Abstract
Papillon–Lefevre syndrome (PALS) is a rare, autosomal recessive disorder characterized by periodontitis and hyperkeratosis over the palms and soles. Mutations in the cathepsin C gene (CTSC) have been recognized as the cause of PALS since the late 1990s. More than 75 mutations in CTSC have been identified, and phenotypic variability between different mutations has been described. Next generation sequencing is widely used for efficient molecular diagnostics in various clinical practices. Here we investigated a large consanguineous Saudi family with four affected and four unaffected individuals. All of the affected individuals suffered from hyperkeratosis over the palms and soles and had anomalies of both primary and secondary dentition. For molecular diagnostics, we combined whole-exome sequencing and genome-wide homozygosity mapping procedures, and identified a recurrent homozygous missense mutation (c.899G>A; p.Gly300Asp) in exon 7 of CTSC. Validation of all eight family members by Sanger sequencing confirmed co-segregation of the pathogenic variant (c.899G>A) with the disease phenotype. This is the first report of whole-exome sequencing performed for molecular diagnosis of PALS in Saudi Arabia. Our findings provide further insights into the genotype–phenotype correlation of CTSC pathogenicity in PALS.
Keywords: Molecular diagnostics, Papillon–Lefevre syndrome, Whole-exome sequencing, Homozygosity mapping, CTSC gene, Saudi Arabia
1. Introduction
Papillon–Lefevre syndrome (PALS, MIM 245000) is a rare, autosomal recessive genodermatosis. The disease was first described by two French scientists, Papillon and Lefevre, in 1924 (Papillon and Lefevre, 1924). It mainly affects skin and teeth, leading to hyperkeratoderma over the palms and soles, also known as palmoplantar hyperkeratosis, and premature loss of primary or secondary dentition (Haneke, 1979, Papillon and Lefevre, 1924). Hyperkeratosis first appears in the early stages of childhood (prior to age 3–4); however, late-onset alterations have also been reported (Pilger et al., 2003). In general, hyperkeratotic features are not severe in PALS, and as reviewed recently, the diffuse type is more common than the punctuate type in most cases (Nagy et al., 2014). Lesions over the elbow, knee, and knuckles resembling psoriasis may also develop in some cases (Toomes et al., 1999). Recurrent mild pus-generating skin infections with self-healing are also observed (Gorlin et al., 1964, Haneke, 1979).
Periodontitis and gingivitis are associated with both primary and secondary dental anomalies, and appear at the time of the two episodes of tooth eruptions: one at ∼3 years of age and the second at ∼15 years old (Fardal et al., 1998, Lundgren and Renvert, 2004, Toomes et al., 1999). Loss-of-function mutations in the cathepsin C gene (CTSC, MIM 602365), encoding a lysosomal exo-cysteine proteinase, at 11q14.2 have been assigned to PALS (Hart et al., 1999, Toomes et al., 1999). To date, more than 75 pathogenic variants from various ethnic groups have been identified as causing PALS and overlapping phenotypes (Nagy et al., 2014). Haim–Munk syndrome (HMS, MIM 245010) is characterized by palmoplantar keratoderma, periodontitis, arachnodactyly, acroosteolysis, pesplanus, and onychogryposis, and is caused by recessive mutations in CTSC (Hart et al., 2000b). Similarly, aggressive periodontitis, which is characterized by severe periodontal inflammation, leading to tooth loss, without the involvement of skin abnormalities, is another overlapping phenotype of PALS and is also caused by mutations in CTSC (Hart et al., 2000c, Hewitt et al., 2004).
Here we present clinical and molecular analysis of a consanguineous five-generation family from Saudi Arabia segregating an autosomal recessive PALS phenotype.
2. Materials and methods
2.1. Ethical approval
Ethical approval for this study was obtained from the King Abdulaziz University (KAU), Jeddah, Saudi Arabia (ref # 24-14), according to the Helsinki Declaration. The parents of the affected children signed informed written consent for their willingness to participate and to publish the results for academic research purposes.
2.2. Study subjects
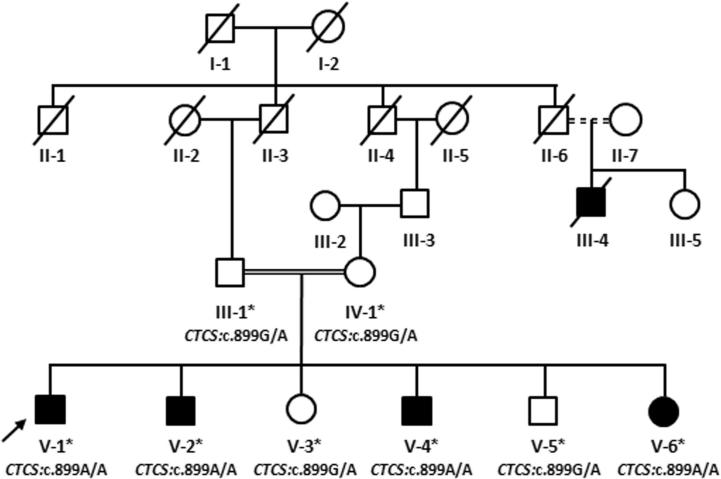
The five-generation family (Fig. 1) resided in a remote southwestern region of Saudi Arabia. Family information for the pedigree was obtained by interviewing the parents of the affected children. All family members, including the four affected siblings, were thoroughly examined in the Department of Oral and Maxillofacial Prosthodontics, Faculty of Dentistry, KAU, Jeddah Saudi Arabia. Venous blood samples from four affected (V-1, V-2, V-4, V-6) and four unaffected (III-1, IV-1, V-3, V-5) family members were collected in EDTA tubes, and genomic DNA was extracted and quantified using standard methods (Ahmed et al., 2015).
Figure 1.
Pedigree analysis of a five-generation consanguineous family presenting with autosomal recessive inheritance of Papillon–Lefevre syndrome.
2.3. Homozygosity mapping
Genomic DNA of three affected (V-1, V-2, V-6) and one unaffected (V-3) individual was subjected to 300 K HumanCytoSNPs12 microarray analysis using an iScan platform (Illumina, USA) following the manufacturer’s protocols. The common regions of homozygosity were identified using GenomeStudio Genotyping Module v1.0 (Illumina).
2.4. Whole-exome sequencing
Two micrograms of genomic DNA from the index patient (V-1) were used for human whole-exome analysis with paired-end-sequencing at 100× resolution. Libraries were constructed using a 51-Mb SureSelect library kit (Agilent Technologies, USA). The target regions with average throughput depths of more than 120 and100-bp paired-ends reads were sequenced using a HiSeq2000 platform (Illumina). BWA (http://bio-bwa.sourceforge.net/) and SAMTOOLS (http://samtools.sourceforge.net/) were used for alignment of sequences and copy number variants, and small indel detection, respectively. The obtained reads were mapped to the UCSC human genome database hg19 (http://genome.ucsc.edu/), and were compared with 1000 genomes (http://www.1000genomes.org/data) and dbSNP (http://www.ncbi.nlm.nih.gov/snp/) databases. Pathogenicity of the obtained variants was predicted using LRT (http://www.genetics.wustl.edu/jflab/lrt_query.html), Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/), SIFT (http://sift.jcvi.org/), MutationTaster (http://www.mutationtaster.org/), and PROVEAN (http://provean.jcvi.org/genome_submit_2).
2.5. Sanger sequencing
The potential candidate variant was validated by Sanger sequencing in all eight family members. The CTSC reference sequence (ENSG00000109861) was obtained from the Ensembl genome browser (http://www.ensembl.org/Homo_sapiens/). Primer3Plus (http://www.bioinformatics.nl/) was used to design the upstream (5′-TCAGGGGTAACATGCAAAGA-3′) and downstream (5′-TTTGCATGGAGAATCAGTGC-3′) primers for PCR amplification of the c.899G>A region from genomic DNA of each subject. PCR products were sequenced using a Big Dye Terminator v3.1 Cycle Sequencing Kit and an ABI 3500 Genetic Analyzer (Life Technologies, USA). Sequence variants were identified using BioEdit sequence alignment editor version 6.0.7 (www.mbio.ncsu.edu/bioedit.html).
3. Results
3.1. Clinical features
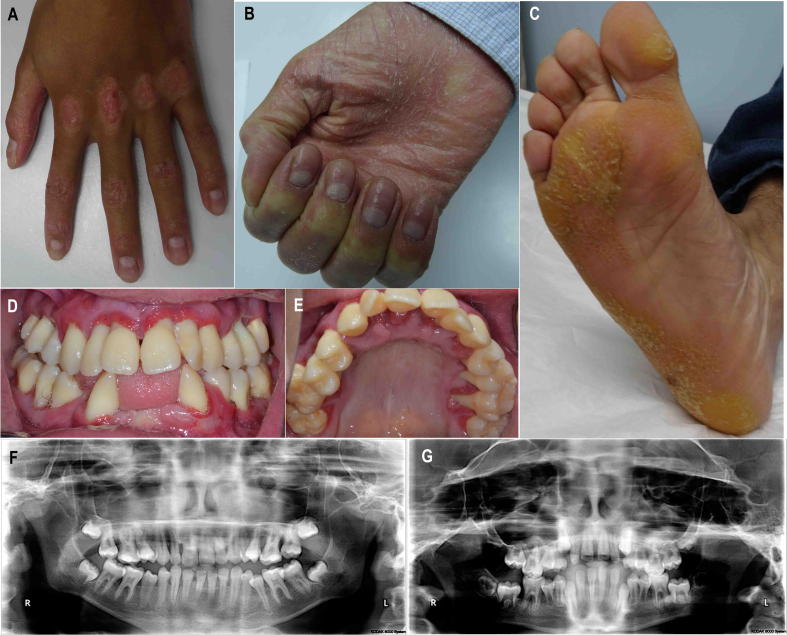
All four affected individuals presented with classical PALS symptoms, including psoriasiform lesions over knuckles, hyphidrosis, palmoplantar hyperkeratosis, and periodontal inflammation (Fig. 2). Detailed dental examinations revealed mild differences among the affected siblings. The index patient (V-1), a 17-year-old boy, had extensive loss of alveolar bone in the lower jaw, leading to loss of the lower anterior teeth. The patient also had generalized severe periodontitis that affected the secondary dentition, leading to multiple instances of furcation involvement and tooth mobility. Likewise, the periodontal health of patient V-2, a 15-year-old boy, was affected, with several instances of grade III furcation involvement and tooth mobility. The third affected individual (V-4), an 11-year-old boy, had generalized mild to moderate bone loss that included furcational involvement of the first molars, and again contributed to the loss of multiple teeth. The youngest affected patient (V-6), a 6-year-old girl, had generalized mild bone loss, spacing, and crowding in the lower anterior teeth. She had also mobility in the primary dentition. The hair and nails appeared normal in all affected individuals, and all were otherwise healthy without involvement of any of the vital organs. The unaffected siblings (V-3, V-5) had normal dentition with no phenotypic indications of PALS, and were clinically indistinguishable from healthy individuals.
Figure 2.
Clinical presentation of the affected individuals. The index patient (V-1) showed psoriasiform lesions over the knuckles (A), hyphidrosis and hyperkeratosis over the palm (B) and sole (C), and periodontal inflammation (D and E). Radiological examination of patients V-1 and V-4 showing extensive loss of the alveolar bone in the lower jaw leading to loss of the lower anterior teeth.
3.2. Genetic analysis
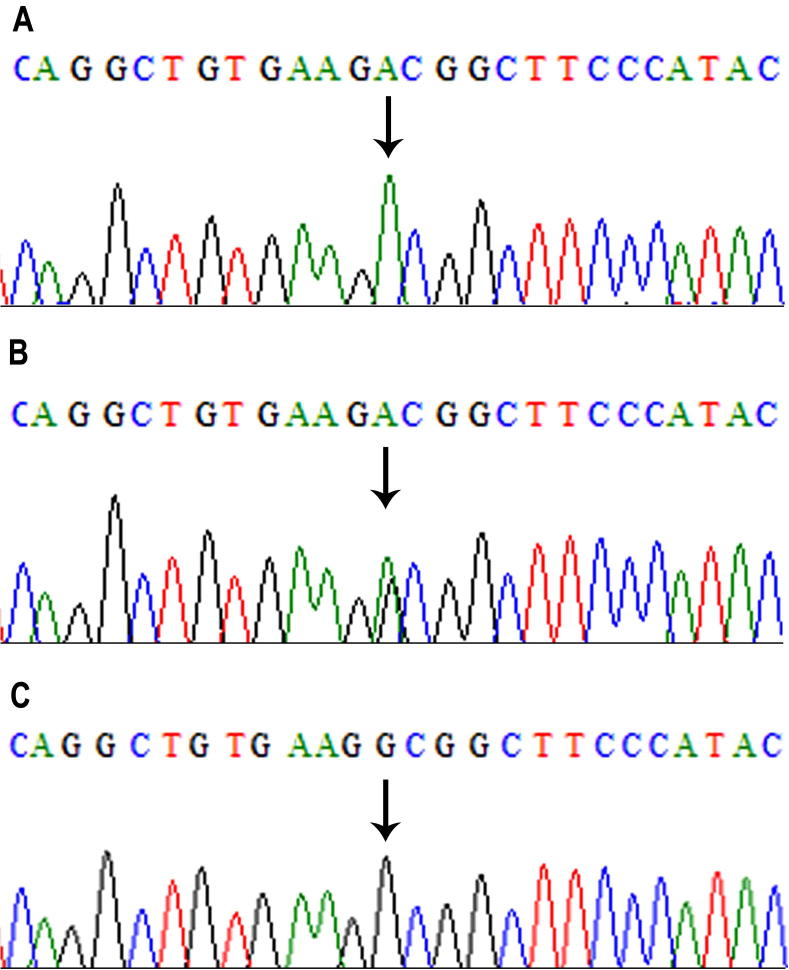
We combined the results of microarray SNP genotyping with the whole-exome sequencing data. We found that the three affected individuals (V-1, V-2, and V-6) shared a common region of homozygosity, at chromosome 11q13.2–11q23.2 (chr11: 69,953,729–112,031,624 bp); however, it was not shared by the unaffected individual (V-3). This region corresponded to a 42-Mb region on human genome Map-Viewer (annotation release 106; http://www.ncbi.nlm.nih.gov/mapview/), which contained 518 labeled genes and 670 variants. As the affected individuals were the products of a consanguineous union, the possibility of homozygous mutation was more likely. We identified 55 variants for further investigation after screening all 670 variants using the criteria: only homozygous single-nucleotide variations (SNV) with altered nucleotide depth of more than 24, present in the exonic or splice-site regions, and having non-synonymous, frame-shift, or stop-gain effects. We further narrowed the list to seven variants by removing minor allele frequency greater than 0.05. Of these, three SNVs (XRRA1: NM_182969, c.622G>A, p.Val208Ile; CTSC: NM_001814, c.899G>A, p.Gly300Asp; and ELMOD1: NM_001130037, c.952G>A, p.Ala318Thr) were not reported in the 1000 genome database (Oct. 2011 release) (Supplementary Table 1). Prediction software analyses and Sanger sequencing validation of all family members confirmed the mutation within CTSC as a pathogenic variant. The mutation was a single nucleotide transition from guanine to adenine in exon 7 of CTSC at cDNA position 899, leading to a single amino acid substitution from glycine to aspartate at amino acid position 300. The obligate carriers were heterozygous (Fig. 3). This variant was not detected in 212 ethnically matched control chromosomes, and the possibility of neutral polymorphism was excluded. A second homozygous missense variant (c.458T>C, p.Ile153Thr) in exon 3 of CTSC was also identified; however, it corresponded to a single nucleotide polymorphism (rs217086) with a minor allele frequency of >90% in the 1000 genome database, and was therefore not likely to be pathogenic.
Figure 3.
Mutation analysis of CTSC (c.899G>A). Electropherogram of a single base-pair G>A substitution at nucleotide 899 in an affected patient (A), a carrier parent (B), and a wild-type unaffected individual (C).
4. Discussion
With the recent advances in next-generation sequencing technologies, whole-exome analysis has significantly improved pathogenic variant identification, especially in hereditary skin disorders with inter- and intra-familial phenotypic variability (Lai-Cheong and McGrath, 2011, Salam et al., 2014). In the current study, we combined genome-wide homozygosity mapping with whole-exome analysis for a successful and efficient molecular diagnosis.
The PALS disease locus was first mapped to 11q14 in 1997, and 2 years later, the causative mutations in CTSC were identified (Fischer et al., 1997, Hart et al., 1999, Toomes et al., 1999). Interestingly, CTSC involvement has been ruled out in several PALS patients through traditional DNA sequencing, despite establishing linkage to 11q14 (Hart et al., 2000a, Khan et al., 2014). More recently, the mutation spectrum of PALS, HMS, and AP1 phenotypes has been widely studied; however, a clear genotype–phenotype correlation could not be established. In an attempt to summarize the genotype–phenotype correlation, a recent review outlined all the known mutations in CTSC (Nagy et al., 2014). Only one is listed for the HMS phenotype, while seven cause more than one phenotype, including PALS (Nagy et al., 2014). Several polymorphisms were associated with the PALS phenotype; however, in our family, one of these polymorphisms (rs217086) was also detected in unaffected healthy individuals.
Two missense mutations, including c.899G>A in an affected sibling from one family and c.815G>C in affected siblings from four unrelated families, have previously been associated with the PALS phenotype in patients from Saudi Arabia (Zhang et al., 2001). The mutation c.815G>C was identified as possible evidence of the founder effect being present in the four families, whose distant relationships were not known (Zhang et al., 2001). Here we report further evidence of the founder effect in CTSC from the same population. As the clinical features of the c.899G>A mutation have not previously been clearly described, the current study included a detailed clinical description of our patients who carried the same mutation as previously reported (Zhang et al., 2001). Our findings expand the knowledge on CTSC pathogenicity in PALS, and will provide a basis for genotype–phenotype correlations in this rare disorder.
5. Conclusions
This study describes the complete clinical and molecular assessment of PALS in a consanguineous family of Saudi origin. Next generation sequencing has been widely adopted for efficient molecular diagnosis because of its precision and cost effectiveness. The Saudi population is unique, and to date there is no publically available reference genome database for this population. Whole-exome sequencing may provide better data for constructing reference databases in Saudi Arabia, and may replace traditional methods of variant detection.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Acknowledgements
This project was funded by the Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah, under Grant No. (4/165/1435/HiCi). The authors, therefore, acknowledge with thanks DSR for technical and financial support.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.sjbs.2015.06.007.
Appendix A. Supplementary data
This file contains supplementary table.
References
- Ahmed S., Jelani M., Alrayes N., Mohamoud H.S., Almramhi M.M., Anshasi W., Ahmed N.A., Wang J., Nasir J., Al-Aama J.Y. Exome analysis identified a novel missense mutation in the CLPP gene in a consanguineous Saudi family expanding the clinical spectrum of Perrault Syndrome type-3. J. Neurol. Sci. 2015;353:149–154. doi: 10.1016/j.jns.2015.04.038. [DOI] [PubMed] [Google Scholar]
- Fardal O., Drangsholt E., Olsen I. Palmar plantar keratosis and unusual periodontal findings. Observations from a family of 4 members. J. Clin. Periodontol. 1998;25:181–184. doi: 10.1111/j.1600-051x.1998.tb02425.x. [DOI] [PubMed] [Google Scholar]
- Fischer J., Blanchet-Bardon C., Prud’homme J.F., Pavek S., Steijlen P.M., Dubertret L., Weissenbach J. Mapping of Papillon–Lefevre syndrome to the chromosome 11q14 region. Eur. J. Hum. Genet. 1997;5:156–160. [PubMed] [Google Scholar]
- Gorlin R.J., Sedano H., Anderson V.E. The syndrome of palmar-plantar hyperkeratosis and premature periodontal destruction of the teeth. A clinical and genetic analysis of the Papillon-Lefevre syndrome. J. Pediatr. 1964;65:895–908. doi: 10.1016/s0022-3476(64)80014-7. [DOI] [PubMed] [Google Scholar]
- Haneke E. The Papillon–Lefevre syndrome: keratosis palmoplantaris with periodontopathy. Report of a case and review of the cases in the literature. Hum. Genet. 1979;51:1–35. doi: 10.1007/BF00278288. [DOI] [PubMed] [Google Scholar]
- Hart T.C., Hart P.S., Bowden D.W., Michalec M.D., Callison S.A., Walker S.J., Zhang Y., Firatli E. Mutations of the cathepsin C gene are responsible for Papillon–Lefevre syndrome. J. Med. Genet. 1999;36:881–887. [PMC free article] [PubMed] [Google Scholar]
- Hart P.S., Zhang Y., Firatli E., Uygur C., Lotfazar M., Michalec M.D., Marks J.J., Lu X., Coates B.J., Seow W.K., Marshall R., Williams D. Identification of cathepsin C mutations in ethnically diverse Papillon–Lefevre syndrome patients. J. Med. Genet. 2000;37:927–932. doi: 10.1136/jmg.37.12.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T.C., Hart P.S., Michalec M.D., Zhang Y., Firatli E., Van Dyke T.E., Stabholz A., Zlotogorski A., Shapira L., Soskolne W.A. Haim–Munk syndrome and Papillon–Lefevre syndrome are allelic mutations in cathepsin C. J. Med. Genet. 2000;37:88–94. doi: 10.1136/jmg.37.2.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T.C., Hart P.S., Michalec M.D., Zhang Y., Marazita M.L., Cooper M., Yassin O.M., Nusier M., Walker S. Localisation of a gene for prepubertal periodontitis to chromosome 11q14 and identification of a cathepsin C gene mutation. J. Med. Genet. 2000;37:95–101. doi: 10.1136/jmg.37.2.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt C., McCormick D., Linden G., Turk D., Stern I., Wallace I., Southern L., Zhang L., Howard R., Bullon P., Wong M., Widmer R. The role of cathepsin C in Papillon–Lefevre syndrome, prepubertal periodontitis, and aggressive periodontitis. Hum. Mutat. 2004;23:222–228. doi: 10.1002/humu.10314. [DOI] [PubMed] [Google Scholar]
- Khan F.Y., Jan S.M., Mushtaq M. Papillon–Lefevre syndrome (PLS) without cathepsin C mutation: a rare early onset partially penetrant variant of PLS. Saudi Dent. J. 2014;26:25–28. doi: 10.1016/j.sdentj.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai-Cheong J.E., McGrath J.A. Next-generation diagnostics for inherited skin disorders. J. Invest. Dermatol. 2011;131:1971–1973. doi: 10.1038/jid.2011.253. [DOI] [PubMed] [Google Scholar]
- Lundgren T., Renvert S. Periodontal treatment of patients with Papillon–Lefevre syndrome: a 3-year follow-up. J. Clin. Periodontol. 2004;31:933–938. doi: 10.1111/j.1600-051X.2004.00591.x. [DOI] [PubMed] [Google Scholar]
- Nagy N., Valyi P., Csoma Z., Sulak A., Tripolszki K., Farkas K., Paschali E., Papp F., Toth L., Fabos B., Kemeny L., Nagy K. CTSC and Papillon–Lefevre syndrome: detection of recurrent mutations in Hungarian patients, a review of published variants and database update. Mol. Genet. Genomic Med. 2014;2:217–228. doi: 10.1002/mgg3.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papillon P.H., Lefevre P. Two cases of symmetrically familial palmar and plantar hyperkeratosis (Meleda disease) within brother and sister combined with severe dental alterations in both cases. Bull. Soc. Fr. Dermatol. Syphiligr. 1924:82–87. [Google Scholar]
- Pilger U., Hennies H.C., Truschnegg A., Aberer E. Late-onset Papillon–Lefevre syndrome without alteration of the cathepsin C gene. J. Am. Acad. Dermatol. 2003;49:S240–3. doi: 10.1016/s0190-9622(03)01558-5. [DOI] [PubMed] [Google Scholar]
- Salam A., Simpson M.A., Stone K.L., Takeichi T., Nanda A., Akiyama M., McGrath J.A. Next generation diagnostics of heritable connective tissue disorders. Matrix Biol. 2014;33:35–40. doi: 10.1016/j.matbio.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Toomes C., James J., Wood A.J., Wu C.L., McCormick D., Lench N., Hewitt C., Moynihan L., Roberts E., Woods C.G., Markham A., Wong M. Loss-of-function mutations in the cathepsin C gene result in periodontal disease and palmoplantar keratosis. Nat. Genet. 1999;23:421–424. doi: 10.1038/70525. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Lundgren T., Renvert S., Tatakis D.N., Firatli E., Uygur C., Hart P.S., Gorry M.C., Marks J.J., Hart T.C. Evidence of a founder effect for four cathepsin C gene mutations in Papillon–Lefevre syndrome patients. J. Med. Genet. 2001;38:96–101. doi: 10.1136/jmg.38.2.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains supplementary table.