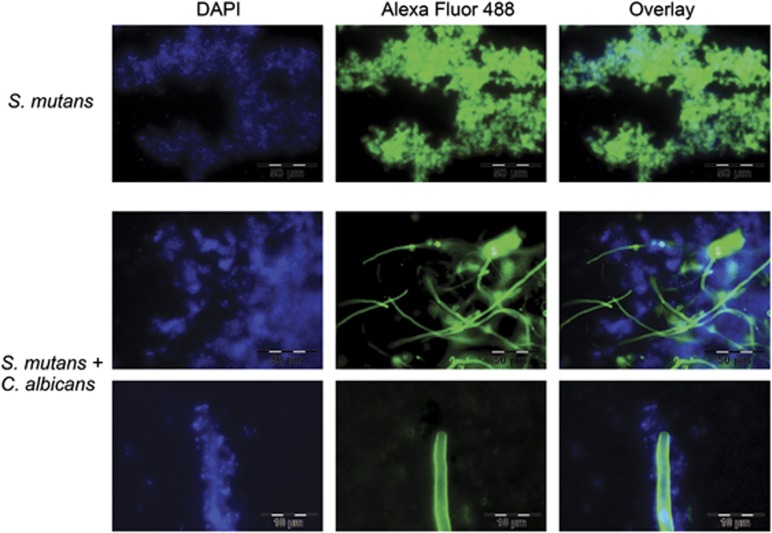
Scanning electron micrographs (Figure 1e) showed that S. mutans formed clumps in mono-species biofilms, which were embedded in extracellular polymeric substance (EPS). By contrast, in dual-species biofilms (Figure 1f) little EPS was visible, and many cells of S. mutans appeared to be ‘naked', that is, lacking the EPS matrix. The EPS matrix of S. mutans consists mainly of glucan and fructan (Trautner et al., 1981) and has been thoroughly studied because it has an important role for caries development (Koo et al., 2013). To visualize the EPS matrix under the fluorescent microscope, we stained the biofilms with the glucan-binding lectin concanavalin A labelled with the green fluorescent dye AlexaFluor 488 (Figure 2). To visualize the DNA of the cells, we used the DNA-binding blue fluorescing dye DAPI. Single-species biofilms of S. mutans showed strong green fluorescence of S. mutans, indicating that EPS had been formed. No such green fluorescence of S. mutans could be observed in dual-species biofilms. The hyphae of C. albicans also contain glucan and therefore can clearly be seen on the pictures. These findings confirm the scanning electron micrographs and show that EPS was not formed by S. mutans in dual-species biofilms.
Figure 2.
EPS matrix in single- and dual-species biofilms. Biofilms (10-h old) were stained with two fluorescent dyes: The lectin concanavalin A labelled with AlexaFluor 488 binds specifically to termical sugar moieties of glycans and fluoresces green. The dye DAPI (4′,6-diamidino-2-phenylindole) binds to DNA and fluoresces blue. Green and blue fluorescence are shown separately (left and middle panel) and overlaid. Scale bar (upper two panels) 50 μm, (bottom panel) 10 μm.
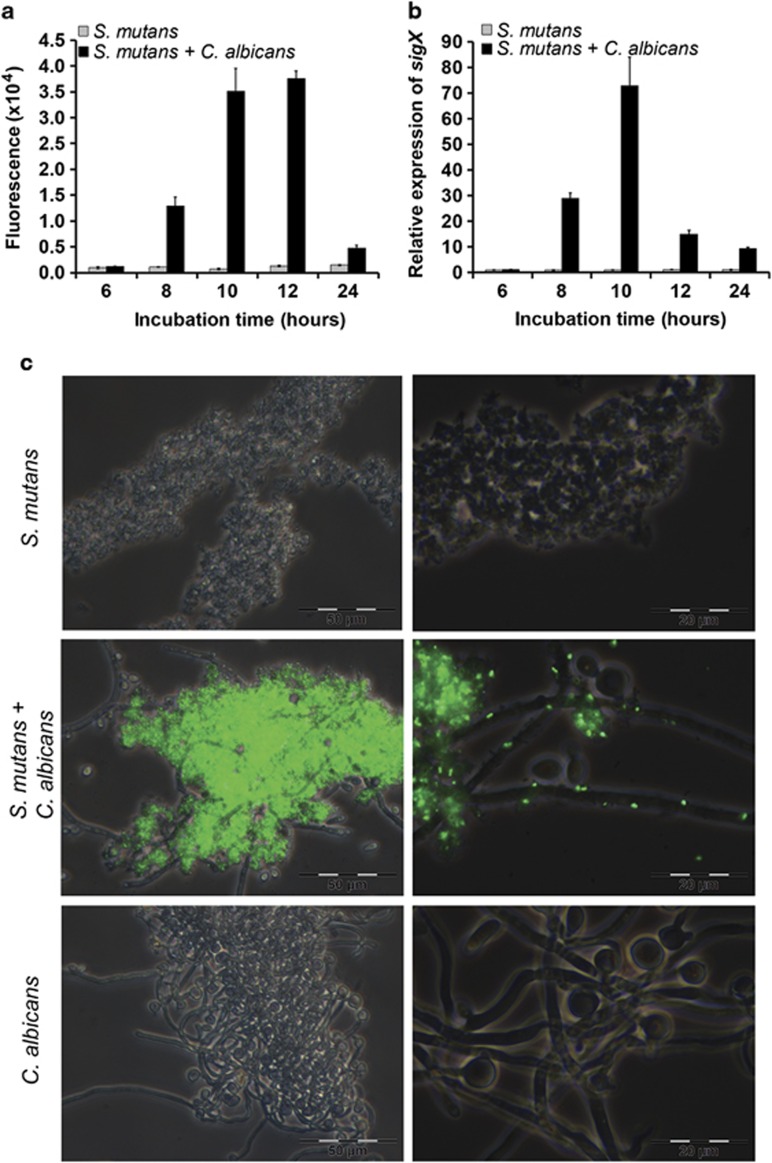
The sigX promoter is induced in dual-species biofilms
To determine whether quorum sensing of S. mutans was induced in our biofilms, we used the S. mutans reporter strain SMPsigXGFP. It carries a plasmid where the promoter of the alternative sigma factor SigX is fused to GFP. Upon induction of SigX green fluorescence is observed. It was quantified using a fluorescence plate reader (Figure 3a), by reverse transcriptase (RT)-PCR of sigX in the wild type (Figure 3b) and observed directly under the fluorescence microscope (Figure 3c). Strong induction of PsigX was observed in dual-species biofilms with C. albicans (Figure 3a), showing a maximum between 8 and 12 h of biofilm growth. There was no detectable activation of PsigX in mono-species biofilms of S. mutans. Gene expression of sigX was quantified using S. mutans wild type by quantitative RT-PCR (Figure 3b). A 73-fold increase of sigX expression was observed in co-culture with C. albicans at 10 h in comparison with S. mutans cultivated alone. At 12 h, sigX expression already decreased. Such a fast decrease could not be observed with the SMPsigXGFP reporter strain owing to the stability of gfp. PsigX induction was confirmed by microscopical analysis. The majority of S. mutans cells revealed strong green fluorescence (Figure 3c) in co-culture with C. albicans. Cells that were attached to C. albicans as well as more distant biofilm cells were fluorescing. Such unimodal induction of sigX has previously only been demonstrated in pure cultures of S. mutans in a peptide-free chemically defined medium (Son et al., 2012). Here, we show that in a semi-defined medium, unimodal activation of SigX occurs if the two species C. albicans and S. mutans are grown together.
Figure 3.
Induction of the alternative sigma factor SigX of S. mutans in dual-species biofilms. (a) Fluorescence intensity of SMPsigXGFP, a gfp-reporter for sigX expression in S. mutans, grown as a single-species biofilm (grey bars) or together with C. albicans as a dual-species biofilm (black bars) quantified using the VictorWallac 1420 fluorescence plate reader. (b) Quantitative RT-PCR of sigX expression in S. mutans wild-type biofilms grown alone (grey bars) or with C. albicans (black bars). The data were normalized to sigX expression in a S. mutans biofilm after 6 h. Mean and s.d. from four independent experiments are shown. (c) Fluorescence microscopy of single- and dual-species biofilms after 10 h of growth. Phase contrast and gfp channel are overlaid. Scale bars (c, left) 50 μm, (c, right) 20 μm.
Conditioned media of mixed biofilms activate the sigX promoter in reporter strain biofilms and comS is required for this activation
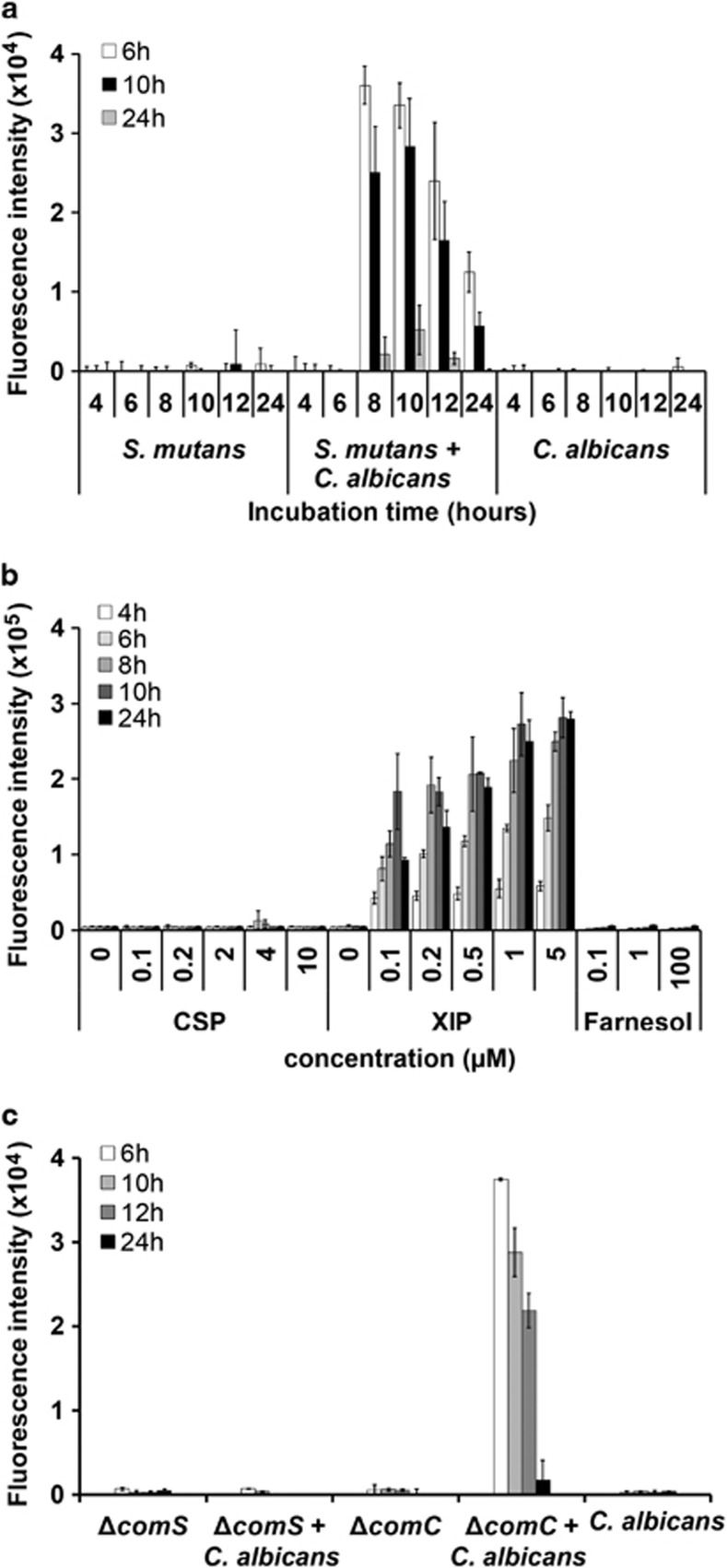
We wanted to know whether (1) an unknown molecule present in the cultivation medium or (2) one of the known quorum sensing signalling molecules of S. mutans or C. albicans, respectively, might be responsible for the induction of sigX in co-culture. Finally, we also tested whether (3) the activating compound was present in the spent medium, or whether activation occurred indirectly by stimulating S. mutans to synthesize the inducer. Sterile-filtered supernatants from single- and dual-species biofilms (4, 6, 8, 10, 12 and 24 h) were applied to biofilms of the reporter strain SMPsigXGFP grown for 6, 10 and 24 h, respectively. Supernatants of dual-species biofilms that had been cultivated at least 8 h caused activation of PsigX, whereas single-biofilm culture supernatants either of S. mutans or of C. albicans had no effect (Figure 4a). This activation was stronger in younger reporter strain biofilms (6 h) than in older ones (10 h), but even 24-h-old reporter strain biofilms could be induced by dual-species biofilm supernatants. The strongest induction was seen for 6-h-old reporter strain biofilms challenged with supernatants from 8-h mixed biofilms. The data show that an external factor produced exclusively in dual-species biofilms was responsible for activating the sigX promoter.
Figure 4.
Activation of sigX-gfp by culture supernatants and pheromones and role of the autoinducer synthases ComC and ComS. (a) Culture supernatants were obtained from biofilms of S. mutans and C. albicans cultivated separately or together for 4–24 h and added to 6, 10 and 24 h-old-test biofilms of the reporter strain SMPsigXGFP. Fluorescence intensity was determined after 2 h of incubation. (b) Activation of sigX-gfp in reporter strain biofilms of S. mutans by the quorum sensing pheromones CSP (competence-stimulating peptide) and XIP (sigX-inducing peptide) produced by S. mutans and by farnesol produced by C. albicans. The autoinducers were added as pure compounds at the indicated concentrations. Fluorescence was determined after 2–24 h of biofilm growth. (c) Same experiment as in (a), except that culture supernatants from deletion mutants for comC and comS of S. mutans were tested. comC encodes the synthesis of the CSP precursor, whereas comS encodes the synthesis of the XIP precursor. Mean and s.d. of four experiments are shown in all cases.
We then tested whether reporter strain biofilms could be activated by the known quorum sensing pheromones of S. mutans, namely XIP and CSP, or by the quorum sensing signal of C. albicans, farnesol. These autoinducers were added at various concentrations to biofilms of the reporter strain, which were then grown for 4, 6, 8, 10 and 24 h. A robust activation of PsigX-GFP after addition of XIP was observed (Figure 4b). The activation was particularly strong for 8- and 10-h-old biofilms and was proportional to the concentration of XIP up to a final concentration of 5 μM XIP. No or only neglectable activation by CSP or farnesol was observed even at concentrations of 10 and 100 μM, respectively. These data suggest that XIP might be responsible for PsigX activation in dual-species biofilms.
To test whether XIP was present in the supernatants or whether it was produced by S. mutans, we constructed deletion mutants for comS and comC and used these mutants for single- and dual-species biofilm growth. Culture supernatants of single-species biofilms of the ΔcomS mutant as well as from dual-species biofilms of the ΔcomS mutant with C. albicans were unable to induce PsigX in the reporter strain (Figure 4c). By contrast, conditioned media from dual-species biofilms of S. mutans ΔcomC and C. albicans clearly induced PsigX. These results suggest that comS, but not comC, is indispensable for activation of PsigX. Therefore, we hypothesize that S. mutans was induced to produce XIP in co-culture with C. albicans.
EPS of S. mutans are not produced in conditioned media from dual-species biofilms
To test whether a factor suppressing EPS synthesis was secreted into the medium, sterile conditioned media were prepared and S. mutans biofilms were cultivated in them. Electron microscopy showed that the EPS matrix was clearly present when S. mutans was grown in culture supernatants from 10-h single-species biofilms of C. albicans or S. mutans, respectively (Figure 5 right and left panels). By contrast, in culture supernatants of 10-h dual-species biofilms, the test biofilm was more dispersed and lacked EPS and the cells of S. mutans appeared to be naked, similar to those grown in co-culture with C. albicans (Figure 1f). This effect was already observed after 4 h of growth of test biofilms in dual-species biofilm extract (Supplementary Figure S1). We conclude that supernatants of dual-species biofilms prevented EPS synthesis of S. mutans. We tested whether XIP, the quorum sensing molecule excreted by S. mutans, might be able to suppress EPS formation, but this was not the case (Supplementary Figure S2).
Figure 5.
EPS formation of S. mutans biofilms in conditioned media. S. mutans biofilms were cultivated in conditioned media from 10-h-old single- (left and right panels) and dual-species biofilms (middle panel) for 10 h and analysed by scanning electron microscopy. Scale bar, 5 μm.
To determine whether an extracellular enzyme might be present that is able to destroy the polysaccharides constituting the EPS matrix, for example, glucan, we added sterile-filtered culture supernatants from dual-species biofilms (grown for 6, 10, 12 and 24 h) to a 10-h-old established biofilm of S. mutans. No significant change in biofilm morphology and EPS thickness occurred as determined by scanning electron microscopy after 2 and 4 h (data not shown).
Transcriptome analysis of S. mutans growing in single- and dual-species biofilms
A whole-genome microarray (Xue et al., 2010) was used to analyse the transcriptome of S. mutans in dual-species biofilms with C. albicans before (6 h), during (10 h) and after (24 h) sigX activation in comparison with gene expression of S. mutans in mono-species biofilms. We found 510 genes that were differentially expressed. They were sorted into six different groups according to their expression profile using the c-means algorithm (Kumar and Futschik, 2007) (Supplementary Figure S3), and putative biological functions were assigned based on clusters of orthologous groups (Supplementary Figure S4). Several microarray data were confirmed by quantitative RT-PCR (Supplementary Table S3). All transcripts showing high abundance in the microarray data revealed similar or higher gene expression levels in quantitative RT-PCR, in accordance with the higher dynamic range of quantitative RT-PCR, which has often been observed.
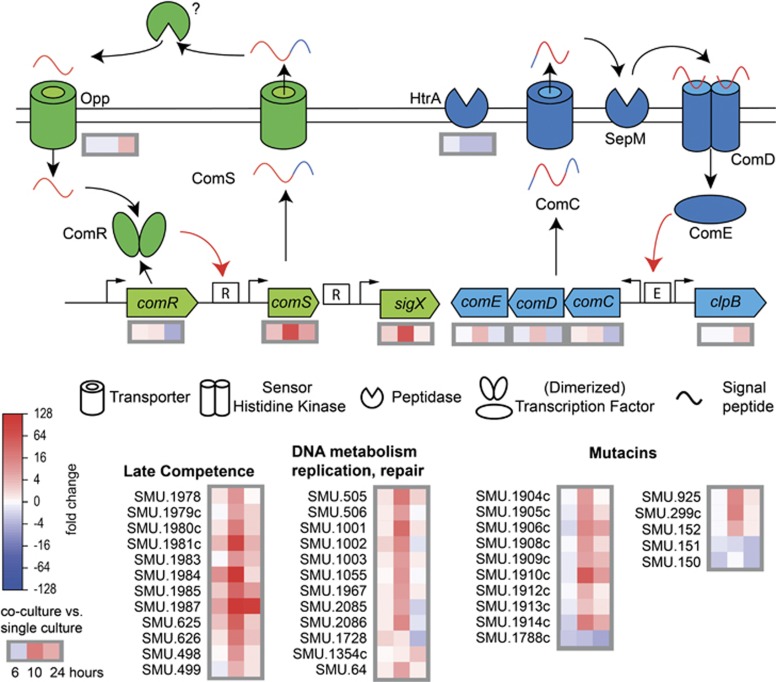
The most abundant transcripts belonged to the quorum sensing regulon
At 10 h of growth in mixed biofilms, the expression of 84 genes was significantly increased (groups 3 and 4). Among them the competence-related transcripts were the most abundant ones (Figure 6; Supplementary Figure S5; Supplementary Table S4). Particularly, comS, sigX and the late competence genes (comYA and comYC) were the most abundant transcripts and revealed fold change values of 68, 56 and above 200, respectively. These data confirm the biological experiments and indicate that sigX was activated by the proximal ComRS system. Genes downstream of sigX (late competence genes) were also highly upregulated, including those belonging to the ComY operon, the transformasome, natural transformation (SMU.1001—SMU.1003), and genes encoding enzymes for excision, insertion, inversion and translocation of DNA (SMU. 2085—recA and SMU. 2086—cinA).
Figure 6.
Induction of the quorum sensing regulon and late competence genes in dual-species biofilms. Upper panel: schematic view of the quorum sensing regulon of S. mutans (modified from (Perry et al., 2009; Lemme et al., 2011)) and differential gene expression after 6, 10 and 24 h of biofilm growth in dual-species biofilms with C. albicans compared with single-species biofilms of S. mutans alone. The ComRS system is shown in green, the ComCDE system is shown in blue. Black and red arrows correspond to processing of the signalling peptide and transcriptional regulation by ComR/E, respectively. Pictograms are explained below the scheme. Lower panel: differential gene expression of the late competence genes, genes related to DNA metabolism and repair and mutacins of S. mutans in dual-species biofilms.
The genes constituting the CSP-driven quorum sensing circuit were also upregulated but much more weakly. The genes of the comCDE operon revealed fold change values of 1.8, 3.2 and 3.4, respectively. The genetic competence of S. mutans can be modulated by HtrA, an enzyme degrading CSP. The gene encoding this protein (SMU.2164) was downregulated likely resulting in weak degradation of CSP and possible induction of competence development via the CSP pathway as well.
Mutacins were strongly upregulated, especially nlmAB (SMU.150 and SMU.151, mutacin IV), cipB (SMU.1914c, nlmC and mutacinV) and several other putative bacteriocins (Supplementary Figure S5). The mutacin IV immunity protein (SMU.152) and two genes, which display some sequence homology to SMU.152 (SMU.1909 and SMU.1913), were also upregulated. However, the mutacin V immunity protein (SMU.925) was slightly downregulated. In addition to the CSP-driven mutacin synthesis controlled by the ComDE TCS, bacteriocin synthesis is regulated by three other TCS, HdrRM, BrsRM and VicRK (Merritt and Qi, 2012). The HdrRM (SMU.1854 and SMU.1855) and BrsRM (SMU.2080 and SMU.2081) systems did not show significant changes in gene expression. However, the response regulator VicR (SMU.1517) was slightly upregulated in all dual-species biofilms. It was reported that it negatively affects transcription of comC, comDE and sigX (Senadheera et al., 2012) and in this way modulates the CSP-driven signalling system of S. mutans. The data show that mutacins were coordinately induced with the development of genetic competence through CSP and XIP, and that even the negative regulator VicR was not able to decrease this effect in dual-species biofilms.
Competent cells of S. pneumoniae produce cell wall hydrolases, so-called fratricins, which are secreted by the competent subpopulation and kill the non-competent brothers—hence the name (Berg et al., 2012; Wei and Havarstein, 2012). In S. mutans, the gene SMU.836 is 100% identical to cbpD, the key fratricin of S. pneumoniae. It was recently shown that this cell wall hydrolase actually acts as a fratricin in S. mutans (Dufour and Levesque, 2013). Interestingly, it was strongly upregulated in 10-h-old dual-species biofilms, that is, during sigX activation, together with the adjacent gene SMU.837, which encodes a putative reductase. To protect themselves against their own fratricins, S. pneumoniae cells produce an immunity protein termed ComM (Berg et al., 2011). In S. mutans the two genes murN (SMU.716) and murM (SMU.717) show homology to ComM but they were not upregulated. It remains to be studied whether a subpopulation suffered cell death as a result of the highly expressed murein hydrolases.
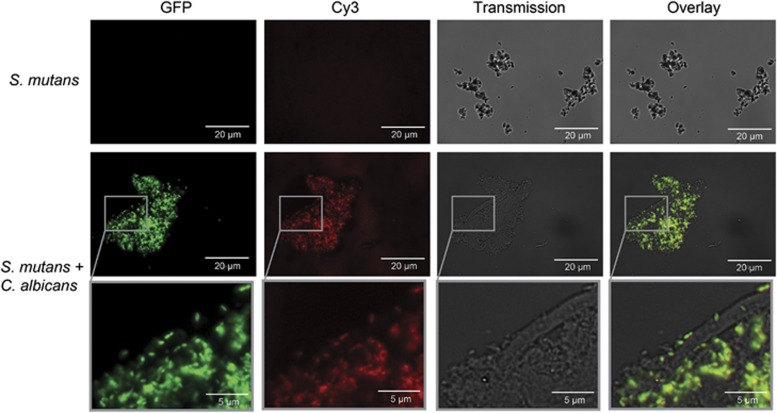
To summarize, the complete quorum sensing regulon of S. mutans was highly induced in co-culture with C. albicans, starting from the two signalling pathways for CSP and XIP, and including the corresponding downstream genes, that is, genetic competence, mutacins and fratricins. To test whether indeed genetic competence was functional, we studied the uptake of fluorescently labelled DNA in biofilms of the reporter strain SMPsigXGFP. The middle panel of Figure 7 shows that in co-culture with C. albicans the cells of S. mutans are not only fluorescing green, but they have taken up labelled DNA, indicated by red fluorescence of the same cells. Both views can be overlaid resulting in yellow fluorescence. The lower panel in Figure 7 shows a magnification of the indicated window from the middle panel. Single cells of S. mutans can clearly be observed as they attach to the hyphae of C. albicans or form flocs and are fluorescing both green and red, indicating induction of sigX as well as uptake of DNA. None of this can be seen in single-species biofilms of S. mutans (upper panel).
Figure 7.
Uptake of DNA in dual-species biofilms of C. albicans and S. mutans. The reporter strain SMPsigXGFP was cultivated for 10 h alone (top panel) or together with C. albicans (middle and bottom panel). DNA labelled with Cy3 was added, and after incubation for 30 min excess DNA was removed by DNAse treatment. See Methods for experimental details. The four rows show (from left to right) the green channel for GPF, the red channel for Cy3, phase contrast and the overlay of red and green channels. Scale bar, 20 μm (top and middle panels) and 5 μm (bottom panel).
Polysaccharide synthesis shifted from the extracellular EPS component glucan to the intracellular storage polymer glycogen
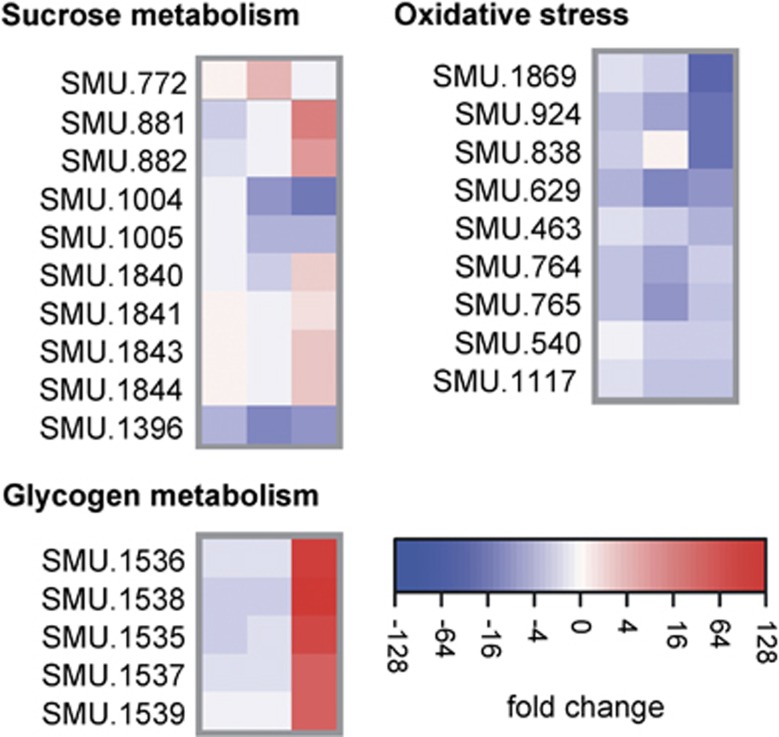
The microarray data suggested fundamental changes in sugar metabolism in dual-species biofilms (Figure 8; Supplementary Table S4). The main component of EPS in S. mutans biofilms are glucans and fructans, which are synthesised by extracellular glucosyltransferases (Gtts) and fructosyltransferases, respectively (Bowen and Koo, 2011). The two main glycosyltansferase genes, gtfB and gtfC, as well as the fructosyltransferase gene scrK showed reduced transcript abundance already at 6 h of dual-biofilm growth (Supplementary Table S4). GtfB expression was reduced 5.6-fold and 9.7-fold at 10 h and 24 h of dual-biofilm growth, respectively. In addition, expression of the glucan-binding protein GbpC was strongly downregulated (up to 8.1-fold). Sucrose is not only used for extracellular glucan synthesis in S. mutans, but also as a carbon source and metabolised intracellularly. However, the genes of the sucrose operon were all downregulated, whereas the sucrose operon repressor ScrR was upregulated. Only in 24-h-old dual-species biofilms, the sucrose phosphorylase GtfA and the sugar-binding transporter MsmK were upregulated (9.2-fold and 6.3-fold, respectively). These data suggest that both extracellular and intracellular sucrose metabolism was suppressed in S. mutans in dual-species biofilms.
Figure 8.
Transcriptional profiling of genes related to sugar metabolism and oxidative stress in dual-species biofilms. Gene expression after 6, 10 and 24 h of biofilm growth of S. mutans in dual-species biofilms with C. albicans compared with expression in single-species biofilms of S. mutans alone.
By contrast, genes encoding enzymes for glycogen synthesis (SMU.1535–SMU.1539) were strongly upregulated in 24-h dual-species biofilms (between 23.3- and 65.9-fold). One can hypothesize that as a result of the high activity of the MsmK transporter an excess of intracellular monosaccharides was present in 24-h-old biofilm cells of S. mutans, which most likely were converted to the storage compound glycogen.
Sugar composition in biofilm supernatants
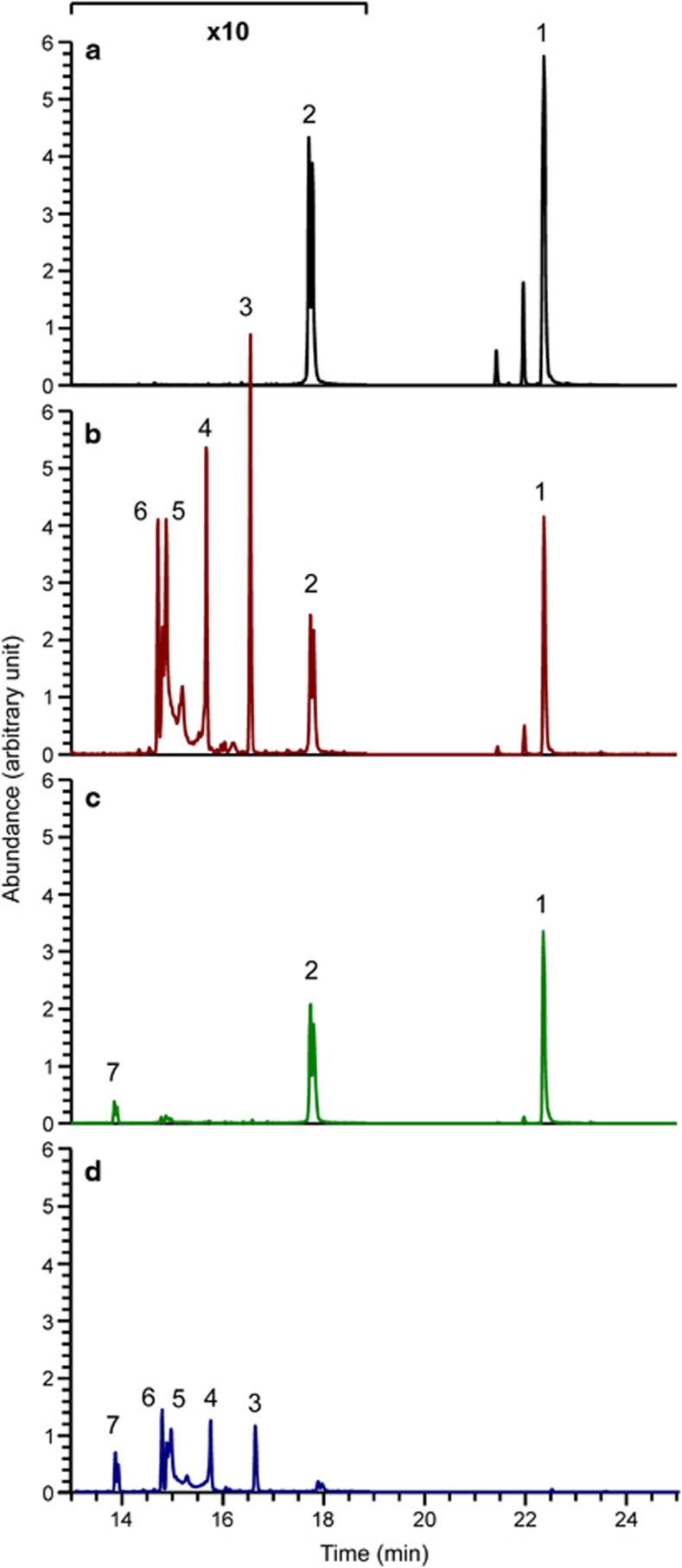
Growth experiments and microarray data suggested major changes in the metabolism of the cultivation medium in dual-species biofilms. Therefore, gas chromatography–mass spectrometry was applied for the analysis of sugars present in the conditioned media (Figure 9; Supplementary Figure S6). Chromatograms from S. mutans alone at 10 h showed a reduction in the peaks for sucrose and GlcNAc in comparison with the cultivation medium, indicating that both compounds were metabolised. New peaks appeared (3,4,5 and 6) that were identified as glucose and fructose. They were already present at 4 h of growth and increased in peak area until 24 h of biofilm growth. C. albicans alone also reduced both the GlcNAc and the sucrose peak; however, it produced only minute amounts of fructose and an unidentified monosaccharide, which might be a pentose or a deoxy-hexose (peak 7). In dual-species biofilms (10 h), strikingly the sucrose peak, which was prominent in single-species biofilms at that time, had already disappeared, and the GlcNAc peak was substantially reduced in comparison with the single-species biofilms. Monosaccharides were also almost completely depleted in dual-species biofilms at 10 h. After 24 h of biofilm growth, only the putative pentose peak remained in the dual-species biofilm supernatant, whereas single-species biofilms still contained large amounts of monosaccharides in the case of S. mutans and GlcNAc in the case of C. albicans. The data show that the kinetics of sugar metabolism differed in single- and dual-species biofilms. Sucrose was lacking from the culture medium after 10 h of growth in dual-species biofilms.
Figure 9.
Sugar composition of the cultivation medium after 10 h of biofilm growth. Biofilm supernatants were sterile filtered and analysed by gas chromatography–mass spectrometry. (a) Cultivation medium, (b) S. mutans biofilm supernatant, (c) C. albicans biofilm supernatant and (d) spent medium from dual-species biofilm of S. mutans and C. albicans. The following peaks were identified: 1, sucrose; 2, N-acetylglucosamine; 3 and 4, glucose; 5 and 6, fructose; 7 possibly C5 sugars (pentose). Peak height shows the maximum ion count for this specific mass (arbitrary units). Note the 10-fold enlargement of the y-axis in the left part of the chromatogram.
Discussion
The data show strong synergism in dual-species biofilms of S. mutans and C. albicans resulting in increased biofilm mass and cell densities, induction of the complete quorum sensing system of S. mutans and lack of the production of EPS, which is fundamental for the cariogenic dental plaque biofilm succession. Thus, in the presence of C. albicans, under the conditions used in our experiments, the cariogenicity of S. mutans was reduced. At the same time, the induction of quorum sensing resulted in phenotypes important for survival (mutacin synthesis) or genetic adaptation (genetic competence). Growth of C. albicans in the virulent hyphal mode was enhanced, thus it was a win-win situation for both pathogens. By contrast, the interaction between P. aeruginosa and C. albicans is detrimental for the fungus. Co-culture with P. aeruginosa results in reduced biofilm formation of C. albicans (Holcombe et al., 2010) and this could be owing to excreted phenanzines (Morales et al., 2013) or to bacterial lipopolysaccharides (Bandara et al., 2013).
It has been reported that S. mutans can improve growth of C. albicans biofilms (Pereira-Cenci et al., 2008) and that C. albicans profits from lactic acid excreted by S. mutans (Metwalli et al., 2013). The gas chromatography–mass spectrometry data show that monosaccharides released by the extracellular glycosyl- und fructosyltransferases of S. mutans from sucrose (Ajdic et al., 2002) were depleted from the spent medium in dual-species biofilms at 10 h, and thus may have been taken up by C. albicans. Moreover, C. albicans may have been more efficient than S. mutans in taking up sucrose, as this sugar was depleted in dual-species biofilms after 10 h, and genes for enzymes for both the extracellular and intracellular sucrose metabolism of S. mutans were downregulated.
There are 14 phosphotransferase systems for sugar uptake in S. mutans, five of which are constitutively expressed independent of the presence of sugars; these five phosphotransferase systems are specific for sucrose (Ajdic and Pham, 2007). A small fraction of sucrose is metabolized by S. mutans extracellularly for the synthesis of fructan and glucan, the main polysaccharides comprising the EPS matrix. The responsible glycosyltransferase enzymes (GtfB, GtfC and GtfD) release fructose, whereas the fructosyltransferases (Ftf) release glucose (Ajdic et al., 2002).
However, the transcriptome analysis showed downregulation of gtfB, gtfC and scrK at all stages of growth of the dual-species biofilm. These enzymes are induced by their substrate sucrose (Shemesh et al., 2007), which was initially present at a high concentration in YNBB, and depleted in 10-h-old biofilms. We hypothesize that these extracellular enzymes were initially present in the biofilms, resulting in synthesis of monosaccharides and glucan, but that their transcription was rapidly downregulated due to depletion of sucrose. The half-life of most proteins is in the range of 24 h, whereas that of mRNA is around 5 min (Moran et al., 2013). Excreted glucosyltransferases have been shown to be active on Candida surfaces (Gregoire et al., 2011) and thus are likely to have been operating although their transcription was rapidly downregulated.
Interestingly, S. mutans failed to produce EPS in co-culture with C. albicans or in spent medium from 10-h dual-species biofilms. We first hypothesized that an inhibitor of glucosyltransferases might have been excreted by C. albicans potentially providing an interesting anti-caries compound (Koo and Jeon, 2009; Koo et al., 2013). For example, farnesol has been shown to inhibit glucosyltransferases (Koo et al., 2003). However, as the biofilms were mostly composed of hyphae, it is unlikely that farnesol, which inhibits the yeast-to-hyphae transition (Lindsay et al., 2012), was present at sufficiently high concentrations. Another explanation for the lack of EPS synthesis in dual-species biofilms might be that the glucosyl- und fructosyltransferase enzymes were inactive owing to a lack of their substrate. The gas chromatography–mass spectrometry data show that sucrose was depleted in in co-culture with C. albicans after 10 h of biofilm growth and no EPS was formed by S. mutans cultivated in such spent media. Accordingly, glucosyl- und fructosyltransferase enzymes were downregulated in dual-species biofilms.
The quorum sensing system of streptococci has encountered a paradigm shift in recent years owing to the discovery of a novel competence-inducing peptide, termed XIP, the proximal response regulator comR and the gene for the synthesis of the XIP precursor comS, all of which are highly conserved in streptococci (Mashburn-Warren et al., 2010; Fontaine et al., 2013). In S. mutans, the heptapeptide XIP induces competence in an unimodal way, whereas the previously studied CSP induces competence only in a fraction of the cells while the majority remains uninduced and a small subpopulation undergoes autolysis (Lemme et al., 2011; Dufour and Levesque, 2013). Interestingly, the cultivation medium controls which of those two circuits is activated: XIP is only active in a peptide-free medium; it has been suggested that peptides might block the Opp transporter required for import of the active XIP heptamer (Federle and Morrison, 2012). Conversely, CSP works only in a peptide-rich medium—here it has been suggested that the membrane-bound HtrA enzyme, which can degrade CSP, must be saturated by peptides to reduce CSP degradation (Desai et al., 2012; Federle and Morrison, 2012). In addition, it has been shown for S. thermophilus that the ComR response regulator is tolerant to small changes in the sequence of XIP and can even be activated by hydrolysis products of casein, that is, octapeptides present in the medium (Fontaine et al., 2013).
These studies were done with pure cultures of streptococci. To the best of our knowledge the induction of the quorum sensing system of S. mutans by another organism, in this case a eukaryotic fungus, has not been observed before. In our data, the sigX-driven competence was strongly induced in mixed biofilms of S. mutans with C. albicans as well as by supernatants from mixed biofilms. If comS was knocked out, no induction occurred. How then was comS synthesis induced 68-fold in dual-species biofilms?
The YNBB medium contained casamino acids, which are obtained by hydrolysis; thus traces of peptides may have been present; however, no induction of the sigX promoter occurred in the sterile medium. comS could have been induced by a novel unknown signal secreted only in dual-species biofilms, either by C. albicans or by S. mutans; several TCS of S. mutans have been shown to induce competence but their signal is not known (Okinaga et al., 2010). Alternatively, C. albicans may have produced XIP by proteolysis of S. mutans proteins and thus triggered the autoinduction of comS. C. albicans grew in the invasive hyphal mode, excreting an array of hydrolases (Sorgo et al., 2013). The secreted aspartatic proteases (Sap) are the largest group, comprising 10 types, Sap1–Sap10, of which Sap4 to Sap6 were enriched at pH 7.4 in cultures grown in the presence of GlcNAc (Sorgo et al., 2010). More data are needed to confirm this hypothesis and other possibilities cannot be ruled out.
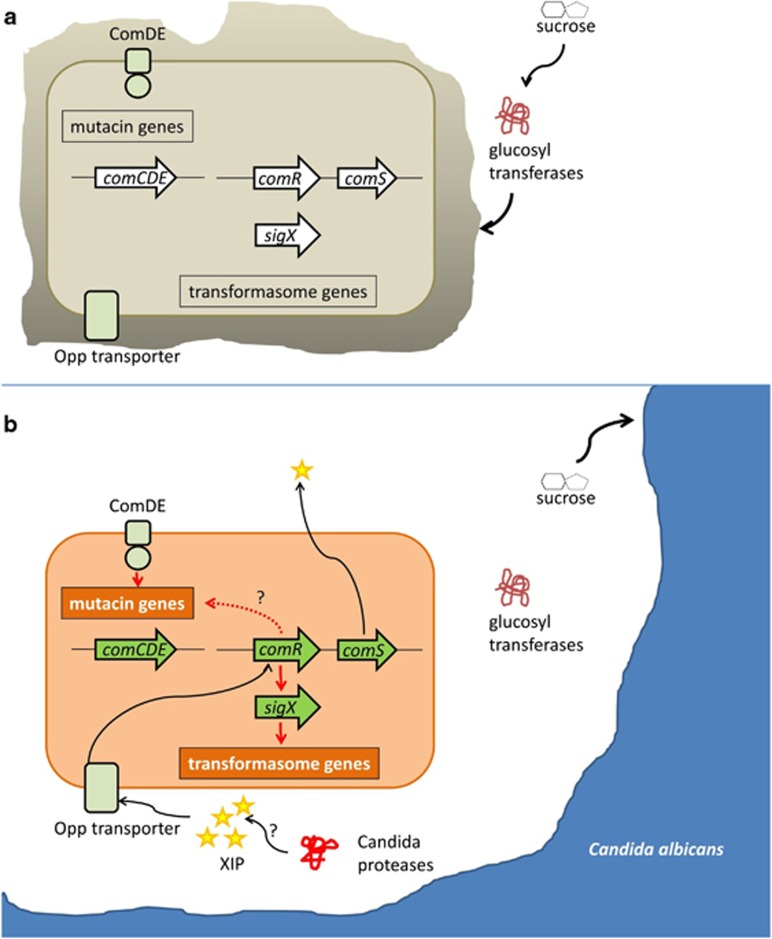
In the dual-species biofilm, the synthesis (comC) and sensing (comDE) of CSP was only weakly activated, but a high abundance of mutacin transcripts was nevertheless observed. The ComDE TCS directly regulates mutacin synthesis through specific promoter-binding sites for the ComE response regulator (Perry et al., 2009; Hung et al., 2011). Recently, it has been shown that mutacin synthesis can also be induced by comR. In S. thermophilus the ComR-box for binding of ComR is not only found upstream of sigX and comA, but also upstream of a number of bacteriocin-encoding genes in the locus blp (Fontaine et al., 2013). In S. mutans two paralogous comR loci are present, of which SMU.61 likely controls competence, whereas SMU.381 controls mutacins (Mashburn-Warren et al., 2010). The mutacin-related loci controlled by comR are distinct from those controlled by the comCDE TCS and could provide a link between comR induction and mutacin synthesis, which does not require induction through CSP. Figure 10 summarizes our main findings and working hypotheses on cross-feeding and interkingdom communication between C. albicans and S. mutans in the biofilms studied here.
Figure 10.
Working hypothesis for cross-feeding and interkingdom communication in dual-species biofilms of S. mutans and C. albicans. S. mutans growing in single culture in a biofilm (a) forms EPS from sucrose due to the glucosyltransferase exoenzymes. The quorum sensing genes are not activated. In the presence of C. albicans (b) sucrose is taken up by C. albicans and no EPS is formed by S. mutans. Extracellular proteases of C. albicans could degrade S. mutans proteins resulting in the production of the heptamer XIP, which is the main quorum sensing signal of S. mutans. XIP is internalized by the Opp transporter and activates the transcriptional regulator comR, thereby triggering activation of the quorum sensing signalling cascade. ComR induces transcription of comS, the precursor of XIP, which is processed and exported to yield active extracellular XIP, resulting in a positive feedback loop. ComR induces expression of the alternative sigma factor sigX, resulting in transcription of the transformasome genes and genetic competence of the cell. Bacteriocin synthesis is induced either through the response regulator ComE or through ComR. Note that the positive feedback loop established through comRS is essential for the observed induction of competence in co-culture, which was not obtained if the comS gene was knocked out in S. mutans.
Conclusion
It is increasingly becoming clear that oral diseases, for example, caries and periodontitis, are polymicrobial and that C. albicans has a crucial role for caries progression. Here, we show that interactions with C. albicans in biofilms result in changes of S. mutans virulence that could not have been anticipated from studying pure cultures. Given the discovery of a conserved core of quorum sensing genes in streptococci, it will be interesting to determine whether C. albicans has a similar role for all of those species. In vivo data are required to observe these interactions in the human host. The synergism between C. albicans and S. mutans shown here could potentially have an important role for early childhood caries and other polymicrobial biofilm infections.
Acknowledgments
We thank Kathrin Opolka, Ina Schleicher and Celine Krajewski for technical assistance. Funding by the DFG TRR51 for JT, by the European Commission COMBACTE project for MRe and by the German Ministry for Economy ZIM program for SPS is gratefully acknowledged. We greatly appreciate the comments of two anonymous reviewers that gave us a chance to make the manuscript stronger.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB et al. (2002). Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci USA 99: 14434–14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajdic D, Pham VT. (2007). Global transcriptional analysis of Streptococcus mutans sugar transporters using microarrays. J Bacteriol 189: 5049–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford CV, d'Mello A, Nobbs AH, Dutton LC, Vickerman MM, Jenkinson HF. (2009). Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun 77: 3696–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandara HM, K Cheung BP, Watt RM, Jin LJ, Samaranayake LP. (2013). Pseudomonas aeruginosa lipopolysaccharide inhibits Candida albicans hyphae formation and alters gene expression during biofilm development. Mol Oral Microbiol 28: 54–69. [DOI] [PubMed] [Google Scholar]
- Banerjee M, Uppuluri P, Zhao XR, Carlisle PL, Vipulanandan G, Villar CC et al. (2013). Expression of UME6, a key regulator of Candida albicans hyphal development, enhances biofilm formation via Hgc1- and Sun41-dependent mechanisms. Eukaryot Cell 12: 224–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg KH, Biornstad TJ, Johnsborg O, Havarstein LS. (2012). Properties and biological role of Streptococcal fratricins. Appl Environ Microbiol 78: 3515–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg KH, Biornstad TJ, Straume D, Havarstein LS. (2011). Peptide-regulated gene depletion system developed for use in Streptococcus Pneumoniae. J Bacteriol 193: 5207–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen WH, Koo H. (2011). Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res 45: 69–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burne RA, Zeng L, Ahn SJ, Palmer SR, Liu Y, Lefebure T et al. (2012). Progress dissecting the oral microbiome in caries and health. Adv Dent Res 24: 77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar-Cruz M, Lopez-Romero E, Villagomez-Castro JC, Ruiz-Baca E. (2012). Candida species: new insights into biofilm formation. Future Microbiol 7: 755–771. [DOI] [PubMed] [Google Scholar]
- Cugini C, Morales DK, Hogan DA. (2010). Candida albicans-produced farnesol stimulates Pseudomonas quinolone signal production in LasR-defective Pseudomonas aeruginosa strains. Microbiology 156(Pt 10): 3096–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho FG, Silva DS, Hebling J, Spolidorio LC, Spolidorio DM. (2006). Presence of mutans streptococci and Candida spp. in dental plaque/dentine of carious teeth and early childhood caries. Arch Oral Biol 51: 1024–1028. [DOI] [PubMed] [Google Scholar]
- Desai K, Mashburn-Warren L, Federle MJ, Morrison DA. (2012). Development of competence for genetic transformation of Streptococcus mutans in a chemically defined medium. J Bacteriol 194: 3774–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PI, Xie Z, Sobue T, Thompson A, Biyikoglu B, Ricker A et al. (2012). Synergistic interaction between Candida albicans and commensal oral Streptococci in a novel in vitro mucosal model. Infect Immun 80: 620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour D, Levesque CM. (2013). Cell death of Streptococcus mutans induced by a quorum-sensing peptide occurs via a conserved streptococcal autolysin. J Bacteriol 195: 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federle MJ, Morrison DA. (2012). One if by land, two if by sea: signalling to the ranks with CSP and XIP. Mol Microbiol 86: 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel JS, Mitchell AP. (2011). Genetic control of Candida albicans biofilm development. Nat Rev Microbiol 9: 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine L, Goffin P, Dubout H, Delplace B, Baulard A, Lecat-Guillet N et al. (2013). Mechanism of competence activation by the ComRS signalling system in streptococci. Mol Microbiol 87: 1113–1132. [DOI] [PubMed] [Google Scholar]
- Gow NA, van de Veerdonk FL, Brown AJ, Netea MG. (2012). Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat Rev Microbiol 10: 112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire S, Xiao J, Silva BB, Gonzalez I, Agidi PS, Klein MI et al. (2011). Role of glucosyltransferase B in interactions of Candida albicans with Streptococcus mutans and with an experimental pellicle on hydroxyapatite surfaces. Appl Environ Microbiol 77: 6357–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan S, Ready D, Jasni AS, Rogers M, Pratten J, Roberts AP. (2010). Transfer of antibiotic resistance by transformation with EDNA within oral biofilms. FEMS Immunol Med Microbiol 59: 345–349. [DOI] [PubMed] [Google Scholar]
- Harriott MM, Noverr MC. (2011). Importance of candida-bacterial polymicrobial biofilms in disease. Trends Microbiol 19: 557–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DA, Vik A, Kolter R. (2004). A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol 54: 1212–1223. [DOI] [PubMed] [Google Scholar]
- Holcombe LJ, McAlester G, Munro CA, Enjalbert B, Brown AJ, Gow NA et al. (2010). Pseudomonas aeruginosa secreted factors impair biofilm development in Candida albicans. Microbiology 156(Pt 5): 1476–1486. [DOI] [PubMed] [Google Scholar]
- Hung DC, Downey JS, Ayala EA, Kreth J, Mair R, Senadheera DB et al. (2011). Characterization of DNA binding sites of the ComE response regulator from Streptococcus mutans. J Bacteriol 193: 3642–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz LM, Deng DM, van der Mei HC, Crielaard W, Krom BP. (2009). Streptococcus mutans competence-stimulating peptide inhibits Candida albicans hypha formation. Eukaryot Cell 8: 1658–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson HF, Lala HC, Shepherd MG. (1990). Coaggregation of Streptococcus sanguis and other streptococci with Candida albicans. Infect Immun 58: 1429–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan R, Rukke HV, Ricomini Filho AP, Fimland G, Arntzen MO, Thiede B et al. (2012). Extracellular identification of a processed type II ComR/ComS pheromone of Streptococcus mutans. J Bacteriol 194: 3781–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sudbery P. (2011). Candida albicans, a major human fungal pathogen. J Microbiol 49: 171–177. [DOI] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Cao S, Chai L, Bottcher T, Kolter R, Clardy J et al. (2012). A self-produced trigger for biofilm disassembly that targets exopolysaccharide. Cell 149: 684–692. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Koo H, Falsetta ML, Klein MI. (2013). The exopolysaccharide matrix: a virulence determinant of cariogenic biofilm. J Dent Res 92: 1065–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Hayacibara MF, Schobel BD, Cury JA, Rosalen PL, Park YK et al. (2003). Inhibition of Streptococcus mutans biofilm accumulation and polysaccharide production by apigenin and Tt-farnesol. J Antimicrob Chemother 52: 782–789. [DOI] [PubMed] [Google Scholar]
- Koo H, Jeon JG. (2009). Naturally occurring molecules as alternative therapeutic agents against cariogenic biofilms. Adv Dent Res 21: 63–68. [DOI] [PubMed] [Google Scholar]
- Kraneveld EA, Buijs MJ, Bonder MJ, Visser M, Keijser BJ, Crielaard W et al. (2012). The relation between oral candida load and bacterial microbiome profiles in dutch older adults. PLoS One 7: e42770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L, Futschik E. (2007). Mfuzz: a software package for soft clustering of microarray data. Bioinformation 2: 5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsch VK, Young DA. (2011). New directions in the etiology of dental caries disease. J Calif Dent Assoc 39: 716–721. [PubMed] [Google Scholar]
- Lemme A, Gröbe L, Reck M, Tomasch J, Wagner-Döbler I. (2011). Subpopulation-specific transcriptome analysis of competence-stimulating-peptide-induced Streptococcus mutans. J Bacteriol 193: 1863–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. (2001). Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol 183: 897–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay AK, Deveau A, Piispanen AE, Hogan DA. (2012). Farnesol and cyclic AMP signaling effects on the hypha-to-yeast transition in Candida albicans. Eukaryot Cell 11: 1219–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant S, Brailsford SR, Twomey AC, Roberts GJ, Beighton D. (2001). The predominant microflora of nursing caries lesions. Caries Res 35: 397–406. [DOI] [PubMed] [Google Scholar]
- Mashburn-Warren L, Morrison DA, Federle MJ. (2010). A novel double-tryptophan peptide pheromone controls competence in Streptococcus Spp. via an Rgg regulator. Mol Microbiol 78: 589–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlester G, O'Gara F, Morrissey JP. (2008). Signal-mediated interactions between Pseudomonas aeruginosa and Candida albicans. J Med Microbiol 57(Pt 5): 563–569. [DOI] [PubMed] [Google Scholar]
- Merritt J, Qi F. (2012). The mutacins of Streptococcus mutans: regulation and ecology. Mol Oral Microbiol 27: 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metwalli KH, Khan SA, Krom BP, Jabra-Rizk MA. (2013). Streptococcus mutans, Candida albicans, and the human mouth: a sticky situation. PLoS Pathog 9: e1003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales DK, Grahl N, Okegbe C, Dietrich LE, Jacobs NJ, Hogan DA. (2013). Control of Candida albicans metabolism and biofilm formation by Pseudomonas aeruginosa phenazines. MBio 4: e00526–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran MA, Satinsky B, Gifford SM, Luo H, Rivers A, Chan LK et al. (2013). Sizing up metatranscriptomics. ISME J 7: 237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Vickerman MM, Jenkinson HF. (2010). Heterologous expression of Candida albicans cell wall-associated adhesins in Saccharomyces cerevisiae reveals differential specificities in adherence and biofilm formation and in binding oral Streptococcus gordonii. Eukaryot Cell 9: 1622–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okinaga T, Xie Z, Niu G, Qi F, Merritt J. (2010). Examination of the HdrRM regulon yields insight into the competence system of Streptococcus mutans. Mol Oral Microbiol 25: 165–177. [DOI] [PubMed] [Google Scholar]
- Peleg AY, Hogan DA, Mylonakis E. (2010). Medically important bacterial-fungal interactions. Nat Rev Microbiol 8: 340–349. [DOI] [PubMed] [Google Scholar]
- Pereira-Cenci T, Deng DM, Kraneveld EA, Manders EM, Del Bel Cury AA, Ten Cate JM et al. (2008). The effect of Streptococcus mutans and Candida glabrata on Candida albicans biofilms formed on different surfaces. Arch Oral Biol 53: 755–764. [DOI] [PubMed] [Google Scholar]
- Perry JA, Jones MB, Peterson SN, Cvitkovitch DG, Levesque CM. (2009). Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol Microbiol 72: 905–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja M, Hannan A, Ali K. (2010). Association of oral candidal carriage with dental caries in children. Caries Res 44: 272–276. [DOI] [PubMed] [Google Scholar]
- Rammohan SN, Juvvadi SR, Gandikota CS, Challa P, Manne R, Mathur A. (2012). Adherence of Streptococcus mutans and Candida albicans to different bracket materials. J Pharm Bioallied Sci 4(Suppl 2): S212–S216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro DG, Pavarina AC, Dovigo LN, Machado AL, Giampaolo ET, Vergani CE. (2012). Prevalence of Candida spp. associated with bacteria species on complete dentures. Gerodontology 29: 203–208. [DOI] [PubMed] [Google Scholar]
- Scannapieco FA. (1994). Saliva-bacterium interactions in oral microbial ecology. Crit Rev Oral Biol Med 5: 203–248. [DOI] [PubMed] [Google Scholar]
- Senadheera DB, Cordova M, Ayala EA, Chavez de Paz LE, Singh K, Downey JS et al. (2012). Regulation of bacteriocin production and cell death by the VicRK signaling system in Streptococcus mutans. J Bacteriol 194: 1307–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh M, Tam A, Steinberg D. (2007). Expression of biofilm-associated genes of Streptococcus mutans in response to glucose and sucrose. J Med Microbiol 56(Pt 11): 1528–1535. [DOI] [PubMed] [Google Scholar]
- Silverman RJ, Nobbs AH, Vickerman MM, Barbour ME, Jenkinson HF. (2010). Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun 78: 4644–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son M, Ahn SJ, Guo Q, Burne RA, Hagen SJ. (2012). Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of ComX. Mol Microbiol 86: 258–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorgo AG, Heilmann CJ, Brul S, de Koster CG, Klis FM. (2013). Beyond the wall: Candida albicans secret(e)s to survive. FEMS Microbiol Lett 338: 10–17. [DOI] [PubMed] [Google Scholar]
- Sorgo AG, Heilmann CJ, Dekker HL, Brul S, de Koster CG, Klis FM. (2010). Mass spectrometric analysis of the secretome of Candida albicans. Yeast 27: 661–672. [DOI] [PubMed] [Google Scholar]
- Sudbery PE. (2011). Growth of Candida albicans hyphae. Nat Rev Microbiol 9: 737–748. [DOI] [PubMed] [Google Scholar]
- Sztajer H, Lemme A, Vilchez R, Schulz S, Geffers R, Yip CY et al. (2008). Autoinducer-2-regulated genes in Streptococcus mutans UA159 and global metabolic effect of the LuxS mutation. J Bacteriol 190: 401–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Nyvad B. (2011). The role of bacteria in the caries process: ecological perspectives. J Dent Res 90: 294–303. [DOI] [PubMed] [Google Scholar]
- Trautner K, Felgenhauer B, Rieder H. (1981). Extracellular polysaccharide synthesized by the oral bacterium Streptococcus mutans of serotype a to e in vitro. Arch Oral Biol 26: 1005–1013. [DOI] [PubMed] [Google Scholar]
- Urzua B, Hermosilla G, Gamonal J, Morales-Bozo I, Canals M, Barahona S et al. (2008). Yeast diversity in the oral microbiota of subjects with periodontitis: Candida albicans and Candida dubliniensis colonize the periodontal pockets. Med Mycol 46: 783–793. [DOI] [PubMed] [Google Scholar]
- Vilchez R, Lemme A, Ballhausen B, Thiel V, Schulz S, Jansen R et al. (2010). Streptococcus mutans inhibits Candida albicans hyphal formation by the fatty acid signaling molecule trans-2-decenoic acid (SDSF). Chembiochem 11: 1552–1562. [DOI] [PubMed] [Google Scholar]
- Wei H, Havarstein LS. (2012). Fratricide is essential for efficient gene transfer between pneumococci in biofilms. Appl Environ Microbiol 78: 5897–5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L, Sissons C. (2001). A comparison of human dental plaque microcosm biofilms grown in an undefined medium and a chemically defined artificial saliva. Arch Oral Biol 46: 477–486. [DOI] [PubMed] [Google Scholar]
- Xue X, Tomasch J, Sztajer H, Wagner-Döbler I. (2010). The delta subunit of RNA polymerase, RpoE, is a global modulator of Streptococcus mutans environmental adaptation. J Bacteriol 192: 5081–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.