Abstract
Single Strand Annealing (SSA) is a DNA double strand break (DSB) repair pathway that uses homologous repeats to bridge DSB ends. SSA involving repeats that flank a single DSB causes a deletion rearrangement between the repeats, and hence is relatively mutagenic. Nevertheless, this pathway is conserved, in that SSA events have been found in several organisms. In this review, we describe the mechanism of SSA and its regulation, including the cellular conditions that may favor SSA versus other DSB repair events. We will also evaluate the potential contribution of SSA to cancer-associated genome rearrangements, and to DSB-induced gene targeting.
Keywords: Single Strand Annealing, Homology Directed Repair, RAD51, RAD52, End Resection, Alternative End Joining
Chromosomal break repair by the Single Strand Annealing (SSA) pathway
A chromosomal double strand break (DSB, see Glossary) can be generated under several circumstances. DSBs can be directly induced by nucleases, such as topoisomerase II, which is important for untangling chromosomes, as occurs during chromosome condensation [1]. Other cellular processes that require induction of DSBs by nucleases include antibody maturation and meiosis [2, 3]. DSBs can also be formed as the result of nucleolytic cleavage of various DNA structures, such as DNA interstrand crosslinks, blocked or reversed DNA replication forks, and three-stranded DNA/RNA hybrids that result from transcription (i.e. R-loops) [4, 5]. Furthermore, DSBs can be induced at defined DNA sequences via site-specific endonucleases [6]. Apart from nucleases, clastogenic (i.e. chromosomal breaking) agents, including small molecules and ionizing radiation, can cause cleavage of phosphodiester bonds and result in DSBs. Sources of clastogens include byproducts of cellular metabolism, and environmental agents [7]. Additionally, clastogens are commonly applied as anti-cancer agents, in the form of small molecule chemotherapeutics and ionizing radiation [8]. Thus, characterizing the pathways that repair DSBs is important to understand both genome stability and the cellular response to clastogenic cancer therapeutics.
The focus of this review is the Single Strand Annealing (SSA) DSB repair pathway. SSA involves annealing of homologous repeat sequences that flank a DSB, which causes a deletion rearrangement between the repeats (Fig 1). SSA events have been demonstrated in mammalian cells and in several model organisms including S. cerevisiae, A. thaliana, D. melanogaster, and C. elegans [9–13]. A study from the Sternberg lab in 1984 provided early evidence for SSA using DNA plasmid substrates harboring homologous repeats transfected into mouse cells, and proposed a model for the steps of this pathway [13]. This model involves a DSB between homologous repeats, followed by DSB end resection that generates 3′ ssDNA, which reveals flanking homologous sequences that are annealed together to form a synapsed intermediate (Fig 1). This intermediate is then processed for ligation, which requires endonucleolytic cleavage of non-homologous 3′ ssDNA tails, and polymerase filling of any gaps (Fig 1). Studies of individual repair factors have provided evidence for many of these proposed steps of SSA.
Figure 1.
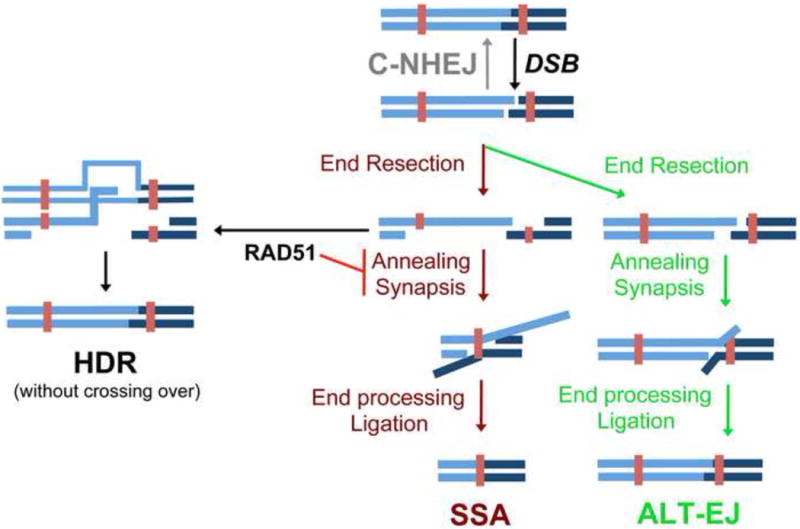
Key Figure. Comparisons between Single Strand Annealing (SSA) and distinct double-strand break (DSB) repair events: Homology Directed Repair (HDR), Alternative End Joining (ALT-EJ), and Canonical Non-Homologous End Joining (C-NHEJ). End resection is depicted as a common intermediate of SSA, HDR, and ALT-EJ. In the subsequent steps of SSA, homologous repeats (depicted as rust colored boxes) anneal to form the synapsis intermediate that is then processed for ligation. The RAD51 recombinase is shown to inhibit SSA, and conversely mediate the strand invasion step of HDR.
Functional analysis of several factors has established that DSB end resection, which refers to the generation of 3′ ssDNA, is a pivotal step of SSA (Fig 1). For example, SSA is dependent on CtIP [14, 15], which is a key end resection factor, based on cell biology measurements of ssDNA formation at sites of DNA damage, as well as physical analysis of site-specific endonuclease-generated chromosomal DSBs [16–18]. Conversely, factors that inhibit end resection have been found to suppress SSA, such as the DNA damage response pathway involving H2AX, RNF168, 53BP1, and RIF1 [14, 18–21]. Beyond these examples, numerous other factors that promote/inhibit end resection have a corresponding effect on SSA [22–37]. Furthermore, this correlation is conserved, since many of the homologs/orthologs of these factors in S. cerevisiae affect both SSA and end resection [9, 38–41]. A corollary of these findings is that SSA assays can be a useful screening tool for genes and/or small molecules that are candidates for affecting end resection. As one example, bortezomib, a small molecule proteasome inhibitor used for treating multiple myeloma [42], disrupts both SSA and end resection [26].
After end resection, the next steps of SSA involve annealing of the flanking repeats, followed by removal of the non-homologous 3′ ssDNA tails, which are mediated by RAD52 and ERCC1, respectively. RAD52 is a DNA binding protein that can mediate annealing of ssDNA substrates [43–45]. ERCC1 associates with XPF to form a protein complex that is proficient at nucleolytic cleavage of 3′ ssDNA tails [46]. Furthermore, this nuclease activity of the ERCC1/XPF complex is enhanced by RAD52 [46]. Both RAD52 and ERCC1 are important for SSA in mammalian cells, as are the homologs of these factors in S. cerevisiae [9, 32, 43, 44, 47, 48]. Following these steps of annealing and 3′ ssDNA tail removal, any gaps are filled by DNA polymerases to generate the substrates for a DNA ligase to complete SSA, as mentioned above. Although, the specific polymerases and ligases required for completion of SSA remain poorly understood.
The outcome of DSB repair via SSA is a deletion rearrangement between homologous repeats, and hence results in loss of genetic information. Given that SSA is a relatively mutagenic repair event, what are the regulatory mechanisms that may limit SSA in favor of less mutagenic repair? Conversely, what are the cellular conditions under which SSA would be a favored repair event? We have sought to consider these questions, which are related to distinctions between SSA and other DSB repair pathways. Furthermore, we will discuss the influence of SSA on chromosomal rearrangements associated with cancer, and on gene targeting.
Contrasting SSA with other DSB repair pathways
In addition to SSA, two other DSB repair pathways are also initiated by end resection: Alternative End Joining (ALT-EJ), and Homology-Directed Repair (HDR). ALT-EJ involves annealing of short homologous repeats (called microhomology) that flank a DSB to bridge the break (Fig 1) [15, 49–54]. The outcome of ALT-EJ is a deletion mutation between the repeats. Accordingly, ALT-EJ is relatively similar to SSA, as both pathways involve annealing of flanking repeats to bridge the DSB. ALT-EJ is also referred to as micro-SSA [55], as well as Microhomology Mediated End Joining [56].
The HDR pathway involves strand invasion of the end resection intermediate into a homologous template, which initiates the synthesis of a nascent DNA strand that can anneal to the other side of the DSB (Fig 1). While SSA and ALT-EJ cause deletions between the annealed homology/microhomology, HDR has the potential to be restorative if the template for recombination is the identical sister chromatid. However, HDR between tandem repeats that is resolved by crossing over causes a deletion rearrangement that is indistinguishable from the outcome of SSA, although HDR with crossing over is apparently infrequent in mitotic cells [57, 58]. Regarding mechanism, the requirement for RAD51 is a key distinction between HDR versus SSA. RAD51 is the eukaryotic RecA homolog that catalyzes the strand invasion step of HDR [59–64]. Conversely, SSA does not require RAD51, and indeed disruption of RAD51, or RAD51 mediator proteins (e.g. BRCA2 and RAD54), causes a substantial increase in the frequency of SSA in several organisms [9, 10, 12, 32, 65, 66].
In contrast to the above repair events, Canonical Non-Homologous End Joining (C-NHEJ) is a repair pathway that does not necessarily require the formation of an annealed intermediate to bridge the DSB before ligation [2]. The C-NHEJ pathway is particularly critical for repair of the DSB-induced rearrangements during antibody maturation, and is mediated by several factors, which include KU70/80, XRCC4, and DNA Ligase IV [2]. C-NHEJ repair has the potential to restore the original DNA sequence, but if the DSB ends are processed prior to ligation via nucleases/polymerases, C-NHEJ can cause insertion or deletion mutations [67]. C-NHEJ does not require end resection, and indeed suppresses end resection [18, 68], along with each of the three repair events described above that involve end resection (SSA, ALT-EJ, and HDR) [15, 32, 69, 70]. Conversely, components of the Fanconi anemia core complex, which both inhibits C-NHEJ and promotes CtIP recruitment to DNA crosslink damage, are important for SSA, ALT-EJ, and HDR [26, 71–74]. Accordingly, inhibition of C-NHEJ to enable end resection appears a common step of SSA, ALT-EJ, and HDR.
Specific distinctions between SSA and ALT-EJ
While the SSA and ALT-EJ pathways both involve annealing of flanking homology to bridge a DSB, these events show key mechanistic distinctions (Fig 2). In particular, the factors that mediate synapsis of the annealing intermediate appear to be distinct. As described above, RAD52 plays a conserved mediator role in SSA. In contrast, RAD52 is dispensable for ALT-EJ in both S. cerevisiae and mammalian cells, based on events that involve ≤10 bp of microhomology [15, 47, 69, 75]. Thus, RAD52 appears to be specifically important for repeat-mediated rearrangements that involve long regions of homology.
Figure 2.
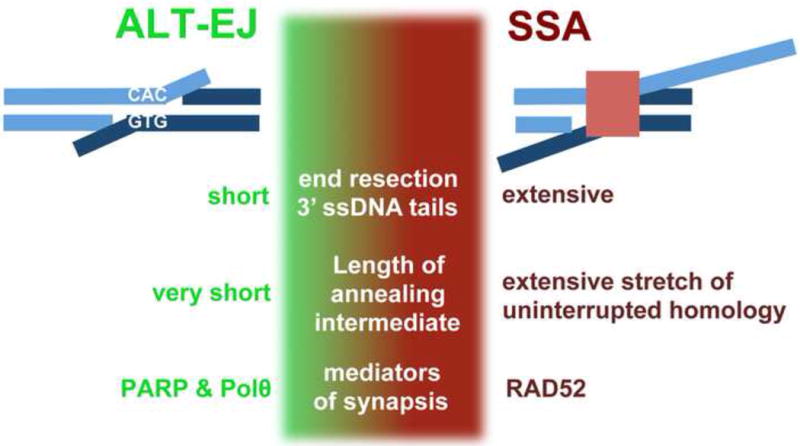
Shown are key distinctions between ALT-EJ and SSA, with diagrams of the annealed/synapsed intermediate for each pathway. Distinct features include the extent of end resection involved and hence the length of 3′ ssDNA that is cleaved during processing of this intermediate, the length of the homologous repeat involved in the annealing, and the relative role of PARP and Polθ versus RAD52. The precise boundaries of these features that define ALT-EJ versus SSA remain unclear. Indeed, we represent the possibility that these features exist along a spectrum with a middle region that could potentially be repaired by either pathway. For example, it is conceivable that intermediate lengths of annealed homology are repaired by either ALT-EJ or SSA (i.e. mediated by PARP and Polθ, or RAD52). Similarly, a moderate extent of resection could be sufficient to reveal such intermediate lengths of flanking homology. However, it is also possible one or more of these features have defined boundaries that distinguish repair by ALT-EJ versus SSA.
In contrast to RAD52, poly-ADP-ribose polymerase (PARP) and DNA polymerase theta (Polθ) appear to be specifically important for ALT-EJ. Disrupting PARP function, either by depleting PARP1 or inhibiting PARP activity with small molecules (e.g. Olaparib treatment), causes ALT-EJ defects in multiple assay systems [26, 52, 53, 76]. Furthermore, PARP appears to specifically mediate ALT-EJ, since Olaparib treatment shows no clear effect on SSA [26]. The precise role of PARP in ALT-EJ remains unclear, but this factor has been shown to promote DNA end synapsis [53], and furthermore is important for recruitment of Polθ to LASER induced DNA damage [77]. Polθ plays a conserved role in mediating ALT-EJ, likely through its capacity to extend templates that are stabilized by annealed microhomology, although it may also play a direct role in end synapsis [78–82].
Understanding the relative role of PARP, Polθ, and RAD52 on repair events involving repeats of varying lengths of uninterrupted homology remains an important mechanistic question for distinguishing ALT-EJ from SSA (Fig 2). Specifically, it will be important to define the length of annealed homology involved in these two pathways. However, such a precise definition may prove difficult. Namely, it is conceivable that intermediate lengths of homology might be synapsed and repaired by either pathway (Fig 2). Alternatively, there may be a clear boundary of homology length that demarcates repair by SSA versus ALT-EJ.
Regarding other factors, the ERCC1/XPF nuclease and the mismatch repair (MMR) machinery also influence SSA and/or ALT-EJ, but their precise role depends on the context of the analysis. As mentioned above, the ERCC1/XPF nuclease plays a major and conserved role in SSA. In contrast, the requirement for this nuclease during ALT-EJ depends on the context: it has a limited role in chromosomal ALT-EJ and plasmid EJ assays in mammalian cells, however it is important for EJ repair in C-NHEJ-deficient (i.e. Ku86−/−) cells [15, 83]. Furthermore, in S. cerevisiae, the homologous nuclease complex, RAD10/RAD1, is important for ALT-EJ [69].
The influence of the MMR (Mismatch Repair) machinery on SSA and ALT-EJ is also complex. In S. cerevisiae, the MSH2/MSH3 complex appears to mediate 3′ ssDNA processing in concert with RAD10/RAD1, and is important for SSA [48], but dispensable for ALT-EJ [84]. However, for SSA between repeats that are not perfectly homologous (called homeologous sequences), the MMR machinery (e.g. MSH6) appears important to suppress such events, via a process called heteroduplex rejection [85–87]. Thus, the MMR machinery appears to play a dual role during SSA: heteroduplex rejection of divergent sequences, and processing of the annealed/synapsed intermediate [85–87]. In contrast, MSH2 appears dispensable for SSA in mammalian cells [25]. Thus, MMR appears to influence SSA at multiple steps in S. cerevisiae, whereas the role of MMR on SSA and ALT-EJ in mammalian cells is not entirely understood.
Cellular contexts that may favor SSA versus other repair events
In addition to involving different factors, distinct repair pathways can be affected by various cellular contexts, including cell cycle phase and the presence of the sister chromatid (Fig 3). Cell cycle phase affects DSB repair, because end resection is specifically activated in the S/G2-phases, which is mediated in part by cyclin-dependent kinase phosphorylation of CtIP (or SAE2 in S. cerevisiae) [16, 17, 20, 88, 89]. As mentioned above, such end resection is important for ALT-EJ, SSA, and HDR, but not C-NHEJ. In contrast, the presence of the sister chromatid is only important for HDR. Namely, the sister chromatid is the preferred template for HDR [90], but is not involved in repair by ALT-EJ or SSA.
Figure 3.
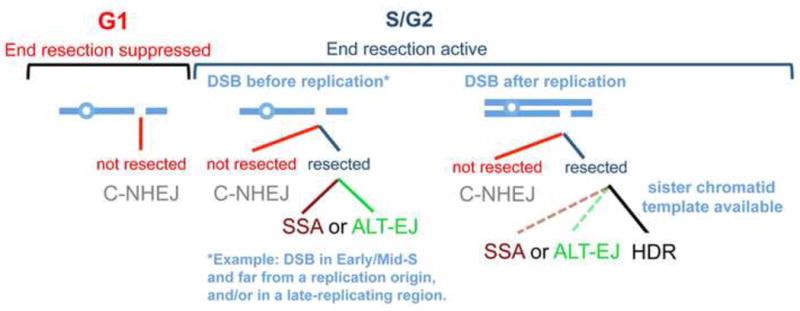
Shown is a model for the influence of cell cycle phase and the presence of the sister chromatid on DSB repair pathways. G1 phase is shown as being suppressed for end resection, such that a DSB in this context would likely be repaired by C-NHEJ. S/G2 phases are shown as permissive for end resection. The middle chromosome diagram shows a DSB occurring in S-phase but before this particular chromosome (or at least the region at the DSB) has been replicated. In this context, the sister chromatid is unavailable for HDR, such that if this DSB is resected, it will likely be repaired by SSA or ALT-EJ. In contrast, the chromosome on the right has been replicated prior to DSB formation, such that HDR with the sister chromatid template is feasible. Thus, HDR is depicted as favored in this context over SSA or ALT-EJ (i.e. the solid line versus the dashed lines). Also shown is the notion that DSBs are not necessarily end resected in S/G2, such that C-NHEJ is active throughout the cell cycle.
Importantly, these two cellular contexts are not necessarily coincident. Specifically, prior sister chromatid synthesis is not required for end resection, based on evidence that expressing a mutant of CtIP that mimics cyclin-dependent kinase phosphorylation (T847E) appears sufficient to support end resection in G1 phase cells [88]. Thus, a DSB that occurs in S phase may be resected, even in the absence of the sister chromatid. Since a resected DSB in this context lacks the preferred template for HDR [90], such DSBs are likely repaired by SSA or ALT-EJ (Fig 3).
Another circumstance that likely affects the relative frequency of SSA, HDR, or ALT-EJ is related to the extent of end resection. Namely, extensive resection increases the probability of revealing long stretches of uninterrupted homology flanking a DSB. Thus, extensive end resection may favor SSA (Fig 2). Regarding such extensive resection, the upper limit on this process is unknown, although SSA events causing deletions >25 kb are readily detected in S. cerevisiae [91]. In contrast, limited resection may be sufficient to uncover short flanking microhomology for ALT-EJ. In between these extremes, a moderate extent of resection could reveal the homology required for RAD51-mediated strand invasion for HDR. Such moderate resection could also uncover flanking homologous regions that are of an intermediate length, which could potentially be repaired by either ALT-EJ or SSA, as described above (Fig 2). Thus, factors that regulate the extent of resection likely cause differential effects on these pathways. Consistent with this notion, the DNA damage response pathway involving the end resection inhibitor 53BP1 appears important for the balance of repair in favor of HDR versus SSA [14, 19, 110].
Finally, additional cellular contexts have been demonstrated to facilitate HDR beyond cell cycle phase and the presence of the sister chromatid. Specifically, DSB chromatin state and capacity for re-localization to the nuclear periphery have been shown to influence the formation of HDR intermediates (i.e. RAD51 recruitment to DSBs) [92, 93]. Whether such conditions are specifically important for HDR, or may also facilitate SSA and/or ALT-EJ, is unknown.
The influence of SSA on chromosomal rearrangements
While SSA is capable of restoring a broken chromosome to avoid its loss during cell division, overreliance on SSA could be catastrophic to mammalian genomes, given the high density of repetitive elements. For example, the human genome contains approximately 1 million copies of Alu-type Short Interspersed Elements (SINEs) [94]. Furthermore, several cancer-associated genes are enriched for repetitive elements [95]. Indeed, the BRCA1 gene alone contains 129 Alu elements, which represent approximately 40% of the gene, and these Alu elements appear at the junctions of several inherited cancer-associated rearrangements in BRCA1 [96]. Alu elements also contribute to several other cancer-associated rearrangements [94], including in the MLL gene [97]. As well, SSA involving multiple DSBs can cause larger rearrangements. For example, SSA between repeats that flank two DSBs on different chromosomes can cause translocations [98, 99]. Furthermore, ionizing radiation exposure can induce chromosomal rearrangements mediated by repeat sequences, i.e. Ty retrotransposon-element mediated rearrangements in S. cerevisiae [100].
Apart from such individual examples of repeat-mediated rearrangements, it is difficult to fully assess the influence of SSA on chromosomal rearrangements, because of two features of SSA. For one, next generation sequencing approaches are often insufficient for a comprehensive detection of rearrangements involving repetitive elements [101], which are the likely outcome of SSA. Nevertheless, a recent genome-wide analysis of human copy number variations identified several inherited chromosomal rearrangements that appear mediated by long interspersed element (LINE) repeat sequences [102]. This advance in identifying inherited structural variants that are repeat-mediated provides promise for identifying such rearrangements in somatic cells, including malignant cells.
The second confounding feature of SSA relates to the influence of sequence divergence on repair. Namely, SINE-mediated rearrangements can involve repeats with substantial sequence divergence [94], which limits the length of uninterrupted homology that could form a stable annealing/synapsis intermediate. A recent study using divergent repeats as substrates for DSB-induced deletion rearrangements in human cells has provided a framework to define the influence of SSA on such rearrangements [47]. Sequence divergence was introduced as dispersed mutations between two Alu elements, but also included analysis of natural diverged Alu elements. Increased divergence caused a progressive reduction in the percentage of deletion rearrangements that resulted in one complete chimeric Alu element (i.e. the outcome consistent with SSA). Similarly, an earlier study demonstrated that divergent Alu elements show reduced DSB-induced SSA in mouse cells, compared to identical Alu elements [98]. In the more recent study, RAD52 was shown to be important to mediate the rearrangements between the identical Alu elements, but showed a reduced influence on the rearrangements mediated by divergent repeats [47]. Thus, rearrangements between divergent repeats could be caused by ALT-EJ, or possibly a composite mechanism of ALT-EJ and SSA.
Regarding such a composite mechanism, this study raised a model for a repair event termed homeology-influenced NHEJ [47]. In this model, divergent Alu elements form a transient synaptic SSA intermediate that is a substrate for endonucleolytic cleavage involving the MMR machinery. Subsequently, the synaptic structure is dissolved, and the cleaved DSB ends are resolved by EJ. In support of this model, this study found that the presence of flanking divergent Alu elements enhanced the frequency of large deletions mediated by EJ [47]. This model reinforces the notion that examining repair outcomes alone may be insufficient to fully understand the mechanistic steps that cause rearrangements. Thus, further defining the factors that influence the frequency of rearrangements between sequences with varying lengths of uninterrupted homology will provide insight into the etiology of chromosomal rearrangements.
Multiple repair pathways influence gene targeting
SSA may also contribute to gene targeting under certain circumstances. Gene targeting, or homology-directed gene editing, refers to site-specific modification of genomic DNA using an exogenous donor template that is homologous to the target site. Spontaneous homologous gene targeting is infrequent in mammalian cells, but is enhanced substantially by a DSB at the target site [103]. Such targeted DSBs have become technically straightforward with the development of modular site-specific nucleases such as the RNA-guided nuclease, Cas9 [6]. Furthermore, mutant forms of Cas9 enable induction of single-strand nicks, which can also induce gene targeting [6].
The relative contribution of SSA, HDR, and ALT-EJ to gene targeting appears to be influenced by the context of the targeting event, including the inducing lesion (DSB vs. single-strand nick), and the nature of the template. A typical design for gene targeting involves DSB induction and use of a dsDNA template that contains extensive homology to the target site [103]. Such targeting events appear to utilize HDR, based on the requirement for RAD51, as demonstrated both by RAD51 depletion [104], and RAD51 overexpression [105].
However, not all targeting events require RAD51, and hence may involve a form of SSA and/or ALT-EJ. For example, gene targeting in human cells using a short ssDNA donor and induced by a single-strand nick was suppressed by RAD51 [104]. Similarly, gene targeting in S. cerevisiae using a short ssDNA donor, although in this case induced by a DSB, was also suppressed by RAD51 [106]. In the latter study, this gene targeting event was shown to be promoted by RAD52, and hence is consistent with SSA [106]. In addition to HDR and SSA, DSB-induced gene targeting events involving ALT-EJ have also been demonstrated, using dsDNA donor templates with short stretches of homology (i.e. 5–25 bp) to the target site [107]. Future studies investigating the role of individual factors (e.g. RAD51, RAD52, PARP, and Polθ) on multiple distinct gene targeting events, will help elucidate the relative contribution of HDR, SSA and ALT-EJ to this process. Such studies are important for developing novel methods to improve gene targeting efficiency, and thereby broaden its therapeutic applications.
Concluding Remarks and Future Perspectives
While relatively mutagenic in terms of causing a rearrangement between repeat elements, SSA may be important to restore a broken chromosome with DSB ends that have undergone extensive end resection, but are unable to be resolved by HDR or ALT-EJ. We have proposed some scenarios that may favor SSA, which include the state of the cell cycle, absence of the sister chromatid, and the length of uninterrupted homology. In addition, genetic deficiencies or impaired expression of factors important for RAD51 function, such as BRCA2 mutations associated with breast/ovarian cancer risk, could also cause increased reliance on SSA [32, 65]. Accordingly, the influence of SSA and repeat-mediated rearrangements in cancer etiology could be specific for particular subsets of malignancies. Furthermore, with the advances in identifying small molecules targeting RAD52 [108, 109] and end resection [26], understanding the reliance of individual tumors on SSA will be important for continued development of these agents as therapeutics. In summary, important future directions include defining the precise cellular conditions that favor SSA versus other DSB repair events, along with determining the influence of repeat-mediated rearrangements and SSA in individual malignancies.
OUTSTANDING QUESTIONS BOX.
What are the precise mechanisms that distinguish SSA and ALT-EJ, including the factors that mediate these events, which is connected to defining how the length of uninterrupted homology affects the mechanisms involved?
What are the factors and conditions that regulate the extent of end resection, and thereby affect the relative frequency of SSA, HDR, and ALT-EJ?
Apart from cell cycle phase and the presence of the sister chromatid, what other cellular contexts influence SSA, HDR, and/or ALT-EJ? Are there specific mechanisms for detecting the presence of the sister chromatid to favor HDR versus other repair pathways?
Chromatin context adjacent to DSBs, as well as the capacity for DSBs to mobilize to the nuclear periphery appear important for HDR. Do these features of DSBs similarly affect SSA and/or ALT-EJ?
To what extent are somatic chromosomal rearrangements mediated by repeat elements?
While most gene targeting events are likely mediated by HDR, what are the various targeting scenarios that instead involve SSA or ALT-EJ?
TRENDS BOX.
The SSA DSB repair pathway causes a rearrangement between two homologous repeats.
SSA and ALT-EJ are similar, as they both involve an annealed intermediate to synapse a DSB, but show mechanistic distinctions, including differential requirements for RAD52 versus PARP and Polθ.
End resection to generate 3′ ssDNA is a pivotal step of SSA, and is a shared intermediate with HDR and ALT-EJ, but each of these events likely require distinct degrees of end resection. Also, if end resection occurs prior to sister chromatid synthesis, which is the preferred template for HDR, then SSA or ALT-EJ may be required for repair.
The relative role of SSA versus ALT-EJ on repeat-mediated rearrangements is affected by the degree of divergence between the repeats.
Most gene targeting events are mediated by HDR (i.e. RAD51-dependent), however some targeting approaches appear to involve SSA or ALT-EJ.
Acknowledgments
Funding support provided by the National Cancer Institute of the National Institutes of Health [R01CA120954 and R01CA197506 to J.M.S.]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
GLOSSARY
- Alternative End Joining (ALT-EJ)
a DSB repair event that is independent of the C-NHEJ pathway, and is often mediated by short stretches of homology to bridge the DSB prior to ligation
- Canonical Non-Homologous End Joining (C-NHEJ)
a major pathway of DSB repair that does not require any homology to bridge the DSB prior to ligation. This pathway is critical for the repair of DSBs that are induced during antibody maturation, particularly V(D)J recombination and class switch recombination
- Double strand break (DSB)
a chromosomal DSB is the cleavage of two phosphodiester bonds on opposing strands that are in close enough proximity to cause loss of the continuous DNA double helix
- End resection
the processing of chromosomal DSB ends to generate 3′ single stranded DNA (ssDNA)
- Gene targeting
the method of editing chromosomal DNA to match the sequence of an exogenous donor, which shares homology with the chromosomal target site. Gene targeting is substantially enhanced by causing a DNA break at the chromosomal target
- Homology-Directed Repair (HDR)
a DSB repair event that involves invasion of a homologous template by at least one strand from the DSB, which templates nascent DNA synthesis to form an extended strand that can form a bridge to the other DSB end. The homologous strand invasion step is catalyzed by the RAD51 recombinase
- Single Strand Annealing (SSA)
a DSB repair event that uses flanking homology to bridge the DNA lesion, causing a deletion between the repeats
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 4.Hanada K, et al. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. Embo J. 2006;25:4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sollier J, et al. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell. 2014;56:777–785. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins A, et al. The comet assay as a tool for human biomonitoring studies: the ComNet project. Mutat Res Rev Mutat Res. 2014;759:27–39. doi: 10.1016/j.mrrev.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Thoms J, Bristow RG. DNA repair targeting and radiotherapy: a focus on the therapeutic ratio. Semin Radiat Oncol. 2010;20:217–222. doi: 10.1016/j.semradonc.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Ivanov EL, et al. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Do AT, et al. Double-strand break repair assays determine pathway choice and structure of gene conversion events in Drosophila melanogaster. G3 (Bethesda) 2014;4:425–432. doi: 10.1534/g3.113.010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orel N, et al. Different pathways of homologous recombination are used for the repair of double-strand breaks within tandemly arranged sequences in the plant genome. Plant J. 2003;35:604–612. doi: 10.1046/j.1365-313x.2003.01832.x. [DOI] [PubMed] [Google Scholar]
- 12.Pontier DB, Tijsterman M. A robust network of double-strand break repair pathways governs genome integrity during C. elegans development. Curr Biol. 2009;19:1384–1388. doi: 10.1016/j.cub.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 13.Lin FL, et al. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984;4:1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munoz MC, et al. RING finger nuclear factor RNF168 is important for defects in homologous recombination caused by loss of the breast cancer susceptibility factor BRCA1. J Biol Chem. 2012;287:40618–40628. doi: 10.1074/jbc.M112.410951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennardo N, et al. Alternative-NHEJ Is a Mechanistically Distinct Pathway of Mammalian Chromosome Break Repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, et al. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, et al. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 2014;42:e19. doi: 10.1093/nar/gkt1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie A, et al. Control of sister chromatid recombination by histone H2AX. Mol Cell. 2004;16:1017–1025. doi: 10.1016/j.molcel.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Escribano-Diaz C, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Hakim O, et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 2012;484:69–74. doi: 10.1038/nature10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sturzenegger A, et al. DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA end resection in human cells. J Biol Chem. 2014;289:27314–27326. doi: 10.1074/jbc.M114.578823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang YG, et al. Conditional deletion of Nbs1 in murine cells reveals its role in branching repair pathways of DNA double-strand breaks. EMBO J. 2006;25:5527–5538. doi: 10.1038/sj.emboj.7601411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gunn A, et al. Correct end use during end joining of multiple chromosomal double strand breaks is influenced by repair protein RAD50, DNA-dependent protein kinase DNA-PKcs, and transcription context. J Biol Chem. 2011;286:42470–42482. doi: 10.1074/jbc.M111.309252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennardo N, et al. Limiting the Persistence of a Chromosome Break Diminishes Its Mutagenic Potential. PLoS Genet. 2009;5:e1000683. doi: 10.1371/journal.pgen.1000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howard SM, et al. DNA damage response factors from diverse pathways, including DNA crosslink repair, mediate alternative end joining. PLoS Genet. 2015;11:e1004943. doi: 10.1371/journal.pgen.1004943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karanja KK, et al. DNA2 and EXO1 in replication-coupled, homology-directed repair and in the interplay between HDR and the FA/BRCA network. Cell Cycle. 2012;11:3983–3996. doi: 10.4161/cc.22215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costelloe T, et al. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature. 2012;489:581–584. doi: 10.1038/nature11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X, et al. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature. 2012;489:576–580. doi: 10.1038/nature11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onyango DO, et al. Tetratricopeptide repeat factor XAB2 mediates the end resection step of homologous recombination. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen J, et al. ARID1A Deficiency Impairs the DNA Damage Checkpoint and Sensitizes Cells to PARP Inhibitors. Cancer Discov. 2015;5:752–767. doi: 10.1158/2159-8290.CD-14-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stark JM, et al. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scully R, et al. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 34.Schlegel BP, et al. BRCA1 promotes induction of ssDNA by ionizing radiation. Cancer Res. 2006;66:5181–5189. doi: 10.1158/0008-5472.CAN-05-3209. [DOI] [PubMed] [Google Scholar]
- 35.Cruz-Garcia A, et al. BRCA1 accelerates CtIP-mediated DNA-end resection. Cell Rep. 2014;9:451–459. doi: 10.1016/j.celrep.2014.08.076. [DOI] [PubMed] [Google Scholar]
- 36.Tkac J, et al. HELB Is a Feedback Inhibitor of DNA End Resection. Mol Cell. 2016;61:405–418. doi: 10.1016/j.molcel.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 37.Coleman KA, Greenberg RA. The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. J Biol Chem. 2011;286:13669–13680. doi: 10.1074/jbc.M110.213728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Z, et al. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lazzaro F, et al. Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. Embo J. 2008;27:1502–1512. doi: 10.1038/emboj.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferrari M, et al. Functional interplay between the 53BP1-ortholog Rad9 and the Mre11 complex regulates resection, end-tethering and repair of a double-strand break. PLoS Genet. 2015;11:e1004928. doi: 10.1371/journal.pgen.1004928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caravita T, et al. Bortezomib: efficacy comparisons in solid tumors and hematologic malignancies. Nat Clin Pract Oncol. 2006;3:374–387. doi: 10.1038/ncponc0555. [DOI] [PubMed] [Google Scholar]
- 43.Mortensen UH, et al. Rad52. Curr Biol. 2009;19:R676–677. doi: 10.1016/j.cub.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 44.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rothenberg E, et al. Human Rad52-mediated homology search and annealing occurs by continuous interactions between overlapping nucleoprotein complexes. Proc Natl Acad Sci U S A. 2008;105:20274–20279. doi: 10.1073/pnas.0810317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Motycka TA, et al. Physical and functional interaction between the XPF/ERCC1 endonuclease and hRad52. J Biol Chem. 2004;279:13634–13639. doi: 10.1074/jbc.M313779200. [DOI] [PubMed] [Google Scholar]
- 47.Morales ME, et al. The contribution of alu elements to mutagenic DNA double-strand break repair. PLoS Genet. 2015;11:e1005016. doi: 10.1371/journal.pgen.1005016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugawara N, et al. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc Natl Acad Sci U S A. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guirouilh-Barbat J, et al. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 50.Haber JE. Alternative endings. Proceedings of the National Academy of Sciences. 2008;105:405–406. doi: 10.1073/pnas.0711334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu C, et al. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 52.Wang M, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Audebert M, et al. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 54.Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol. 2010;17:410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Decottignies A. Microhomology-mediated end joining in fission yeast is repressed by pku70 and relies on genes involved in homologous recombination. Genetics. 2007;176:1403–1415. doi: 10.1534/genetics.107.071621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McVey M, Lee SE. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends in Genetics. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.LaRocque JR, et al. Interhomolog recombination and loss of heterozygosity in wild-type and Bloom syndrome helicase (BLM)-deficient mammalian cells. Proc Natl Acad Sci U S A. 2011;108:11971–11976. doi: 10.1073/pnas.1104421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ira G, et al. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benson FE, et al. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. Embo J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 61.Ogawa T, et al. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 62.Lambert S, Lopez BS. Characterization of mammalian RAD51 double strand break repair using non-lethal dominant-negative forms. Embo J. 2000;19:3090–3099. doi: 10.1093/emboj/19.12.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen JJ, et al. BRCA1, BRCA2, and Rad51 operate in a common DNA damage response pathway. Cancer Res. 1999;59:1752s–1756s. [PubMed] [Google Scholar]
- 64.Stark JM, et al. ATP hydrolysis by mammalian RAD51 has a key role during homology-directed DNA repair. J Biol Chem. 2002;277:20185–20194. doi: 10.1074/jbc.M112132200. [DOI] [PubMed] [Google Scholar]
- 65.Tutt A, et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. Embo J. 2001;20:4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dronkert ML, et al. Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol Cell Biol. 2000;20:3147–3156. doi: 10.1128/mcb.20.9.3147-3156.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Betermier M, et al. Is non-homologous end-joining really an inherently error-prone process? PLoS Genet. 2014;10:e1004086. doi: 10.1371/journal.pgen.1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee SE, et al. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 69.Ma JL, et al. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol Cell Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pierce AJ, et al. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang YG, et al. The Fanconi anemia group A protein modulates homologous repair of DNA double-strand breaks in mammalian cells. Carcinogenesis. 2005;26:1731–1740. doi: 10.1093/carcin/bgi134. [DOI] [PubMed] [Google Scholar]
- 72.Pace P, et al. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329:219–223. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- 73.Adamo A, et al. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 74.Murina O, et al. FANCD2 and CtIP cooperate to repair DNA interstrand crosslinks. Cell Rep. 2014;7:1030–1038. doi: 10.1016/j.celrep.2014.03.069. [DOI] [PubMed] [Google Scholar]
- 75.Yu X, Gabriel A. Ku-dependent and Ku-independent end-joining pathways lead to chromosomal rearrangements during double-strand break repair in Saccharomyces cerevisiae. Genetics. 2003;163:843–856. doi: 10.1093/genetics/163.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mansour WY, et al. The absence of Ku but not defects in classical non-homologous end-joining is required to trigger PARP1-dependent end-joining. DNA Repair (Amst) 2013;12:1134–1142. doi: 10.1016/j.dnarep.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 77.Mateos-Gomez PA, et al. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature. 2015;518:254–257. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ceccaldi R, et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature. 2015;518:258–262. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kent T, et al. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nat Struct Mol Biol. 2015;22:230–237. doi: 10.1038/nsmb.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zahn KE, et al. Human DNA polymerase theta grasps the primer terminus to mediate DNA repair. Nat Struct Mol Biol. 2015;22:304–311. doi: 10.1038/nsmb.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yousefzadeh MJ, et al. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 2014;10:e1004654. doi: 10.1371/journal.pgen.1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu AM, McVey M. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res. 2010;38:5706–5717. doi: 10.1093/nar/gkq379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ahmad A, et al. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol Cell Biol. 2008;28:5082–5092. doi: 10.1128/MCB.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee K, Lee SE. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 2007;176:2003–2014. doi: 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sugawara N, et al. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc Natl Acad Sci U S A. 2004;101:9315–9320. doi: 10.1073/pnas.0305749101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.George CM, Alani E. Multiple cellular mechanisms prevent chromosomal rearrangements involving repetitive DNA. Crit Rev Biochem Mol Biol. 2012;47:297–313. doi: 10.3109/10409238.2012.675644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chakraborty U, et al. A Delicate Balance Between Repair and Replication Factors Regulates Recombination Between Divergent DNA Sequences in Saccharomyces cerevisiae. Genetics. 2016;202:525–540. doi: 10.1534/genetics.115.184093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fu Q, et al. Phosphorylation-regulated transitions in an oligomeric state control the activity of the Sae2 DNA repair enzyme. Mol Cell Biol. 2014;34:778–793. doi: 10.1128/MCB.00963-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. Embo J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vaze MB, et al. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell. 2002;10:373–385. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- 92.Aymard F, et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:366–374. doi: 10.1038/nsmb.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ryu T, et al. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat Cell Biol. 2015;17:1401–1411. doi: 10.1038/ncb3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Belancio VP, et al. All y’all need to know `bout retroelements in cancer. Seminars in Cancer Biology. 2010;20:200–210. doi: 10.1016/j.semcancer.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang W, et al. Alu distribution and mutation types of cancer genes. BMC Genomics. 2011;12:157. doi: 10.1186/1471-2164-12-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pavlicek A, et al. Evolution of the tumor suppressor BRCA1 locus in primates: implications for cancer predisposition. Hum Mol Genet. 2004;13:2737–2751. doi: 10.1093/hmg/ddh301. [DOI] [PubMed] [Google Scholar]
- 97.Strout MP, et al. The partial tandem duplication of ALL1 (MLL) is consistently generated by Alu-mediated homologous recombination in acute myeloid leukemia. Proc Natl Acad Sci U S A. 1998;95:2390–2395. doi: 10.1073/pnas.95.5.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Elliott B, et al. Chromosomal translocation mechanisms at intronic alu elements in mammalian cells. Mol Cell. 2005;17:885–894. doi: 10.1016/j.molcel.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 99.Manthey GM, Bailis AM. Rad51 inhibits translocation formation by non-conservative homologous recombination in Saccharomyces cerevisiae. PLoS One. 2010;5:e11889. doi: 10.1371/journal.pone.0011889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Argueso JL, et al. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc Natl Acad Sci U S A. 2008;105:11845–11850. doi: 10.1073/pnas.0804529105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weckselblatt B, Rudd MK. Human Structural Variation: Mechanisms of Chromosome Rearrangements. Trends Genet. 2015;31:587–599. doi: 10.1016/j.tig.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Startek M, et al. Genome-wide analyses of LINE-LINE-mediated nonallelic homologous recombination. Nucleic Acids Res. 2015;43:2188–2198. doi: 10.1093/nar/gku1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rouet P, et al. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Davis L, Maizels N. Homology-directed repair of DNA nicks via pathways distinct from canonical double-strand break repair. Proc Natl Acad Sci U S A. 2014;111:E924–932. doi: 10.1073/pnas.1400236111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Song J, et al. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat Commun. 2016;7:10548. doi: 10.1038/ncomms10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Storici F, et al. Conservative repair of a chromosomal double-strand break by single-strand DNA through two steps of annealing. Mol Cell Biol. 2006;26:7645–7657. doi: 10.1128/MCB.00672-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sakuma T, et al. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat Protoc. 2016;11:118–133. doi: 10.1038/nprot.2015.140. [DOI] [PubMed] [Google Scholar]
- 108.Chandramouly G, et al. Small-Molecule Disruption of RAD52 Rings as a Mechanism for Precision Medicine in BRCA-Deficient Cancers. Chem Biol. 2015;22:1491–1504. doi: 10.1016/j.chembiol.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huang F, et al. Targeting BRCA1- and BRCA2-deficient cells with RAD52 small molecule inhibitors. Nucleic Acids Res. 2016;44:4189–4199. doi: 10.1093/nar/gkw087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ochs F, et al. 53BP1 fosters fidelity of homology-directed DNA repair. Nat Struct Mol Biol. 2016 doi: 10.1038/nsmb.3251. 10.1038/nsmb.3251. [DOI] [PubMed] [Google Scholar]


