Abstract
A new species of Venturia (V. chinensis) is described and illustrated from the leaves of Lonicera praeflorens collected from Lesser Khingan Mountains, the northeast China. It is characterized by habitat saprobic; ascomata small-sized, solitary or scattered, superficial, subglobose to citriform, wall black, papillate, ostiolate, covered with setae; peridium thin; hamathecium evanescent in mature ascomata; asci 8-spored, bitunicate, fissitunicate, oblong to obclavate, with or without a short, knob-like pedicel; ascospores ellipsoidal, olivaceous pale brown, 1-septate, ascospore wall thin, smooth. Comparisons of V. chinensis with V. lonicerae (another species on Lonicera caerulea) and other species of Venturia lead to the conclusion that collected taxon is new. Its relationships with other species of Venturia are discussed based on morphology and 28S nrDNA and ITS nrDNA sequence comparisons.
Keywords: China, DNA sequences, Foliicolous fungi, Morphology, Taxonomy
1. Introduction
Species of Venturia De Not. are widely distributed in north temperate area of the world, which are saprobic or parasitic on a large variety of plants. Some species of Venturia are notorious plant pathogens, such as the apple scab caused by V. inaequalis (Cooke) G. Winter and pear scab by V. pyrina Aderh. (Barr, 1968, Sivanesan, 1977). Venturia was first described by De Notaris (1844) to accommodate V. rosea De Not. and V. dianthi De Not. with no type designated. Subsequently, Cesati and De Notaris (1863) described another two species, i.e. V. dickiei (Berk. & Broome) Ces. & De Not. and V. eres (Berk. & Broome) Ces. & De Not. Saccardo (1882) emended the description of Venturia, excluded both V. rosea and V. dianthi, while accepted V. dickiei and V. eres. Venturia Sacc. was widely accepted, and was neotypified by V. inaequalis (Korf, 1956, Sivanesan, 1977). The diagnostic characteristics of Venturia Sacc. include habitat parasitic or saprobic on dicotyledonous leaves; ascomata small-sized, solitary, scattered, or gregarious, initially immersed, becoming erumpent, globose, subglobose, wall black, papillate, ostiolate; peridium thin, composed of a few layers of pigmented cells of textura angularis; hamathecium rare, evanescent in mature ascomata; asci 8-spored (rarely 4-spored), bitunicate, fissitunicate dehiscence unknown, oblong to obclavate, with a short, thick pedicel or pedicel lacking, with an inconspicuous ocular chamber; ascospores obliquely uniseriate and partially overlapping to biseriate, especially at the base, ellipsoidal, with broadly rounded ends, pale brown, 1-septate, slightly constricted at the septum, the upper cell shorter than the lower one, smooth-walled; having Fusicladium Bonord., Pollaccia E. Bald. & Cif. or Spilocaea Fr. ananmorphs (Zhang et al., 2011).
One hundred and ninety-three species names are listed in the Index Fungorum, which were estimated to comprise 57 species (Kirk et al., 2008). Early studies of Venturia in China were conducted by Zhu (1927), and in the general summary of Chinese fungi by Tai, 1941, Tai, 1979, five species were reported, i.e. V. geranii (Fr.) G. Winter, V. inaequalis, V. microseta Pat., V. pyrina and V. tremulae Aderh. Subsequently, Zhang (2003) revised the Fusicladium in China with 15 species of Fusicladium reported. In the summary of phytopathogens of woody plants in China conducted by Xu and He (2008), about 30 venturiaceous species were listed.
In the course of an ongoing survey of biodiversity of venturialean ascomycete in China initiated in 2014, a venturiaceous fungus was collected that appeared to fit Venturia s. s. well in morphological traits. This was supported by the comparisons of LSU and ITS nrDNA sequences of this new species with DNA sequences of other venturiaceous species deposited in GenBank. Based on the combination of subtle morphological and molecular differences a new taxon, Venturia chinensis is proposed.
2. Materials and methods
2.1. Morphological study
Leaf samples were collected from growing Lonicera praeflorens in August 2014, from Lesser Khingan Mountains in Heilongjiang province, Yichun, Wuyiling forestry station. Leaf samples were dried with absorbent paper in herbarium press, and studied directly under an Olympus SZ 61 dissecting microscope after preliminary incubation in a moist chamber. Microscopic observations of ascomatal contents were carried out from material mounted in water. Photomicrographs were taken on a Nikon Eclipse E600 Microscope fitted with a Nikon Digital Sight DS FI1 digital camera and processed with NIS-Elements F 3.2 software. Measurements of asci, hamathecial hyphae and ascospores were made from water mounts.
Isolations were made from single ascospores and grown on 2% water agar (WA) (Biolab, S.A.), and subsequently transferred to malt extract agar (MEA) with sterilized pine needles. Isolates were placed at ambient temperatures (about 26–28 °C) in the dark condition for establishing colony characteristics. Fungal isolates and herbarium specimens have been deposited at Beijing Forestry University (BJFU) with duplicates in the Mycological Herbarium of the Institute of Microbiology, Chinese Academy of Sciences (HMAS).
2.2. DNA extraction, PCR, sequencing
Sequencing of portions of rDNA was attempted from DNA extracted from the mycelium from the surface of MEA plates with CTAB plant genome DNA fast extraction kit (Aidlab Biotechnologies Co., Ltd, Beijing, China). The LSU (large subunit, 28S) nrDNA region was amplified and sequenced with primers LROR and LR5. The ITS nrDNA amplifications and sequencing used primers ITS-1 and ITS-4. Comparisons to other nrDNA sequences were conducted with BLAST 2.2.24 queries (National Center for Biotechnology Information, National Institute of Health, Bethesda, Maryland). Representative sequences were deposited in GenBank.
2.3. Sequence alignment and phylogenetic analysis
Sequences generated were analysed with other sequences obtained from GenBank (Table 1). All of the other species included in the phylogeny were chosen based on the phylogeny of Zhang et al. (2011). A Multiple alignment was done in Mega 5 (Tamura et al., 2011) and analyses were performed in PAUP V. 4.0b10 (Swofford, 2002). Prior to phylogenetic analysis, ambiguous sequences at the start and the end were deleted and gaps manually adjusted to optimize alignment. ITS rDNA dataset was analysed in this study. Maximum likelihood (ML) and maximum parsimony (MP) were conducted using heuristic searches as implemented in PAUP, with the default options method. For the ML analysis, best-fit model of nucleotide evolution (GTR + I + G) was selected by Akaike information criterion (AIC) (Posada and Buckley, 2004) in MrModeltest 2.3. Bootstrap analysis with 1000 replicates was used to test the statistical support of the branches. With model parameters estimated from the data, a heuristic search with ten random taxon addition sequences and TBR branch swapping was performed. For MP analysis, clade stability was assessed in a bootstrap (BS) analysis with 1000 replicates, random sequence additions with maxtrees set to 5000 and other default parameters as implemented in PAUP. Trees were viewed in TREEVIEW. The nucleotide sequences reported in this paper were deposited in GenBank.
Table 1.
Species and sequences database accession numbers used in this study (newly generated sequences are indicated in bold).
| Species | Strain | GenBank accession number |
|
|---|---|---|---|
| 28S rDNA | ITS | ||
| V. chlorospora | CBS 466.61 | EU035453 | EU035453 |
| V. chinensis | CGMCC 3.17685 | KP689595 | KP689596 |
| V. helvetica | CBS 474.61 | EU035458 | EU035458 |
| V. lonicera | CBS 445. 54 | EU035461 | EU035461 |
| V. minuta | CBS 478.61 | EU035464 | EU035464 |
| V. polygoni-vivipari | CBS 114207 | EU035466 | EU035466 |
3. Results
Taxonomy
Venturia chinensis Y. Zhang ter & J.Q. Zhang, sp. nov. MycoBank: MB 811424 (Fig. 1, Fig. 2, Fig. 3).
Fig. 1.
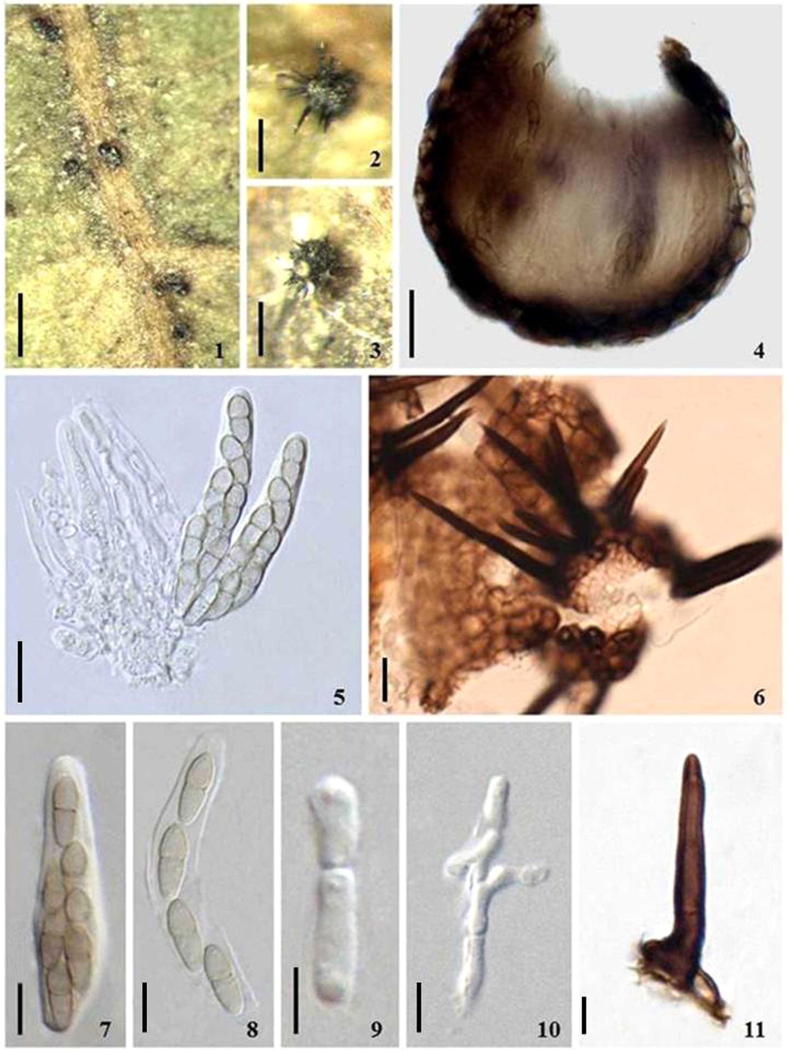
Venturia chinensis (holotype HMAS246485). (1–3) Ascomata on the host surface. Note the long and black setae. (4) Section of an ascoma. (5,7) Obclavate asci. (6) Crushed ascoma showing the thick walled setae. (8) Ascus releasing ascospores. (9–10) Decomposing pseudoparaphyses. (11) Seta. Scale bars: (1) = 300 μm; (2–3) = 100 μm; (4–5) = 20 μm; (6–8) = 10 μm; (9–11) = 5 μm.
Fig. 2.
Venturia chinensis (extype CGMCC 3.17685). (1) Colony growing on MEA after two weeks. (2–9) Chlamydospores and hyphae. Scale bars: (1) = 1 mm; (2), (4), (6), (9) = 10 μm; (3), (5), (7–8) = 20 μm.
Fig. 3.
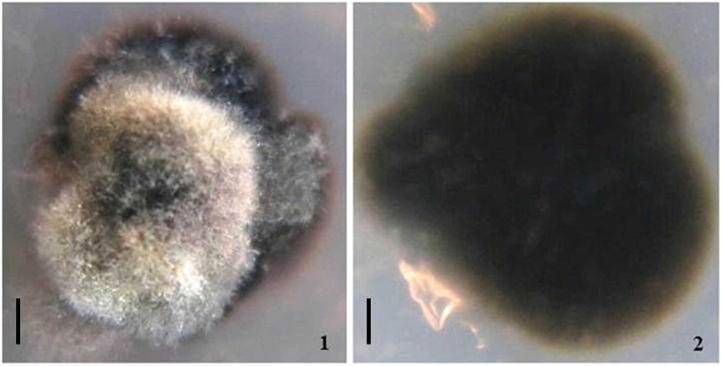
Venturia chinensis (extype CGMCC 3.17685). Upper (1) and reverse (2) view of colony on MEA 6 months after inoculation. bars: 1 mm.
Etymology: — The epithet “chinensis” refers to China, the country from which it is described.
Known distribution: — China.
Asexual state: Unknown.
Ascomata 40–100 μm diam., solitary, scattered, initially immersed or slightly erumpent, becoming superficial, globose or citriform with a small papilla, ostiolate, wall black, upper one third of the ascomata covered with setae (Fig. 1, Fig. 2, Fig. 3). Setae dark brown, 0–1 septate, 23–61 × 5–7 μm, setae wall 1–1.5 μm (Fig. 1: 6, 11). Peridium 1-layered, composed of (1-) 2–3 layers of pigmented cells of textura angularis, cells 9–5 μm diam., cell wall 0.8–1 μm thick. Hamathecium rare, evanescent in mature ascomata (Fig. 1: 9–10). Asci 34–59 × 10–13 μm ( = 48.3 × 10.9 μm, n = 10), 8-spored, bitunicate, fissitunicate, oblong to obclavate, with or without a short, knob-like pedicel or pedicel lacking, with an inconspicuous ocular chamber (Fig. 1: 5, 7). Ascospores 11–15(–20) × 4–5 μm ( = 13.4 × 4.5 μm, n = 20), obliquely overlapping to biseriate at the base, ellipsoidal, with broadly rounded ends, olivaceous pale brown, 1-septate, slightly constricted at the septum, the upper cell shorter and wider than the lower one (length of upper/lower cell = (7:13–)5:7–1:1), ascospore wall thin, smooth (Fig. 1: 8).
Culture characteristics: Ascospore germinating on MEA (malt extract agar) after 2–3 d; colony growth is extremely slow, on MEA reaching up to 0.5 mm diameter in 14 days at 26–28 °C in darkness, reaches 0.8–1.1 mm after 4 weeks and reaches 1.2–2.3 mm after 3 months. Colony first blackish, turning olivaceous black, reverse blackish; hyphae olivaceous brown, branched, 6–11 μm wide, constricted at septa, wall thickened, 1–1.5 μm, often giving rise to dark brown ellipsoid chlamydospores, chlamydospores 11–18 × 7–14 μm, wall 1–2 μm; hyaline to pale brown thin-walled hyphae rarely produced, 4–7 μm wide, straight to coiled, and hyphae tips usually swollen or forming a pale brown to brown lemon-shaped cell, 19–30 × 10–18 μm; odour not detected. No conidiogenous structures were observed (see Fig. 2, Fig. 3).
Type: CHINA. Heilongjiang province, Yichun, Wuyiling district, Wuyiling forestry station, 48° 33′N, 129° 30′E, ca. 330 m, on leaves of L. praeflorens, 26 August 2014 (HMAS246485, holotype).
Diagnosis: V. chinensis differs from other known species in the genus by the striking superficial ascomata covered with dark brown setae on the upper one-third part, and different ITS and LSU sequences.
Note: Ascomata scattered on decaying the leaf surface of L. praeflorens with no scab symptom observed. Thus V. chinensis should be a saprobic fungus.
4. Discussion
Venturia chinensis is characterized by saprobic habitat, small-sized, superficial ascomata, subglobose to citriform, with a striking papilla and covered with setae; evanescent hamathecium; oblong to obclavate asci with or without a short, knob-like pedicel; ellipsoidal, olivaceous pale brown, 1-septate and thin-walled ascospore. These morphological characteristics fit Venturia well, which are also strongly supported by DNA sequences data (Fig. 4).
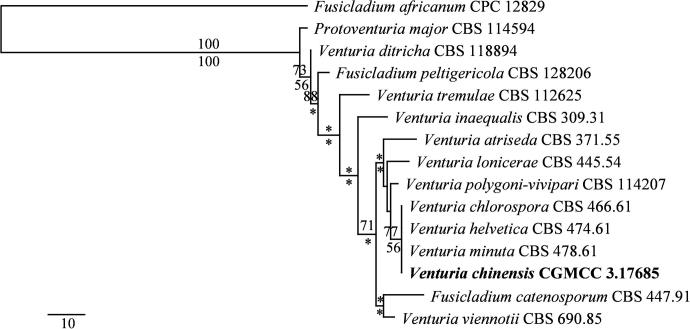
Fig. 4.
Maximum likelihood tree generated from sequence analysis of the ITS rDNA dataset. Designated outgroup taxon is Fusicladium africanum. Maximum likelihood bootstrap support values above 50% are shown under the branches and based on 1000 replicates. Maximum parsimony bootstrap support values above 70% are shown at the nodes.
Host specificity has been considered as one of the distinguishing characteristics of Venturiaceae, and one particular venturiaceous species usually restricted to one or a few closely related species of a host genus, or to members of closely related genera (Barr, 1989). Host species of one genus may bear more than one member of the Venturia, such as three species of Venturia has been reported in Epilobium, and 5 species of Venturia in Salix (Nüesch, 1960). Venturia lonicerae Sacc. has been reported from Lonicera xylosteum (Saccardo, 1882). CBS 445.54, an isolate of V. lonicera from Lonicera caerulea, significantly differs from V. chinensis in ITS (identical rate: 98.6%) and LSU (identical rate: 99.8%) sequences (Table 2). In particular, the strikingly superficial and citriform ascomata of V. chinensis can be readily distinguished from V. lonicerae and other species of Venturia. Based on a megablast search of NCBIs GenBank nucleotide database, the closest hits using the ITS sequence are V. chlorospora (Ces.) P. Karst. (strain CBS 466.61, GenBank EU035453), V. helvetica Nüesch (strain CBS 474.61, GenBank EU035458), and V. minuta M.E. Barr (strain CBS 478.61, GenBank EU035464) with identity rate as 655/656 (100%). What is amazing is that the ITS and LSU sequences of all these three strains (CBS 466.61, CBS 474.61, CBS 478.61) show no difference at all. All of these three strains are isolated from Salix in Switzerland by E. Müller. Both V. chlorospora and V. helvetica have been reported from Salix (Nüesch, 1960). The length/width index of V. chinensis is 2.97, which is much higher than that of V. chlorospora (2.1) and V. helvetica (2.4). In particular, the striking superficial ascomata of V. chinensis can be readily distinguished from the erumpent ascomata of V. chlorospora and V. helvetica. The identification status of these three isolates however, needs to be verified yet. In addition, the small-sized (40–60 μm diam.), immersed to erumpent ascomata of V. minuta can be readily distinguished from those of V. chinensis.
Table 2.
Polymorphic nucleotides from sequence data of the ITS and 28S rDNA to show the relationship between Venturia chinensis and V. lonicerae and other related species.
| Identity | Strain | ITS |
28S rDNA |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 178 | 217 | 227 | 269 | 283 | 284 | 473 | 569 | 581 | 179 | 180 | ||
| V. lonicerae | CBS 445.54 | C | C | G | C | C | C | T | C | C | T | G |
| V. chinensis | CGMCC 3.17685 | T | T | – | A | T | T | G | T | T | C | T |
| V. minuta | CBS 478.61 | T | T | G | A | T | T | G | T | T | C | T |
| V. helvetica | CBS 474.61 | T | T | G | A | T | T | G | T | T | C | T |
| V. chlorospora | CBS 466.61 | T | T | G | A | T | T | G | T | T | C | T |
| V. polygoni-vivipari | CBS 114207 | T | T | G | A | T | A | G | A | C | T | G |
Venturia chinensis is saprobic, and the ascomata of which is readily produced on the growing leaves of L. praeflorens in natural environment, and more ascomata could be obtained by inoculation in the lab. These characteristics differ from some parasitic species, the ascomata of which are usually produced during or after winter time (Nüesch, 1960). Comparable with other species of venturiaceous fungi, the colony of V. chinensis grows rather slowly in culture. The microstructures produced on the surface of media of V. chinensis derives from the normal asexual stage of Fusicladium or Spilocaea, Pollaccia in hyphae which are rather thick, heavily pigmented, producing chlamydospores and hyphal tips swollen. These characteristics, however, are unlike those encountered in saprobic fungi.
Acknowledgements
This study is supported by the Fundamental Research Funds for the Central Universities (NO. YX2015-05), National Natural Science Foundation of China (General Program) (31370063), NSFC Projects of International Cooperation and Exchanges (31461143028) and National Science and Technology Foundation Project (2014FY210400). Prof. Dr. Pedro W. Crous (the Centraalbureau voor Schimmelcultures, the Netherlands (CBS)) is also acknowledged for his suggestions concerning the colony characteristics of Venturia chinensis.
Footnotes
Peer review under responsibility of King Saud University.
References
- Barr M.E. The Venturiaceae in Noth America. Can. J. Bot. 1968;46:799–864. [Google Scholar]
- Barr M.E. The Venturiaceae in North America: Revisions and additions. Sydowia. 1989;41:25–40. [Google Scholar]
- Cesati V., De Notaris G. Schema di classificazione degli sferiacei italici aschigeri. Comm. Soc. Crittog. Ital. 1863;1:177–240. [Google Scholar]
- De Notaris G. Cenno sulla tribu de’pirenomiceti sferiacei e descrizione di alcuni nuovi generi. Giorn. Bot. Ital. l. 1844:322–335. [Google Scholar]
- Kirk P.M., Cannon P.F., Minter D.W., Staplers J.A. 10th ed. CABI Bioscience; UK: 2008. Dictionary of the Fungi. [Google Scholar]
- Korf R.P. Nomenclatural Notes. I. Misuse of Neotypes for Venturia and Phaeosphaerella. Mycologia. 1956;48:591–595. [Google Scholar]
- Nüesch J. Beitrag zur Kenntnis der weidenbewohnenden Venturiaceae. Phytopathol. Z. 1960;39:329–360. [Google Scholar]
- Posada D., Buckley T.R. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Systemat. Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Saccardo P.A. Syll. Fung. 1882;1:1–768. [Google Scholar]
- Sivanesan A. Lubrecht & Cramer Ltd; Vaduz: 1977. The taxonomy and pathology of Venturia species. [Google Scholar]
- Swofford, D.L., 2002. PAUP∗. Phylogenetic Analysis Using Parsimony (∗and other methods). Version 4.0b10. Sinauer Associates, Sunderland, Massachusetts.
- Tai F.L. A preliminary survey of Plant pathogens of economical plants in Yunnan provence. Bull. Agric. Res. Inst. Nat. Tsing Hua Univ. 1941;6:1–36. (in Chinese) [Google Scholar]
- Tai F.L. Science Press; Beijing, China: 1979. Sylloge Fungorm Sinicorum; pp. 1–1527. (in Chinese) [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M.Q., He P.X. Press of Northeast Forestry University; Harbin: 2008. Sylloge of Phytopathogens on Woody Plants in China. 1–1640 (in Chinese) [Google Scholar]
- Tai F.L. Science Press; Beijing, China: 2003. Flora fungorum sinicorum. Vol. 14. Cladosporium, Fusicladium, Pyricularia; pp. 1–324. (in Chinese) [Google Scholar]
- Zhang Y., Crous P., Schoch C., Bahkali A., Guo L. A molecular, morphological and ecological re-appraisal of Venturiales – a new order of Dothideomycetes. Fung. Div. 2011;51:249–277. doi: 10.1007/s13225-011-0141-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.M. Preliminary study phytopathogens in China. Chin. Agric. Sci. Bull. 1927;54:23–43. (in Chinese) [Google Scholar]