Abstract
2,5-Dimethylfuran (DMF) and furfuryl alcohol (FFA) are two substituted furans that are formed during the processing of foods and have also been used as food flavorings. DMF and FFA are proposed to be bioactivated by human sulfotransferases (SULTs) which are not expressed in conventional cell lines used for genotoxicity testing. Therefore, in addition to the standard V79 cell line, we used a transfected V79 derived cell line co-expressing human cytochrome P450 (CYP) 2E1 and human SULT1A1 to assess the genotoxicity of DMF and FFA. The alkaline single cell gel electrophoresis (SCGE) assay was used to detect DNA damage in the form of single strand breaks and alkali-labile sites after exposure to DMF (0.5h; 0.5, 1, 1.5 or 2mM) or FFA (3h; 1, 3, 6 or 15mM). DMF induced DNA damage in V79 cells in a concentration-dependent manner irrespective of the expression of human CYP2E1 and SULT1A1. Almost no increase in the level of DNA damage was detected after exposure to FFA, except for a weak effect at the highest concentration in the transfected cell line. The results suggest that DNA damage in V79 cells from exposure to DMF detected by the alkaline SCGE assay is independent of human CYP2E1 and SULT1A1, and the genotoxic effect of FFA, as assessed by SCGE, is minimal in V79 cells.
Introduction
Substituted furans are formed when sugar-containing foods are heated or dehydrated during processing to prevent spoilage and reduce the risk of foodborne disease. 2,5-Dimethylfuran (DMF) and furfuryl alcohol (FFA) are examples of substituted furans found in foodstuffs that are frequently consumed by the general population. Some substituted furans, including the two mentioned, are added to foods as flavoring. DMF has been detected in coffee beans (217 μg/kg) and canned food (67 μg/kg) (1). Detected levels of FFA in foods and beverages range from high amounts in coffee (267–564 μg/g) (2) to relatively low amounts in many other types of foods such as cooked meats and dairy products (2–4). The daily intake of DMF (0.012 μg/capita/day) and FFA (180 μg/capita/day) from flavorings is considered by the European Food Safety Authority (EFSA) to be underestimated (5). Approval for use of DMF as a flavoring was on hold due to toxicological concerns over suspected reactivity towards DNA, but was in 2015 no longer supported as a flavoring substance by industry (6). The estimated intake of FFA is low enough to justify approval of FFA as a flavoring (5), but these estimates are rough estimates based on production volumes and do not include the much higher FFA levels formed during food processing.
Some of the substituted furans, including DMF and FFA, are anticipated to be activated to genotoxic metabolites (7). In vitro exposure to DMF led to the induction of micronuclei in murine bone marrow cells (8). DMF tested negative in the Ames tests (9,10), but tested positive in the rec-assay using Bacillus subtilis bacteria (11). Chromosomal aberration test results of exposure to DMF in CHO and V79 were mixed (12,13). The only published article on the in vivo effects of DMF did not show definitive evidence of DMF mediated genotoxicity in the in vivo alkaline single cell gel electrophoresis assay (14).
There has been no substantial evidence of genotoxicity caused by FFA using conventional in vitro Ames test systems (15). It is possible that DMF and FFA are bioactivated by SULTs within the cell and involve an intermediate metabolite that reacts with DNA at the site of activation (7). Many target cells of in vitro genotoxicity test systems do not include SULT and liver preparations lack the cofactor 3′-phosphoadenosine-5′-phosphosulfate (PAPS), and therefore may have provided false negative results on the genotoxicity of compounds activated by SULTs.
Previously, a genetically engineered Chinese hamster V79-derived cell line transfected with human cytochrome P450 (CYP) 2E1 and SULT1A1 (V79-hCYP2E1-hSULT1A1) has been used to detect hCYP2E1 and hSULT1A1-dependent promutagens (16–18). Genotoxicity tests report positive findings after in vitro exposure to FFA in a modified Ames test with relevant enzymes (19). FFA exposure produced both positive (20) and negative (21) results in chromosomal aberration tests of metabolically competent cells. The detection of DNA adducts in recent in vivo studies on the effect of FFA could explain the mechanism leading to positive results reported in modified test models that incorporate human sulfotransferases (SULTs) (22,23). Furthermore, there is evidence of the inhalation of FFA causing carcinogenic activity in rats (21). Moreover, DNA adducts of FFA have been detected in ten (four males and six females) non-tumorous human lung biopsies from lung tumor patients using isotope-dilution LC-MS/MS (24). Only DNA adducts of a handful of other xenobiotics have been detected in any human tissues with this most reliable technique.
In this study, we investigated the genotoxic potential of DMF and FFA and the role of hCYP2E1 and hSULT1A1 in the bioactivation of these compounds. This was accomplished by using a metabolically competent V79 cell line expressing hCYP2E1 and hSULT1A1. DNA from DMF and FFA exposed cells was analyzed using the alkaline single cell gel electrophoresis assay (SCGE).
Materials and methods
Cells and reagents
The Chinese hamster V79 cell line (V79, clone V79-MZ) (25) and genetically engineered V79-derived cell line expressing hCYP2E1 and hSULT1A1 (V79-hCYP2E1-hSULT1A1) (25,26) have previously been described.
2,5-Dimethylfuran (DMF, ≥99% purity, CAS no. 625-86-5), furfuryl alcohol (FFA, ≥98% purity, CAS no. 98-00-0) and dimethylsulfoxide (DMSO) were purchased from Sigma-Aldrich (St. Louis, MO, USA).
The cell lines were grown in Gibco® Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen Life Technologies™, Carslbad, CA, USA) containing 25mM D-Glucose, 4mM l-glutamine, 25mM HEPES and further supplemented with 5% fetal calf serum (FCS, Biochrome, Berlin, Germany), 1mM sodium pyruvate (Sigma-Aldrich), 100U/ml penicillin and 100U/ml streptomycin (Lonza, Basel, Switzerland). Prior to treatment, 7.5×105 cells were grown overnight as a monolayer culture in vented flasks at 37ºC in a humidified atmosphere with 5% CO2.
Treatment of V79 and V79-hCYP2E1-hSULT1A1 cells
V79 and V79-hCYP2E1-hSULT1A1 cells were exposed to either DMF (0.5, 1.0, 1.5 or 2.0mM) for 0.5h or FFA (1.0, 3.0, 6.0 or 15.0mM) for 3h in flasks (25cm2) and incubated in medium (10ml) at 37ºC. Due to the difference in toxicity between DMF and FFA (Figures 1 and 2), different treatment (concentration and time) were selected. The flasks were sealed tight during exposure due to the volatile nature of the substituted furans. DMF was dissolved in DMSO immediately before exposure; the final concentration of DMSO was ≤0.2%, whereas FFA was dispersed directly into the medium. The cells were washed with Dulbecco’s PBS (pH 7.4) without Ca2+ or Mg2+ prior to adding 1ml of pre-warmed trypsin (170 000U/L)/EDTA (Lonza) and incubated for 7min. Trypsination was arrested by adding ice-cold cell medium. Cells were centrifuged (300 × g, 2min) and the pellets were re-suspended in DMEM without FCS and placed on ice. A preparation was made of the cell suspension (20 µl) and Trypan Blue Stain (0.4%, Lonza, Basel, Switzerland) (1:1) to determine cell count and viability using an automated cell counter (BioRad Laboratories, Hercules, CA, USA). The positive assay control consisted of cell samples from each cell line (V79; V79-hCYP2E1-hSULT1A1) being subjected to 225 KeV X-rays filtered through 0.5mm Cu for a total dose of 10 Gy (3.07 Gy/min).
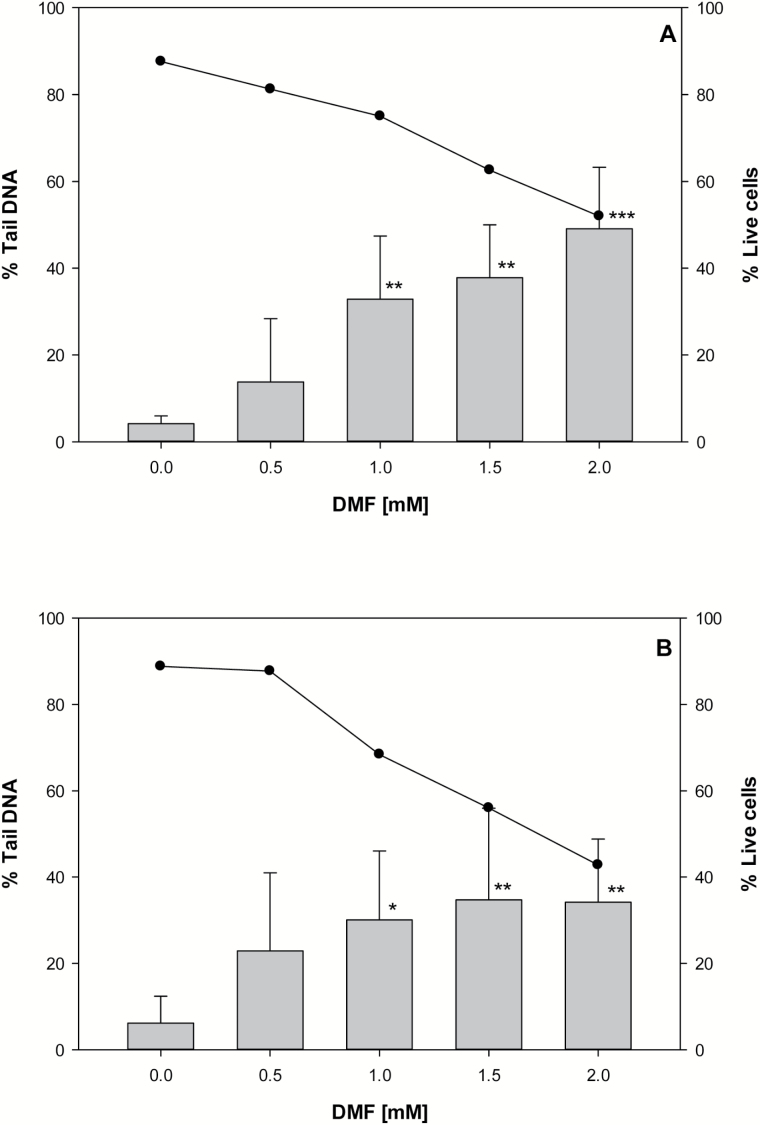
Figure 1.
Alkaline SCGE assay and cell viability of V79 cells (A) and V79-hCYP2E1-hSULT1A1 cells (B) exposed to DMF. DNA damage was detected after exposure to DMF for 30min. Vertical bars represent the mean from five experiments and whiskers indicate the standard deviation. The cell viability (•) is expressed as the mean of four experiments with (2,3) technical replicates within an experiment. Statistically significant differences against control; *P < 0.05, **P < 0.01 and ***P < 0.001.
Alkaline SCGE assay
The alkaline single-cell gel electrophoresis (SCGE) assay (27) was conducted according to an established protocol with some modification (28). The harvested cell suspensions were mixed with 0.75% low melting point agarose (NuSieve® GTG®Agarose, Lonza) using a ratio of 1:10. A series of four gel replicates (4 µl) were then embedded on cold Gelbond® hydrophilic polyester films (Lonza). The films were then immersed in a lysis buffer solution (2.5M NaCl, 0.1M Na2 EDTA, 0.01M Tris, 0.2M NaOH, 0.034M N-lauroylsarcosine, 10% DMSO, 1% Triton X-100, pH 10) overnight at 4ºC. Electrophoresis solution (0.3M NaOH, 0.001M Na2EDTA, pH > 13) was used to unwind (40min) the nucleoid double-stranded DNA into single strands prior to electrophoresis for 20min (0.8–1.0V/cm) at 8ºC with circulation. Films were then immersed in neutralization solution (0.32M Tris–HCl, 0.08M Tris-base, pH 7.5) before fixation in ethanol and dried. The DNA was stained using SYBR®Gold Nucleic Acid Gel Stain (Invitrogen Life Technologies™) and diluted (10 000×) in TE-buffer (10mM Tris–HCl, 1mM Na2EDTA, pH 8.0).
SCGE assay results were scored blind. For each category of exposure, thirty nuclei were scored from each of the four replicate gels using an Olympus BX51 fluorescent microscope (Olympus, Tokyo, Japan) connected to a CCD camera (BASLER A312f-VIS, Basler, Ahrensburg, Germany), and employing the Comet Assay IV image analysis system (Perceptive Instruments Ltd., Bury St. Edmunds, UK). DNA damage was defined as the tail intensity (TI) of the light captured from the migrated DNA in the tail as a percentage of the total intensity. Any overlapping DNA or DNA fragments were visually excluded.
Immunodetection of hCYP2E1 and hSULT1A1
The presence of heterologous enzymes (hCYP2E1 and hSULT1A1) was confirmed by Western blot. Cell lysis, protein measurement and Western blotting were performed as previously described with minor modifications (29). In short, frozen cells were thawed and mixed with 200 µl of lysis buffer (20mM Tris–HCl, 150mM NaCl, 1mM EDTA, 1mM EGTA, 2.4mM Na-pyrophosphate, 1.0mM orthovanadate, 1.0mM NaF, 21 µM leupeptin, 1.5 µM aprotinin, 15 µM pepstatin A, 1mM phenylmethylsulfonyl fluoride and 1% Triton-X) and sonicated using a BioRuptor (Diagenode, Seraing, BE, USA) prior to centrifugation (3000 × g for 15min).
Protein was measured using BioRad DC Protein Assay (BioRad Life Science, CA, USA). Glycerol, l-mercaptoethanol and SDS were added to the samples and dilutions were made to arrive at similar protein concentrations in both cell lines by adding more lysis buffer. Proteins (35 µg/well) were separated on 10% SDS-PAGE gel (100V, 30–45min; 200V, 30min) and transferred to a nitrocellulose membrane by blotting in circulated cold buffer (70V, 1.5h). Membranes were blocked using 3% bovine serum albumin (BSA) dissolved in Tris-buffer (10mM Tris, 0.15M NaCl, 0.1% tween-20, pH 7.6) and incubated with human monoclonal anti-CYP2E1 antibody (Cat. no. SAB1405688, Sigma-Aldrich) or anti-SULT1A1 antibody (Cat. no. SAB1406500, Sigma-Aldrich) overnight at 4ºC. Blots were washed before treatment with the secondary conjugated polyclonal Rabbit Anti-Mouse Immunoglobulin/horseradish peroxidase antilbody (Cat. no. P0260, Dako, Glostrup, Denmark). The Super-Signal® West Dura chemiluminescence system (Pierce, Perbio Science, Sweden) was used for developing the blots and chemiluminescence was captured using a CCD camera (Fuji LAS-1000, Raytest, Straubenhardt, Germany).
Statistical analysis
The mean of the median % tail DNA intensity (TI) from four technical replicate gels (30 nuclei analysed per gel) was calculated for each biological sample (n = 4 or 5). Data analysis was completed using SigmaPlot 12.0 (Systat Software, San Jose, CA, USA). A Two-Way ANOVA test was used to compare the treatment groups, followed by a post hoc Holm–Šídák test on groups against the respective control groups (P < 0.05).
Results
DNA damage and cell viability in DMF-treated V79 and V79-hCYP2E1-hSULT1A1 cells
Results from the alkaline SCGE assay showed an increase in DNA damage corresponding to increasing concentrations of DMF exposure in V79 and V79-hCYP2E1-hSULT1A1 cells (Figure 1). In the V79 cell line, DMF concentrations of 1.0, 1.5 and 2mM resulted in a significant 8-fold (P = 0.006), 9-fold (P = 0.002) and 12-fold (P < 0.001) increase in DNA damage compared to the unexposed cells, respectively. Likewise, the transfected V79 cells exposed to the same concentrations of DMF had a significantly 5-fold (P =0 .024), 6-fold (P = 0.013) and 6-fold (P = 0.011) higher amount of DNA damage than the unexposed cells. The tail intensity was also enhanced at the lowest concentration of DMF (0.5mM) in both cell lines, but this possible effect was not statistically corroborated.
The percentage of live cells decreased as the concentration of DMF exposure increased, with the lowest viability at the 2.0mM DMF for both V79 cells (52%) and V79-hCYP2E1-hSULT1A1 cells (43%) (Figure 1; Supplementary Table 1). On average, the percentage of live control cells were high (>85%) and approximately the same for both cell lines.
DNA damage and cell viability in FFA-treated V79 and V79-hCYP2E1-hSULT1A1 cells
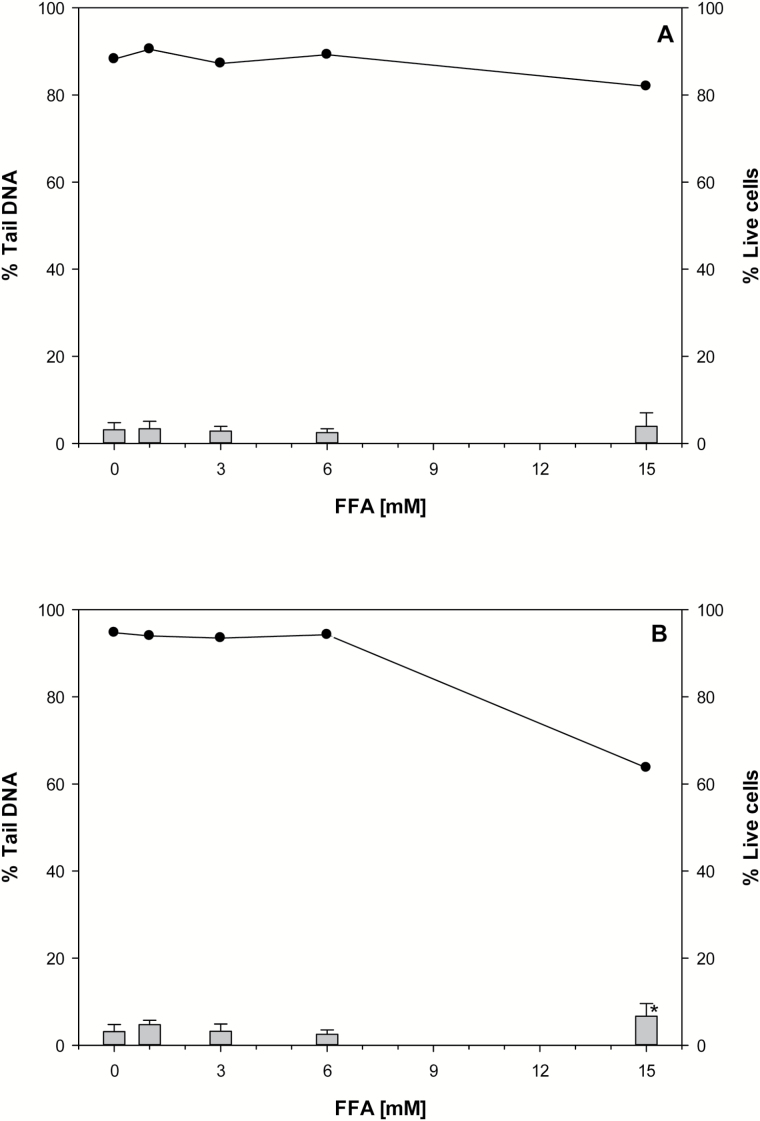
Results from the SCGE assay indicated a statistical significant (P = 0.04) increase in DNA damage in FFA exposed V79-hCYP2E1-hSULT1A1 cells compared to the control only at the highest concentration tested (15mM) (Figure 2).
Figure 2.
Alkaline SCGE assay and cell viability of V79 cells (A) and V79-hCYP2E1-hSULT1A1 cells (B) exposed to FFA. DNA damage was detected after exposure to FFA for 3h. Vertical bars represent the mean from four experiments and whiskers indicate the standard deviation. The cell viability (•) is expressed as the mean of three experiments with (2,3) technical replicates within an experiment. Statistically significant differences against control; *P < 0.05.
Cell viability remained high (>80%) at all the tested concentrations of FFA, except for V79-hCYP2E1-hSULT1A1 cells following exposure to 15mM of FFA which gave a live cell count of 64% (Figure 2; Supplementary Table 1).
Western Blot
The presence of transfected human CYP2E1 and SULT1A1 proteins in V79-hCYP2E1-hSULT1A1 cells was verified by Western blotting (Figure 3). Strong proteins bands corresponding to hCYP2E1 (54.2kDa) and hSULT1A1 (32.5kDa) were detected for the V79-hCYP2E1-hSULT1A1 cell samples, but not in those from the parental V79 cells (Figure 3).
Figure 3.
Detection of human CYP2E1 (A) and SULT1A1 (B) in V79-hCYP2E1-hSULT1A1 and V79 cells by Western Blot. Four samples from each cell line were tested for the presence of each protein. Each protein band is from one independent sample. Bands for hCYP2E1 correspond to MW ~54.2kDa; and bands for hSULT1A1 correspond to MW ~32.5kDa.
Discussion
FFA is able to induce gene mutations in bacteria upon activation by SULTs (19). This mutagenicity is mediated by benzylic DNA adducts at the exocyclic amino group of purine bases. The formation of these adducts has also been detected in mouse models and even in lung tissue in humans in vivo (30–32). Disruption of the Sult1a1 gene in mice led to a drastic decrease in the formation of these DNA adducts in mouse tissues, whereas transgenic human SULT1A1 enhanced the adduct formation in mouse models (30). Therefore, we performed the SCGE assay with FFA not only in control V79 cells, which are SULT-deficient, but also in a line engineered for expression of human SULT1A1. Somewhat surprisingly, FFA induced minimal DNA damage, as detected in the SCGE assay, only at the highest concentration used (15mM), which was considered too high to be biologically relevant. Even at this concentration the effect was weak. It is likely that alkaline SCGE assay may not be able to detect the type of damage induced by FFA, since adducts formed by FFA are shown to be bypassed by DNA polymerase and thereby will not necessarily lead to DNA strand brakes detected by the Comet assay (33).
DMF is not chemically reactive as such. To the best of our knowledge, no metabolism studies have been conducted with this compound. However, the structural similarities with furan and FFA could indicate potential activation pathways. Furan is oxidatively metabolized, under ring-opening, to cis-2-butene-1,4-dial, which is highly reactive (34). This reaction can be catalyzed by CYPs, in particular CYP2E1 (35). A homologous reaction with DMF would yield cis-3-hexene-2,5-dione. Such α,β-unsaturated bis-ketones are markedly less reactive than α,β-unsaturated bis-aldehydes. However, DMF, unlike furan, additionally contains two methyl group. Such benzylic positions are preferred sites for oxidative metabolic attack in numerous molecules. DMF is a very small molecule, and therefore CYP2E1 is a primary candidate that might mediate is side-chain hydroxylation. The expected primary product, 5-methylfurfuryl alcohol had demonstrated mutagenic activity—even stronger than that of FFA—in Salmonella strains expressing human SULTs (7). For these reasons, we performed SCGE assays in V79-hCYP2E1-hSULT1A1 cells as well as V79 control cells.
The results were surprising in two respects: (1) the positive response in control V79 cells and (2) the lack of any impact of the human enzymes tested. However, point 1 has a precedent: Furan induced SCE in V79 and hCYP2E1-hSULT1A1 cells in an equal manner. One may speculate that V79 express some non-CYP enzymes, e.g. peroxidases or oxidases that can mediate the oxidative ring opening of furan and DMF, which could lead to a cross binding of DNA by DMF. This could be a possible explanation for why the SCGE assay detects the DNA damage caused by DMF, but not by FFA (se above). It should also be noted that the toxicity of DMF and FFA in the V79 cells was very different. Alternatively, these compounds may lead to DNA damage through an unidentified mode of action, not requiring reactive metabolites. The missing influence of human enzymes (point 2) may be owed to different factors, most likely in combination. (2a) DMF may be relatively resistant to metabolism. Indeed it is excreted in rat urine as a terminal metabolite of hexane (36); (2b) DMF is only moderately lipohilic and is therefore not expected to accumulate in the cells. This is toxicokinetically unfavorable in cell culture models, in which the volume of the medium usually exceeds the volume of the cells by a factor 1000–100 000; (2c) The phase 1 metabolites of DMF are small, amphiliphilic molecules, expected to readily equilibrate with the extracellular space, i.e. formed by the enormous amount of medium present (2d); CYP2E1 may not be the optimal CYP form for metabolism of DMF; (2e) Potentially endogenous alcohol dehydrogenase expressed in V79 cells, which could further reduce the concentration of 5-methylfurfuryl alcohol, a potential proximate genotoxic; (2f) in the present study we noticed that possible DNA damage produced by FFA (even in SULT-expressing cells) is poorly detected in the SCGE assay; it is likely that the same is true for its congener, 5-methylfurfuryl alcohol. However, the genotoxicity of DMF and FFA in cells transfected with other metabolizing enzymes, like the V79-hCYP1A2-hSULT1A1 cells, could have given different results.
In conclusion, in vitro exposure to DMF causes a concentration-dependent increase in DNA damage in V79 and V79-hCYP2E1-hSULT1A1 cells which were readily detected by the alkaline SCGE assay. Tests using the alkaline SCGE assay indicate that the previously reported genotoxicity of FFA is not accompanied by strand breaks. The alkaline SCGE is a sensitive assay, but should be used in conjunction with other mutagenicity/genotoxicity tests to evaluate the overall potential hazard of substituted furans.
Supplementary data
Supplementary Table 1 is available at Mutagenesis Online.
Funding
This work was funded by the Research Council of Norway (project no. 204487).
Supplementary Material
Acknowledgements
The authors want to thank Tone Rasmussen for her excellent technical assistance, and we greatly acknowledge the experienced guidance of Hildegunn Dahl, Tonje Skuland and Jan Alexander.
Conflict of interest statement: None declared.
References
- 1. Fromberg A. Mariotti M. S. Pedreschi F. Fagt S. and Granby K (2014) Furan and alkylated furans in heat processed food, including home cooked products. Czech. J. Food Sci., 32, 443–448. [Google Scholar]
- 2. Murkovic M. and Swasti Y (2013) 5-Hydroxymethyl-furfural and furfuryl alcohol: occurrence, exposure and detection. Chemical food safety and health. Nova Publishers, New York, NY. [Google Scholar]
- 3. Karagül-Yüceer Y. Cadwallader K. R. and Drake M (2002) Volatile flavor components of stored nonfat dry milk. J. Agric. Food Chem., 50, 305–312. [DOI] [PubMed] [Google Scholar]
- 4. Pérez-Palacios T. Petisca C. Melo A. and Ferreira I. M (2012) Quantification of furanic compounds in coated deep-fried products simulating normal preparation and consumption: optimisation of HS-SPME analytical conditions by response surface methodology. Food Chem., 135, 1337–1343. [DOI] [PubMed] [Google Scholar]
- 5. Larsen J.C. Nørby K.K. Beltoft V.M. Lund P. and Binderup M.-L (2011) EFSA; Scientific Opinion on Flavouring Group Evaluation 66, Revision 1 (FGE. 66Rev1): Consideration of Furfuryl Alcohol and Related Flavouring Substances Evaluated by JECFA (55th meeting). European Food Safety Authority. [Google Scholar]
- 6. EFSA (2015) Scientific Opinion on Flavouring Group Evaluation 67 Revision 2 (FGE.67Rev2): Consideration of 28 furan-substituted compounds evaluated by JECFA at the 55th, 65th and 69th meetings (JECFA, 2001, 2006a, 2009b)1. EFSA J., 13, 4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Glatt H. Schneider H. Murkovic M. Monien B. H. and Meinl W (2012) Hydroxymethyl-substituted furans: mutagenicity in Salmonella typhimurium strains engineered for expression of various human and rodent sulphotransferases. Mutagenesis, 27, 41–48. [DOI] [PubMed] [Google Scholar]
- 8. Fromowitz M. Shuga J. Wlassowsky A. Y. Ji Z. North M. Vulpe C. D. Smith M. T. and Zhang L (2012) Bone marrow genotoxicity of 2,5-dimethylfuran, a green biofuel candidate. Environ. Mol. Mutagen., 53, 488–491. [DOI] [PubMed] [Google Scholar]
- 9. Zeiger E. Anderson B. Haworth S. Lawlor T. and Mortelmans K (1992) Salmonella mutagenicity tests: V. Results from the testing of 311 chemicals. Environ. Mol. Mutagen., 19, 2–141. [DOI] [PubMed] [Google Scholar]
- 10. Lee H., Bian S.S., Chen Y.L. (1994) Genotoxicity of 1, 3-dithiane and 1, 4-dithiane in the CHO/SCE assay and the Salmonella/microsomal test. Mutat. Res./Genetic Toxicol., 321, 213–218. [DOI] [PubMed] [Google Scholar]
- 11. Shinohara K. Kim E.-H. and Omura H (1986) Furans as the mutagens formed by amino-carbonyl reactions. Dev. Food Sci., 13, 353. [Google Scholar]
- 12. Stich H. F. Rosin M. P. Wu C. H. and Powrie W. D (1981) Clastogenicity of furans found in food. Cancer Lett., 13, 89–95. [DOI] [PubMed] [Google Scholar]
- 13. Ochi T., Ohsawa M. (1985) Participation of active oxygen species in the induction of chromosomal aberrations by cadmium chloride in cultured Chinese hamster cells. Mutat. Res. Lett., 143, 137–142. [DOI] [PubMed] [Google Scholar]
- 14. Høie A. H. Svendsen C. Brunborg G. Glatt H. Alexander J. Meinl W. and Husøy T (2015) Genotoxicity of three food processing contaminants in transgenic mice expressing human sulfotransferases 1A1 and 1A2 as assessed by the in vivo alkaline single cell gel electrophoresis assay. Environ. Mol. Mutagen., 56, 709–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mortelmans K. Haworth S. Lawlor T. Speck W. Tainer B. and Zeiger E (1986) Salmonella mutagenicity tests: II. Results from the testing of 270 chemicals. Environ. Mutagen., 8, 1–119. [PubMed] [Google Scholar]
- 16. Liu Y. and Glatt H (2010) Human cytochrome P450 2E1 and sulfotransferase 1A1 coexpressed in Chinese hamster V79 cells enhance spontaneous mutagenesis. Environ. Mol. Mutagen., 51, 23–30. [DOI] [PubMed] [Google Scholar]
- 17. Glatt H., Schneider H., Liu Y. (2005) V79-hCYP2E1-hSULT1A1, a cell line for the sensitive detection of genotoxic effects induced by carbohydrate pyrolysis products and other food-borne chemicals. Mutat. Res., 580, 41–52. [DOI] [PubMed] [Google Scholar]
- 18. Zhang C. Lai Y. Jin G. Glatt H. Wei Q. and Liu Y (2016) Human CYP2E1-dependent mutagenicity of mono- and dichlorobiphenyls in Chinese hamster (V79)-derived cells. Chemosphere, 144, 1908–1915. [DOI] [PubMed] [Google Scholar]
- 19. McGregor D. B. M. Prentice R. D. M. and Riach C. G (1981) Mutagenic activity of 123 compounds with known carcinogenic potential. Presented at 7th International Symposium on Chemical & Toxicological Aspects of Environmental Quality Inveresk Research International Limited, Musselburgh, September 7–10, London. [Google Scholar]
- 20. Stich H. Rosin M. San R. Wu C. and Powrie W (1981) Intake, formation, and release of mutagens by man. Banbury Report. [Google Scholar]
- 21. (1999) National Toxicology Program; Toxicology and Carcinogenesis Studies of Furfuryl Alcohol (CAS No. 98-00-0) in F344/N Rats and B6C3F1 Mice (Inhalation Studies). National Toxicology Program Technical Report Series, 482, 1. [PubMed] [Google Scholar]
- 22. Sachse B. Meinl W. Glatt H. and Monien B. H (2014) The effect of knockout of sulfotransferases 1a1 and 1d1 and of transgenic human sulfotransferases 1A1/1A2 on the formation of DNA adducts from furfuryl alcohol in mouse models. Carcinogenesis, 35, 2339–2345. [DOI] [PubMed] [Google Scholar]
- 23. Høie A. H. Monien B. H. Sakhi A. K. Glatt H. Hjertholm H. and Husøy T (2015) Formation of DNA adducts in wild-type and transgenic mice expressing human sulfotransferases 1A1 and 1A2 after oral exposure to furfuryl alcohol. Mutagenesis, 30, 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Monien B. H. Schumacher F. Herrmann K. Glatt H. Turesky R. J. and Chesné C (2015) Simultaneous detection of multiple DNA adducts in human lung samples by isotope-dilution UPLC-MS/MS. Anal. Chem., 87, 641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Glatt H. Gemperlein I. Setiabudi F. Platt K. L. and Oesch F (1990) Expression of xenobiotic-metabolizing enzymes in propagatable cell cultures and induction of micronuclei by 13 compounds. Mutagenesis, 5, 241–249. [DOI] [PubMed] [Google Scholar]
- 26. Liu Y. and Glatt H (2008) Mutagenicity of N-nitrosodiethanolamine in a V79-derived cell line expressing two human biotransformation enzymes. Mutat. Res., 643, 64–69. [DOI] [PubMed] [Google Scholar]
- 27. Singh N. P. McCoy M. T. Tice R. R. and Schneider E. L (1988) A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res., 175, 184–191. [DOI] [PubMed] [Google Scholar]
- 28. Gutzkow K. B. Langleite T. M. Meier S. Graupner A. Collins A. R. and Brunborg G (2013) High-throughput comet assay using 96 minigels. Mutagenesis, 28, 333–340. [DOI] [PubMed] [Google Scholar]
- 29. Burnette W. N. (1981) “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate–polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem., 112, 195–203. [DOI] [PubMed] [Google Scholar]
- 30. Sachse B. Meinl W. Glatt H. and Monien B. H (2014) The effect of knockout of sulfotransferases 1a1 and 1d1 and of transgenic human sulfotransferases 1A1/1A2 on the formation of DNA adducts from furfuryl alcohol in mouse models. Carcinogenesis, 35, 2339–2345. [DOI] [PubMed] [Google Scholar]
- 31. Monien B. H. Schumacher F. Herrmann K. Glatt H. Turesky R. J. and Chesné C (2015) Simultaneous detection of multiple DNA adducts in human lung samples by isotope-dilution UPLC-MS/MS. Anal. Chem., 87, 641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Høie A. H. Monien B. H. Sakhi A. K. Glatt H. Hjertholm H. and Husøy T (2015) Formation of DNA adducts in wild-type and transgenic mice expressing human sulfotransferases 1A1 and 1A2 after oral exposure to furfuryl alcohol. Mutagenesis, 30, 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ghodke P. P. Gore K. R. Harikrishna S. Samanta B. Kottur J. Nair D. T. and Pradeepkumar P. I (2016) The N(2)-Furfuryl-deoxyguanosine Adduct Does Not Alter the Structure of B-DNA. J. Org. Chem., 81, 502–511. [DOI] [PubMed] [Google Scholar]
- 34. Chen L. J. Hecht S. S. and Peterson L. A (1995) Identification of cis-2-butene-1,4-dial as a microsomal metabolite of furan. Chem. Res. Toxicol., 8, 903–906. [DOI] [PubMed] [Google Scholar]
- 35. Peterson L. A. Cummings M. E. Vu C. C. and Matter B. A (2005) Glutathione trapping to measure microsomal oxidation of furan to cis-2-butene-1,4-dial. Drug Metab. Dispos., 33, 1453–1458. [DOI] [PubMed] [Google Scholar]
- 36. Perbellini L. Amantini M. Brugnone F. and Frontali N (1982) Urinary excretion of n-hexane metabolites. Arch. Toxicol., 50, 203–215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.