Abstract
Alcohol is a human carcinogen. A causal link has been established between alcohol drinking and cancers of the upper aerodigestive tract, colon, liver and breast. Despite this established association, the underlying mechanisms of alcohol-induced carcinogenesis remain unclear. Various mechanisms may come into play depending on the type of cancer; however, convincing evidence supports the concept that ethanol’s major metabolite acetaldehyde may play a major role. Acetaldehyde can react with DNA forming adducts which can serve as biomarkers of carcinogen exposure and potentially of cancer risk. The major DNA adduct formed from this reaction is N 2-ethylidenedeoxyguanosine, which can be quantified as its reduced form N 2-ethyl-dG by LC-ESI-MS/MS. To investigate the potential use of N 2-ethyl-dG as a biomarker of alcohol-induced DNA damage, we quantified this adduct in DNA from the oral, oesophageal and mammary gland tissues from rhesus monkeys exposed to alcohol drinking over their lifetimes and compared it to controls. N 2-Ethyl-dG levels were significantly higher in the oral mucosa DNA of the exposed animals. Levels of the DNA adduct measured in the oesophageal mucosa of exposed animals were not significantly different from controls. A correlation between the levels measured in the oral and oesophageal DNA, however, was observed, suggesting a common source of formation of the DNA adducts. N 2-Ethyl-dG was measured in mammary gland DNA from a small cohort of female animals, but no difference was observed between exposed animals and controls. These results support the hypothesis that acetaldehyde induces DNA damage in the oral mucosa of alcohol-exposed animals and that it may play role in the alcohol-induced carcinogenic process. The decrease of N 2-ethyl-dG levels in exposed tissues further removed from the mouth also suggests a role of alcohol metabolism in the oral cavity, which may be considered separately from ethanol liver metabolism in the investigation of ethanol-related cancer risk.
Introduction
Numerous epidemiological studies have established the relationship between alcohol consumption and various types of cancers, including cancers of the oral cavity, pharynx, oesophagus, colon, rectum, liver, larynx and breast (1,2). A better understanding of mechanisms of alcohol-induced cancer is critical for developing rational approaches to cancer prevention. A variety of mechanisms may contribute to alcohol-mediated carcinogenesis, including: the effect of ethanol in increasing solubility of carcinogens; the production of toxic, reactive oxygen species; the perturbation of methyl transfer and other enzymatic systems; or the abnormal metabolism of vitamin A and its derivative retinoic acid (3). These mechanisms may play different roles depending on the target tissue and cancer type. Convincing evidence increasingly suggests acetaldehyde—the major metabolite of ethanol and a DNA-reactive compound—as being at least partially responsible for the carcinogenic effects of alcohol, in particular for the upper aerodigestive tract. Therefore, acetaldehyde associated with alcohol consumption has been classified recently as ‘carcinogenic to humans’ by the International Agency for Research on Cancer. Although ethanol is mainly metabolised in the liver, the concentration of acetaldehyde in saliva after ethanol ingestion is much higher than in the blood, due to oral microflora metabolism of ethanol, as well as the acetaldehyde content of alcoholic beverages (4,5). Bacteria present in the normal oral flora, such as Streptococcus salivarius and Neisseria, contribute substantially to the production and accumulation of acetaldehyde from ethanol oxidation (5–7).
The oral microflora metabolism of ethanol can result in levels of acetaldehyde above what is considered to be carcinogenic in vivo a few minutes after ingestion of alcohol, providing a specific source of exposure, which could potentially be considered an additional cancer risk beyond the physiological metabolism of ethanol (5).
Acetaldehyde reacts with DNA bases to produce modifications known as DNA adducts, which are critical in the carcinogenic process, because they can cause miscoding, resulting in mutated genes and loss of normal growth control mechanisms. Exposure of DNA to acetaldehyde results in several lesions. N 2-Ethylidene-deoxyguanosine is the major modification and can be detected via LC-mass spectrometry as its reduced and more stable form, N 2-ethyl-dG. The formation of the crotonaldehyde-derived DNA adduct 1, N 2-propano-2′-deoxyguanosine (Cro-dG) has also been observed in acetaldehyde-treated DNA together with DNA interstrand crosslinks (8). These adducts can provide information on the DNA damage resulting from the metabolism of ethanol and can serve as biomarkers of exposure; additionally, due to their miscoding potential, they can provide information on potential of cancer risk.
In a previous study, we have demonstrated a dose–response relationship between amounts of alcohol ingested and N 2-ethyl-dG in human oral cells collected with a mouthwash. Levels of this adduct increased 2h after alcohol consumption and reached a peak in 2–6h (9). The collection of oral cells with a mouthwash has the limitation of providing mostly exfoliating epithelial cells and of being potentially, greatly contaminated by bacterial cells from the oral microflora. In order to confirm whether the alcohol-derived DNA damage we observed would be generated in the mucosa of the oral cavity, we tested samples obtained from the Monkey Alcohol Tissue Research Resource (MATRR) (10). This multi-institutional collaboration is designed to determine the functional consequences of long-term ethanol exposure in non-human primates. The tissues are harvested from animals that chronically self-administer ethanol or a control solution under identical operant conditions (11). The availability of tissues from this model prompted us to extend our investigation to the oesophageal mucosa and the mammary gland, other target organs for which a role of acetaldehyde in the carcinogenic process has been suggested.
Methods
Animal study
Adult Rhesus monkeys (Macaca mulatta) were trained and exposed following the protocol previously reported (10–12). Briefly, after acclimation to the laboratory monkeys are trained on operant panels that allow access to food and fluids via responses controlled by a computerised system. Once trained on the panel, the animals undergo a stepwise induction paradigm during which they are first induced to drink water followed by a series of escalating doses of ethanol (4% w/v in water). Age- and sex-matched controls follow the same protocol but self-administer an isocaloric maltose-dextrin solution rather than ethanol. Following induction, the open access 22-h daily self-administration stage begins and ethanol and water are available ad libitum. After 12 months of daily self-administration, the animals are taken to necropsy for collection of tissues following a state-of-the-art protocol (13). In this study, tissue samples from 29 animals (18 exposed to ethanol and 11 controls) were harvested at necropsy. The cheek mucosa was resected starting from the mouth and cutting back for a couple of centimeters while the oesophageal mucosa was isolated from the cervical portion of the oesophagus, within 5cm below the larynx. Female monkeys were included in the study (five exposed to ethanol and three controls). In these animals, the mammary gland tissue was resected collecting from the ‘upper outer’ quadrant of either side, from the nipple up and laterally, within about 4cm from the nipple. Tissues were then snap frozen and stored at −80°C until DNA isolation.
Chemicals and enzymes
N 2-Ethyl-dG and [15N5] N 2-ethyl-dG were prepared as described (14). Ethanol was obtained from AAPER Alcohol and Chemical Co. (Shelbyville, Ky). Isopropanol was purchased from Acros Organics (Morris Plains, NJ, USA). Puregene DNA purification solutions were obtained from Qiagen (Valencia, CA, USA). Calf thymus DNA was purchased from Worthington Biochemical Corporation (Lakewood, NJ, USA). Alkaline phosphatase (from calf intestine) was obtained from Roche Diagnostics Corporation (Indianapolis, IN, USA). All other chemicals and enzymes were purchased from Sigma–Aldrich (St. Louis, MO, USA).
DNA samples
DNA was extracted from the tissues after separating the mucosa from the basal tissue layer. DNA isolation was performed following the modified Puregene DNA isolation protocol (Qiagen) as reported in the literature (15). DNA was then hydrolysed and purified as previously reported (9). NaBH3CN was used to convert the major acetaldehyde–DNA adduct, N 2-ethylidene-dG, to the more stable N 2-ethyl-dG. [15N5] N 2-Ethyl-dG was added as internal standard. For enzyme hydrolysis, DNA was dissolved in 400 µl of 10mM Tris/5mM MgCl2 buffer (pH 7.5), containing [15N5] N 2-ethyl-dG (50fmol) and NaBH3CN (30mg). After the pH was adjusted to 7 with 0.1N HCl, the DNA was initially digested overnight at room temperature with 1300 units of DNase I (type II, from bovine pancreas). Then to the resulting mixture were added 1300 additional units of DNase I, 0.07 units of phosphodiesterase I (type II, from Crotalus adamanteus venom), and 750 units of alkaline phosphatase. The mixture was incubated at 37°C for 70min and then allowed to stand overnight at room temperature. Enzymes were removed by centrifugation using a centrifree MPS device (MW cutoff of 30000; Amicon, Beverly, MA, USA).
Sample enrichment and purification
N2-Ethyl-dG
Sample enrichment and purification were carried out as reported (9). The DNA hydrolysate, after removal of a 10 µl aliquot for 2′-deoxyguanosine (dG) analysis, was desalted and purified using a solid-phase extraction (SPE) cartridge [Strata-X 33 µm, 30mg/1ml (Phenomenex, Torrance, CA, USA)]. The 70% CH3OH fraction was collected and evaporated to dryness, dissolved in 1ml of H2O, and purified using a mixed mode, anion exchange reversed phase extraction cartridge (Oasis MAX, 30mg/cartridge, Waters). Adducts were eluted with 1ml of 70% CH3OH, and the solution was evaporated to dryness. The residue was dissolved in 20 µl of H2O, and 8 µl aliquots were analysed by LC-ESI-MS/MS. Samples from all animals were processed simultaneously. Samples from each tissue (oral tissue, oesophagus and mammary gland), were processed together as a set, resulting in three sets. Buffer blanks containing internal standard were processed as above and analysed to check the MS instrument baseline and for possible contamination. Calf thymus DNA (0.1mg) with internal standard added as above was used as a positive control to determine inter-day precision and accuracy. Each set of samples was run together with one buffer blank and three positive controls.
Cro-dG
Quantitation of this DNA adduct was performed following the protocol reported in the literature (16). DNA hydrolysis was performed following the procedure described above for the analysis of N 2-ethyl-dG after adding 50fmol of [15N5]Cro-dG as internal standard. The hydrolysate was purified using a SPE [Strata-X, 33 μm, 30mg/1ml (Phenomenex)]. After loading the sample, the cartridge was washed with 1ml H2O and 1ml 15% CH3OH/H2O, and the analyte was eluted with 1ml 70% CH3OH/H2O. The eluants were evaporated to dryness, and dissolved in 20 μl of H2O for LC-ESI-MS/MS analysis. Calf thymus DNA (0.1mg) with internal standard added as above was used as a positive control to determine inter-day precision and accuracy. Each set of samples was run together with one buffer blank and three positive controls.
DNA quantitation
High-performance liquid chromatography-UV analysis
Quantitation of dG was carried out with an Agilent 1100 capillary flow high-performance liquid chromatography (HPLC) with a diode array UV detector set at 254nm (Agilent Technologies, Palo Alto, CA, USA). A 4.6mm × 25cm Luna 5 µm C18 column (Phenomenex) was used with a gradient from 5% to 40% CH3OH in H2O over the course of 35min at a flow rate of 10 µl/min.
Real-time polymerase chain reaction
Genomic DNA from 21 oral mucosa samples was quantified in triplicate using the Qubit Fluorometric Quantification system (Life Technologies, Carlsbad, CA, USA). The samples were then normalised at 200ng/µl, and a 1:40 dilution (5ng) was used for real-time PCR quantification. Non-human primate DNA was quantified using rhesus macaque β-actin primers (F: 5′-CGTGGACATCCGTAAAGAC and R: 5′-GGGCAGTAATCTCCTTCTG). A standard curve was built with five-serial dilutions of blood gDNA (from 25 to 0.13ng/μl). Real-time PCR was performed in the QuantStudio 12K Flex (Life Technologies) using the EVAGREEN dye as recommended by the manufacturer. Each reaction (10 μl) contained 5 μl of the 2x EVAGREEN Master Mix (Biotium, Hayward, CA, USA), 300nM forward and reverse primers and 1 μl (5ng) of gDNA. Reactions were performed in triplicate, including a negative control. In addition the real-time PCR was replicated three times. The reactions were incubated for 2min at 95°C followed by 40 cycles of 5s at 95°C and 30s at 60°C. To verify the specificity of the product, a melting curve analysis was carried out between 50°C and 95°C with a plate read every 0.5°C after holding the temperature for 5s. The efficiency (E) was calculated using the formula: E = [10(− 1/k) − 1], with k = slope. The QuantStudio 12K Flex Software was used to calculate the C t values. Data from the standard dilution series were used to generate the standard curve for β-actin, which was then used to calculate the amount of DNA in the experimental samples. The average from the three replicates was compared to the average obtained after Qubit quantification to estimate the fraction of DNA from a potentially non-non–human primate source.
Statistical analysis
Statistical analysis of adduct levels was performed using Stata software (StataIC 11, College Station, TX, USA). Comparison of adduct levels generated by ethanol ingestion at each site was performed using analysis of variance (ANOVA) with Bonferroni correction for multiple comparisons. P values less than 0.05 were considered statistically significant. When comparing levels of adducts formed between exposed animals and controls, a two sample t-test with equal variances was performed. Also in this case, P values of less than 0.05 were considered statistically significant.
Results
Rhesus monkeys were trained to consume alcohol and then allowed to voluntarily consume an ethanol solution (4% in water) and/or water on a daily basis over the course of 1 year. The animals were organised into three cohorts that also included control animals that self-administered an isocaloric sweetened solution and/or water under identical operant conditions. The first two cohorts included only male monkeys; eight exposed animals and four controls, and five exposed animals and four controls, respectively. The third cohort included only female monkeys; five exposed animals and three controls. The amounts of ethanol consumed over the course of a day by the exposed animals were measured as summarised in Table 1. The exposed monkeys consumed an average of 2.3±0.8g/kg of ethanol per day, corresponding to around nine drink equivalents per day, referring to a common approximation of one drink = 15g of alcohol (11,17). One year after the beginning of the ethanol self-administration period, the animals were humanely euthanised and the tissues were collected.
Table 1.
Summary of the levels of N 2-ethyl-dG measured in the three tissues in monkeys exposed to various amounts of ethanol and in controls
| Sample no. | Gender | Cheek DNA N 2-ethyl-dG (fmol/µmol dG) | Oesophagus DNA N 2-ethyl-dG (fmol/µmol dG) | Mammary gland DNA N 2-ethyl-dG (fmol/µmol dG) | Average EtOH consumption in 22h (g/kg) |
|---|---|---|---|---|---|
| 1 | M | 757 | 396 | – | 1.80 |
| 2 | M | 460 | 292 | – | 3.10 |
| 3 | M | 722 | 152 | – | 2.30 |
| 4 | M | 900 | 633 | – | 2.80 |
| 5a | M | 1503 | 238 | – | 3.00 |
| 6 | M | 310 | 184 | – | 1.90 |
| 7 | M | 427 | 135 | – | 1.90 |
| 8 | M | 556 | 167 | – | 3.30 |
| 9 | M | 164 | 216 | – | 2.40 |
| 10 | M | 401 | 150 | – | 1.40 |
| 11 | M | 690 | 301 | – | 1.80 |
| 12 | M | 109 | 142 | – | 2.06 |
| 13 | M | 116 | 227 | – | 2.26 |
| 14 | F | 115 | 99 | 167 | 1.00 |
| 15 | F | 67 | 74 | 106 | 4.02 |
| 16 | F | 81 | 99 | 91 | 1.77 |
| 17 | F | 226 | 116 | 84 | 2.81 |
| 18 | F | 77 | 91 | 93 | 1.45 |
| 19 | M | 234 | 191 | – | 0.00 |
| 20 | M | 354 | 141 | – | 0.00 |
| 21 | M | 85 | 145 | – | 0.00 |
| 22 | M | 372 | 189 | – | 0.00 |
| 23 | M | 267 | 190 | – | 0.00 |
| 24 | M | 72 | 142 | – | 0.00 |
| 25 | M | 78 | 100 | – | 0.00 |
| 26 | M | 65 | 101 | – | 0.00 |
| 27 | F | 35 | 46 | 129 | 0.00 |
| 28 | F | 55 | 55 | 84 | 0.00 |
| 29 | F | 48 | 128 | 150 | 0.00 |
aSample with the highest amount of N 2-ethyl-dG and quantifiable levels of Cro-dG: 2.1fmol/µmol dG of (6S, 8S) Cro-dG and 2fmol/µmol dG of (6R, 8R) Cro-dG.
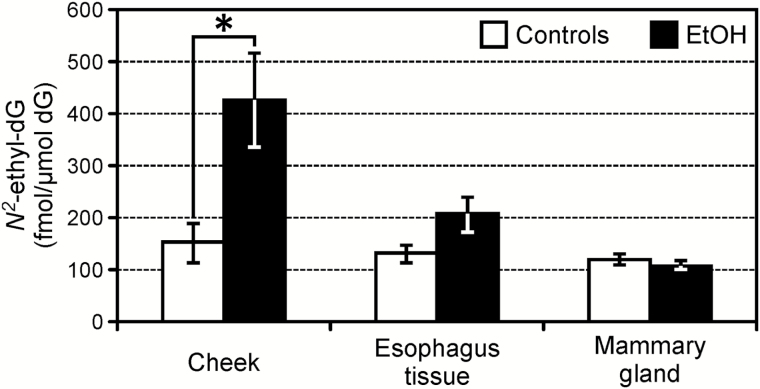
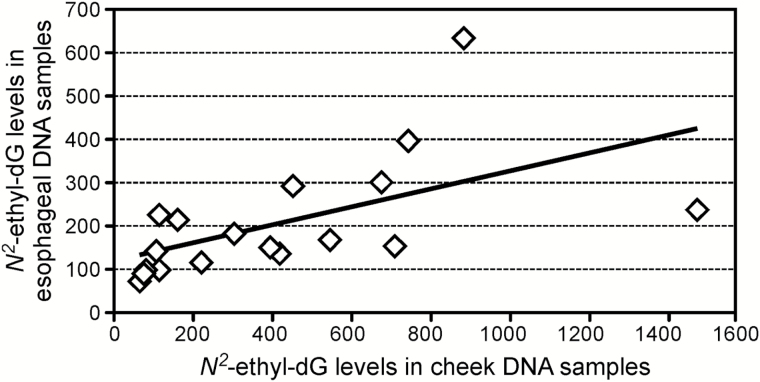
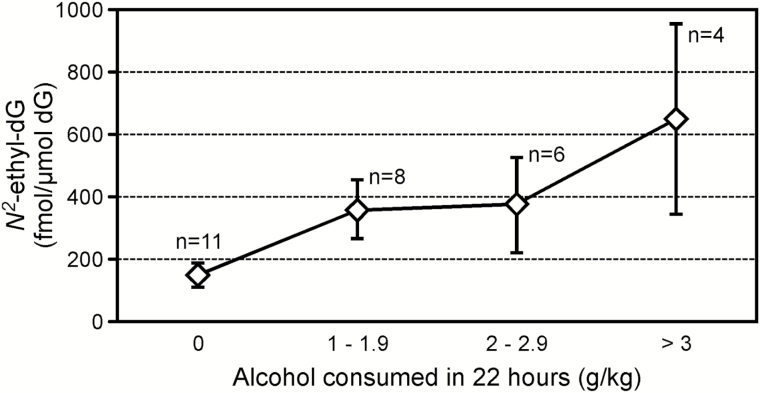
Samples were obtained from 29 cheek tissues and 29 oesophageal tissues, harvested during necropsy, from 11 controls and 18 animals exposed to various amounts of ethanol. DNA was isolated after separating the mucosa from the basal tissue layer. Eight mammary gland tissues were collected from the eight female animals. Average amounts of DNA used for the DNA adducts analysis of the samples from the three tissues (oral mucosa, oesophageal mucosa and mammary gland), were 58±8, 61±11 and 100±12 µg, respectively. The DNA obtained from the cheek mucosa was analysed to investigate bacterial contamination. Samples were quantified using the Qubit Fluorometric Quantification system, then the results were compared to those obtained by performing a qPCR. The difference between the oral mucosa DNA amounts measured with the two methods was 8%, which is within the variability range observed when comparing the two methods (5–11%), indicating a negligible contamination from bacteria DNA of the oral mucosa samples. The acetaldehyde-derived DNA adduct, N 2-ethyl-dG, was detected in all the DNA samples analysed from all tissues. The results obtained from each sample are summarised in Table 1. The average amount of N 2-ethyl-dG measured in the oral mucosa DNA from monkeys exposed to alcohol was 427±90fmol/µmol dG, which was significantly higher than the average level measured in the controls: 151±39fmol/µmol dG (P < 0.02). N 2-Ethyl-dG levels in the oesophageal mucosa DNA from exposed animals were lower, compared to the oral mucosa, with an average of 206±32fmol/µmol dG. Consequently, the difference between these levels and those measured in the controls (130±15fmol/µmol dG) was not statistically significant (P < 0.08). No difference was observed when comparing levels of N 2-ethyl-dG in mammary gland DNA samples from the female animals; 108±8fmol/µmol dG were measured in the exposed group and 121±10fmol/µmol dG in the controls. Average levels of N 2-ethyl-dG were comparable across tissues in animals not exposed to alcohol. The results obtained from this analysis are summarised in Figure 1. As shown in Figure 2, a significant correlation between the N 2-ethyl-dG levels in the oral mucosa DNA and in the oesophageal mucosa DNA was observed in animals consuming alcohol: Spearman’s rho = 0.8 (P = 0.001). The correlation between N 2-ethyl-dG levels in oral mucosa DNA and amounts of alcohol consumed per day was investigated, as shown in Figure 3. Levels of the DNA adducts increased with amounts of alcohol consumed though the trend was not significant. The presence of Cro-dG was also investigated in the oral and oesophageal mucosa DNA samples. No quantifiable levels of this DNA adduct were found in the samples analysed except for the oral mucosa sample number 5 of Table 1. This sample had the highest amount of N 2-ethyl-dG 1503fmol/µmol dG and resulted to have 2.1fmol/µmol dG of (6S, 8S) Cro-dG and 2fmol/µmol dG of (6R, 8R) Cro-dG.
Figure 1.
Average amounts of N 2-ethyl-dG measured in the oral mucosa, oesophagus and mammary gland DNA from animals exposed to alcohol and from controls (*P < 0.02 compared to controls).
Figure 2.
Correlation between the N 2-ethyl-dG levels measured in the oral mucosa DNA and the levels measured in the oesophageal mucosa DNA from alcohol exposed animals.
Figure 3.
Average levels of N 2-ethyl-dG in oral mucosa DNA of monkeys drinking increasing amounts of ethanol per day.
Discussion
The results of this study confirm that alcohol consumption increases levels of the major acetaldehyde-DNA adduct N 2-ethylidene-dG, measured as N 2-ethyl-dG, in the oral cavity mucosa of exposed rhesus monkeys. The effect was significant even when comparing a relatively small number of samples from 18 exposed animals and 11 controls. The presence of bacterial DNA in the samples used for this analysis was investigated by qPCR, and no major contribution from microbial DNA was observed. The rationale for this study originated in our recent observation of a dose–response effect of ethanol consumption on levels of N 2-ethyl-dG in human oral cells (9). The human cell samples collected for that study were obtained using a mouthwash, which provides a heterogeneous mixture consisting mostly of epithelial exfoliating cells, with some bacterial cells. Consequently, contribution of microbial DNA to those results could not be ruled out and the relevance of our findings to alcohol-induced DNA damage in the oral mucosa remained unclear. Alcohol consumption increases the risk for developing oro-oesophageal squamous cell carcinoma. This malignancy can develop from the histopathological evolution of precancerous lesions of the mucosa from hyperplasia to dysplasia and ultimately to squamous cell carcinoma (18). Hence, the aim of this study was to verify whether the DNA damaging effects of alcohol could be observed in the mucosa of the oral cavity and of the proximal oesophagus. Tissue samples were collected from the inside of the cheek and the cervical part of the oesophagus, and the mucosa was separated from the basal muscular tissue before DNA extraction. Our results confirm that the DNA damaging effect of alcohol-derived acetaldehyde is relevant to the oral mucosa and suggest a similar effect on the mucosa of the oesophagus. To our knowledge, this is the first study to investigate the formation of DNA adducts in these tissues in non-human primates exposed to alcohol. Higher levels of N 2-ethyl-dG were measured in the tongue, in the sub-mandibular gland and in the oesophagus of mice exposed to an 8% ethanol solution in drinking water for 8 months. In this experiment, Aldh2-knockout mice (Aldh2 -/-) carrying the genotype encoding for an inactive aldehyde dehydrogenase enzyme were compared to control mice (Aldh +/+) carrying the active enzyme. Consistent with our findings, knockout animals unable to detoxify acetaldehyde and are thus exposed to higher concentrations had higher DNA adduct levels; however, the Aldh +/+ mice exposed to alcohol did not show an increase in N 2-ethyl-dG levels compared to the controls (19). Alcohol metabolism in the oral cavities of rodents may be very different from those of humans. The use of non-human primates allows testing a model with many anatomic, neurophysiological and behavioural characteristics similar to humans. Non-human primates, in particular macaques, have oral structures and oral microbiological characteristics similar to humans (20). Furthermore, monkeys can drink voluntarily and can develop a physical dependence that closely mimics human drinking levels and patterns, increasing the significance of our findings to humans.
Overall, the levels of N 2-ethyl-dG we measured in the oesophageal mucosa of the exposed animals were lower than those observed in the oral cavity. The average level of N 2-ethyl-dG measured in the oesophageal DNA of exposed animals was higher than in the controls, but this difference was not statistically significant; however, these results correlated significantly with those found in the oral cavity tissue, suggesting that a substantial difference between exposed and controls may have been observed with a larger sample size. The relatively small size of our cohort is the major limitation of this study. Models based on non-human primates are particularly expensive, labour-intensive and time-consuming; these factors, combined with ethical considerations, prohibited our access to larger sample sizes.
Female breast cancer is causally related to alcohol consumption, and as for cancers of the upper aerodigestive-tract, acetaldehyde has been hypothesised to play a role in this carcinogenic process (21). This hypothesis is based on studies that have shown that acetaldehyde resulting from even a low dose of alcohol can accumulate in mammary tissue resulting in concentrations higher than those measured in plasma several hours after the alcohol dose (22). The availability of a cohort of female monkeys prompted us to test the levels of N 2-ethyl-dG in mammary tissue. Interestingly, the levels of the DNA adduct in the exposed animals were the same as in the controls, and these levels were consistent with the levels measured in the other tissues in control animals. A comparison of N 2-ethyl-dG levels across tissues from exposed animals seem to indicate that the increase of the DNA adduct levels we measured may be mostly a consequence of the alcohol metabolism in the oral cavity rather than a result from acetaldehyde exposure from liver metabolism and systemic circulation. Although ethanol is mainly metabolised in the liver, the concentration of acetaldehyde in saliva after ingesting ethanol is much higher than in the blood, due to oral microflora metabolism of ethanol (5,7,23).
The conversion of ethanol into acetaldehyde in the oral cavity would seem to have a decreasing effect on target tissues as they are further removed from the site of exposure, inducing a significant increase in N 2-ethyl-dG levels in the oral mucosa, a less pronounced increase in the upper part of the oesophagus and resulting in no effect in the mammary gland tissue.
In our previous study focusing on the effects of alcohol on human oral cell DNA, we observed a dose–response relationship between the levels of N 2-ethyl-dG and amounts of alcohol consumed. In this study, the investigation of a possible relationship between the levels of N 2-ethyl-dG and the amounts of alcohol habitually consumed by each animal did not show a significant trend.
The acetaldehyde-related effects that we observed in human oral cells decreased significantly 6h after exposure. The animal model used for this study collects information on the amounts of alcohol consumed by each animal, which ranged between 1 and 4g/kg per day, comparable to 4–16 drink equivalents per day. No information, however, was available on the time duration between the last sip of alcohol and the tissue harvesting. This period of time, which was estimated to be between 2 and 12h, may have affected the levels of N 2-ethyl-dG leading to an underestimation of the DNA adduct amounts. This observation may help to explain the lack of a significant correlation between N 2-ethyl-dG levels and amounts of alcohol consumed.
The chemical instability of N 2-ethylidene-dG prevents direct investigation of its biological properties, thus the mutagenic consequences of this DNA modification are not completely understood.
The condensation of two molecules of acetaldehyde can also produce a reactive electrophile, 3-hydroxybutanal (crotonaldehyde), which can also form a Schiff base on the same amino group of dG. These crotonaldehyde-derived propano-dG (Cro-dGs) adducts can have multiple biologic effects as a result of their abilities to undergo ring opening reactions. Ring opening yields another aldehyde moiety, which can react with proteins to form DNA–protein cross-links or (in some sequence contexts) with deoxyguanosine on the opposite strand to form DNA-interstrand cross-links. Levels of these DNA adducts were measured in the oral mucosa DNA. Only the sample with the highest levels of N 2-ethyl-dG showed detectable and quantifiable levels of Cro-dG, indicating that the levels of alcohol exposure in this model were probably too low to induce measurable amounts of these DNA adducts. Because of its higher levels, N 2-ethyl-dG was confirmed to be easier to detect and measure than other acetaldehyde DNA adducts and thus a better potential marker for the investigation of acetaldehyde-related DNA damage.
In summary, we present conclusive evidence linking alcohol drinking and acetaldehyde–DNA adduct formation in oral mucosa of Rhesus monkeys, confirming the genotoxic role of acetaldehyde from ethanol metabolism. Our results demonstrate this difference is significant even when considering a relatively small sample size. DNA damage is a crucial step in the carcinogenic process; our results contribute to understanding the DNA-damaging effects of alcohol in the oral mucosa. Additionally, this study suggests that a similar DNA-damaging effect may be relevant to the oesophageal mucosa, while it may not play a role in the female mammary gland although additional studies are warranted on a larger sample size to confirm this observation. Overall, our results support the hypothesis of acetaldehyde exposure from ethanol oral metabolism being an additional cancer risk factor beyond ethanol liver metabolism and confirm the use of N 2-ethyl-dG as a potential relevant biomarker for the investigation of alcohol-related mechanisms of head and neck cancers.
Funding
National Institute of Environmental Health Sciences (ES-11297); National Institute on Alcohol Abuse and Alcoholism (AA019431). Mass spectrometry was carried out in the Analytical Biochemistry core facility of the Masonic Cancer Center, supported in part by Cancer Center Support Grant CA-77598.
Conflict of interest statement: None declared.
References
- 1. IARC working group on the evaluation of carcinogenic risks to humans (2010) Alcohol consuption and ethyl carbamate. IARC Monogr Eval. Carcinog. Risks Hum., 96, 171–602. [PMC free article] [PubMed] [Google Scholar]
- 2. Baan R. Straif K. Grosse Y. Secretan B. El Ghissassi F. Bouvard V. Altieri A. and Cogliano V (2007) Carcinogenicity of alcoholic beverages. Lancet Oncol., 8, 292–293. [DOI] [PubMed] [Google Scholar]
- 3. Seitz H. K. and Mueller S (2010) Mechanisms of tumorgenesis associated with alcohol consumption. Alcohol. Clin. Exp. Res., 34, 47A. [Google Scholar]
- 4. Yokoyama A. Tsutsumi E. Imazeki H. Suwa Y. Nakamura C. Mizukami T. and Yokoyama T (2008) Salivary acetaldehyde concentration according to alcoholic beverage consumed and aldehyde dehydrogenase-2 genotype. Alcohol. Clin. Exp. Res., 32, 1607–1614. [DOI] [PubMed] [Google Scholar]
- 5. Lachenmeier D. W. and Monakhova Y. B (2011) Short-term salivary acetaldehyde increase due to direct exposure to alcoholic beverages as an additional cancer risk factor beyond ethanol metabolism. J. Exp. Clin. Cancer Res., 30, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jokelainen K. Heikkonen E. Roine R. Lehtonen H. and Salaspuro M (1996) Increased acetaldehyde production by mouthwashings from patients with oral cavity, laryngeal, or pharyngeal cancer. Alcohol. Clin. Exp. Res., 20, 1206–1210. [DOI] [PubMed] [Google Scholar]
- 7. Kurkivuori J. Salaspuro V. Kaihovaara P. Kari K. Rautemaa R. Grönroos L. Meurman J. H. and Salaspuro M (2007) Acetaldehyde production from ethanol by oral streptococci. Oral Oncol., 43, 181–186. [DOI] [PubMed] [Google Scholar]
- 8. Brooks P. J. and Theruvathu J. A (2005) DNA adducts from acetaldehyde: implications for alcohol-related carcinogenesis. Alcohol, 35, 187–193. [DOI] [PubMed] [Google Scholar]
- 9. Balbo S. Meng L. Bliss R. L. Jensen J. A. Hatsukami D. K. and Hecht S. S (2012) Kinetics of DNA adduct formation in the oral cavity after drinking alcohol. Cancer Epidemiol. Biomarkers Prev., 21, 601–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daunais J. B. Davenport A. T. Helms C. M. Gonzales S. W. Hemby S. E. Friedman D. P. Farro J. P. Baker E. J. and Grant K. A (2014) Monkey alcohol tissue research resource: banking tissues for alcohol research. Alcohol. Clin. Exp. Res., 38, 1973–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grant K. A. Leng X. Green H. L. Szeliga K. T. Rogers L. S. and Gonzales S. W (2008) Drinking typography established by scheduled induction predicts chronic heavy drinking in a monkey model of ethanol self-administration. Alcohol. Clin. Exp. Res., 32, 1824–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baker E. J. Farro J. Gonzales S. Helms C. and Grant K. A (2014) Chronic alcohol self-administration in monkeys shows long-term quantity/frequency categorical stability. Alcohol. Clin. Exp. Res., 38, 2835–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Davenport A. T. Grant K. A. Szeliga K. T. Friedman D. P. and Daunais J. B (2014) Standardized method for the harvest of nonhuman primate tissue optimized for multiple modes of analyses. Cell Tissue Bank., 15, 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang M. Yu N. Chen L. Villalta P. W. Hochalter J. B. and Hecht S. S (2006) Identification of an acetaldehyde adduct in human liver DNA and quantitation as N 2-ethyldeoxyguanosine. Chem. Res. Toxicol., 19, 319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao L. Balbo S. Wang M. Upadhyaya P. Khariwala S. S. Villalta P. W. and Hecht S. S (2013) Quantitation of pyridyloxobutyl-DNA adducts in tissues of rats treated chronically with ®- or (S)-N′-nitrosonornicotine (NNN) in a carcinogenicity study. Chem. Res. Toxicol., 26, 1526–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang S. Villalta P. W. Wang M. and Hecht S. S (2006) Analysis of crotonaldehyde- and acetaldehyde-derived 1,n(2)-propanodeoxyguanosine adducts in DNA from human tissues using liquid chromatography electrospray ionization tandem mass spectrometry. Chem. Res. Toxicol., 19, 1386–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gunzerath L. Faden V. Zakhari S. and Warren K (2004) National Institute on Alcohol Abuse and Alcoholism report on moderate drinking. Alcohol. Clin. Exp. Res., 28, 829–847. [DOI] [PubMed] [Google Scholar]
- 18. Liu Y. Chen H. Sun Z. and Chen X (2015) Molecular mechanisms of ethanol-associated oro-esophageal squamous cell carcinoma. Cancer Lett., 361, 164–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu H. S. Oyama T. Matsuda T. Isse T. Yamaguchi T. Tanaka M. Tsuji M. and Kawamoto T (2012) The effect of ethanol on the formation of N 2-ethylidene-dG adducts in mice: implications for alcohol-related carcinogenicity of the oral cavity and esophagus. Biomarkers, 17, 269–274. [DOI] [PubMed] [Google Scholar]
- 20. Weinberg M. A. and Bral M (1999) Laboratory animal models in periodontology. J. Clin. Periodontol., 26, 335–340. [DOI] [PubMed] [Google Scholar]
- 21. Seitz H. K. Pelucchi C. Bagnardi V. and La Vecchia C (2012) Epidemiology and pathophysiology of alcohol and breast cancer: Update 2012. Alcohol Alcohol, 47, 204–212. [DOI] [PubMed] [Google Scholar]
- 22. Fanelli S. L. Maciel M. E. Díaz Gómez M. I. Delgado de Layño A. M. Bietto F. M. Castro J. A. and Castro G. D (2011) Further studies on the potential contribution of acetaldehyde accumulation and oxidative stress in rat mammary tissue in the alcohol drinking promotion of breast cancer. J. Appl. Toxicol., 31, 11–19. [DOI] [PubMed] [Google Scholar]
- 23. Jones A. W. and Andersson L (2003) Comparison of ethanol concentrations in venous blood and end-expired breath during a controlled drinking study. Forensic Sci. Int., 132, 18–25. [DOI] [PubMed] [Google Scholar]