Abstract
The activating NK-cell receptor KIR3DS1 has been implicated in the outcome of various human diseases, including delayed HIV-1 disease progression, yet a ligand that accounts for its biological effects remained unknown. We screened 100 HLA-I proteins and found that KIR3DS1 binds HLA-F, which was validated biochemically and functionally. Primary human KIR3DS1+ NK cells degranulated and produced antiviral cytokines upon encountering HLA-F, and inhibited HIV-1 replication in vitro. CD4+ T-cell activation triggered HLA-F transcription and expression and induced KIR3DS1 ligand expression. HIV-1 infection further increased HLA-F transcription, but decreased KIR3DS1 ligand expression, indicating an immune-evasion mechanism. Altogether, we established HLA-F as a ligand of KIR3DS1, and demonstrated cell-context-dependent expression of HLA-F that may explain the widespread influence of KIR3DS1 in human diseases.
INTRODUCTION
Killer-cell immunoglobulin-like receptors (KIRs) are a family of HLA class I-binding receptors expressed on natural killer (NK) cells that are implicated in various areas of human health and disease. The KIR locus contains some of the most highly polymorphic human genes, a diversity comparable to HLA genes in the major histocompatibility complex (MHC) locus1. This highlights a deep-seated evolutionary interplay of receptor-ligand pairs driven mainly by forces of reproduction and infectious disease survival1. Through variegated expression of KIRs and other highly diverse germline-encoded receptors, NK cells are able to discriminate between healthy “self” and a variety of pathological cell states2. As such, it is unsurprising that NK cells and KIRs have a broad involvement in human disease3, especially in light of their increasingly recognized roles in innate and adaptive immune responses4,5.
KIR family receptors are categorized by the number of immunoglobulin-like extracellular domains they contain (2D or 3D) and by whether they have a long (L) or short (S) cytoplasmic tail. KIR-L receptors are inhibitory, as they have immune tyrosine inhibitory motif (ITIM)-bearing cytoplasmic tails, while KIR-S receptors have truncated cytoplasmic tails and possess a transmembrane positive charge that allows association to immune tyrosine activating motif (ITAM)-bearing adaptor molecules such as DAP12. In general, KIR2D receptors bind HLA-C ligands, while KIR3D receptors bind HLA-A and -B ligands. Nevertheless, while many HLA class I (HLA-I) ligands have been identified for multiple KIRs, several have remained more elusive, particularly for activating KIRs.
KIR3DS1 was the first KIR to be associated with the outcome of a viral infection, namely, delayed human immunodeficiency virus (HIV)-1 disease progression in patients with certain HLA-B alleles6. Since then, it has been extensively linked to other viral infections6–9, autoimmune disorders10, cancer development/clearance11–13, and transplantation outcomes14,15, and therefore has become one of the most studied KIRs. KIR3DS1 is an activating receptor that stimulates cytotoxicity and IFN-γ production in NK cells16. It is encoded in the KIR3DL1-KIR3DS1 gene locus, a unique KIR locus because it encodes for functionally divergent alleles1. Remarkably, while sharing >95% homology in their extracellular domain, KIR3DS1 and its inhibitory counterpart KIR3DL1 have different ligand binding profiles. KIR3DL1 has conclusively been shown to bind HLA-A and -B proteins with a Bw4 motif, with variable sensitivity to C-terminal residues of HLA-Bw4–bound peptides and to residues at position 80 of HLA-I17. However, attempts to identify a KIR3DS1 ligand by various groups have repeatedly failed18,19, save for a single recent study demonstrating peptide-dependent binding of KIR3DS1 to HLA-B*57:01 in vitro20. Disease association studies have found both KIR3DL1 and KIR3DS1 to be influential in disease outcomes when co-present with HLA-Bw4, including HIV-1 pathogenesis6,21, but direct KIR3DS1–HLA-Bw4 interactions are a major point of controversy. Indeed, some studies report an independence between KIR3DS1 and HLA-Bw422,23. Furthermore, even in studies linking KIR3DS1 and HLA-Bw4, the vast majority of subjects possessing KIR3DS1 also possessed KIR3DL1, which imposes a confounding variable since HLA-Bw4 is a well-documented ligand for KIR3DL1. Of note, while KIR3DL1 exhibits significant sequence polymorphisms that have been shown to alter surface expression level24 and ligand binding25,26, KIR3DS1 is remarkably conserved and virtually monomorphic. This is despite KIR3DS1 having arisen in the human genome >3 million years ago along with various alleles of KIR3DL1, and being present in almost all human populations worldwide1.
The goal of the present study was to identify ligands of KIR3DS1 and thus uncover the mechanistic basis for its influence in various human diseases. To this end, we employed soluble receptor binding assays and cell-based functional assays to comprehensively screen HLA-I proteins in two biologically relevant conformation states: as HLA-I complexes, which are folded heavy chains bound to β2-microglobulin (β2m) and peptide, and as HLA-I open conformers (OCs), which are HLA-I heavy chains without bound β2m or peptide (reviewed in 27). We identified that HLA-F OCs are high-affinity ligands of KIR3DS1, and also lower affinity ligands of the inhibitory receptors KIR3DL1 and KIR3DL2. We also demonstrated that this interaction is of functional relevance using reporter cell lines, primary KIR3DS1+ NK cells, and primary CD4+ T cells, providing an explanation for the widespread influence of KIR3DS1 in human disease.
RESULTS
HLA-I screen shows KIR3DS1 binds HLA-F
We aimed to systematically assess KIR3DS1 binding to multiple HLA-I allotypes in two conformational states: as HLA-I complexes and as HLA-I OCs. To accomplish this, we tested binding of soluble KIR3DS1-Fc and other KIR-Fc fusion constructs to HLA-I-coated beads individually bearing 97 allotypes of classical HLA-I (i.e. HLA-A, -B, and -C) that were either untreated or acid pulsed. Acid pulsing is a well-established method of stripping away β2m and HLA-I-bound peptides to rapidly generate HLA-I OCs. KIR3DS1-Fc did not bind appreciably to any classical HLA-I allotypes tested either as complexes (untreated) or OCs (acid pulsed) (Fig. 1a). However, KIR3DL1-Fc bound preferentially to HLA-Bw4 complexes, and KIR2DL3-Fc and KIR2DS4-Fc bound to HLA-C complexes (Fig. 1a and Supplementary Fig. 1a), as expected. KIR3DL2-Fc did not have any preferential binding to specific HLA-I proteins. Of note, HLA-I complexes on these beads presented a diverse repertoire of peptides derived from the human cell line the HLA-I proteins were produced in, which precludes any peptide specificity analysis. Thus, KIR3DS1 does not bind to classical HLA-I complexes (presenting a diverse immunopeptidome) or HLA-I OCs.
Figure 1.
KIR-Fc binding to beads coated with classical and non-classical HLA-I proteins. (a, b) Binding of KIR-Fc constructs to untreated (complex) versus acid-pulsed (open conformer; OC) beads coated with classical HLA-I allotypes (in a); HLA-E, -F, or -G (in b); or nothing (negative control beads; red star) was measured and reported as median fluorescence intensity (MdFI). Each dot indicates a single HLA-I allotype, and color coding is used in a to indicate HLA-I allotype grouping (see legend and On-line Methods for further details). (c) Flow cytometry histograms showing KIR3DS1-Fc (25 μg/mL) binding to untreated HLA-E, -F-, and -G-coated beads as plotted in b; NC denotes negative control beads. Data in a are representative of three technical replicates and two independent experiments; b is representative of two independent experiments.
Because non-classical HLA-I proteins (i.e. HLA-E, -F, and -G) were not included in the original HLA-I-coated bead panel, we separately produced these. Streptavidin-beads were coated with biotinylated HLA-E, -F, or -G monomers refolded around β2m and peptide (no peptide in the case of HLA-F given that it does not present peptide28; details in on-line Methods). To confirm that HLA-I proteins were in complex conformation after loading them onto beads, we assessed anti-pan-HLA-I complex antibody binding to HLA-I-coated beads before and after acid pulsing by flow cytometry; this antibody can recognize all HLA-A, -B, -C, -E, -F, and -G proteins but only when they are bound to β2m. As expected, all acid-pulsed beads did not bind anti-pan-HLA-I complex antibody due to stripping of β2m (Supplementary Fig. 1c). However, we found that for untreated beads, while HLA-E and -G readily bound anti-pan-HLA-I complex antibody, HLA-F did not (Supplementary Fig. 1c). We assessed β2m content on the untreated HLA-I-coated beads by anti-β2m antibody staining, and found that HLA-F-coated beads had ~60% less β2m content than HLA-E- and HLA-G-coated beads (Supplementary Fig. 1c). This indicated that unlike other HLA-I proteins, HLA-F readily dissociates from β2m and spontaneously forms HLA-F OCs on artificial surfaces, which is consistent with its unique property of being stable as an open conformer29.
Upon testing KIR-Fc binding to non-classical HLA-I-coated beads, we found that KIR3DS1-Fc strongly bound to HLA-F-coated beads; this was independent of acid treatment, consistent with the fact that both untreated and acid-pulsed beads contained HLA-F OCs (Fig. 1b and Fig. 1c). KIR3DL1-Fc and KIR3DL2-Fc were similarly able to bind HLA-F-coated beads (both acid pulsed and untreated); however, KIR2DL3-Fc and KIR2DS4-Fc did not bind to any non-classical HLA-I-coated beads (Fig. 1b and Supplementary Fig. 1b). Consequently, we determined from this assay that KIR3DS1, along with the functional divergent KIR3DL1 and the phylogenetically related KIR3DL21, bind HLA-F OCs.
Surface plasmon resonance confirms KIR3DS1–HLA-F binding
To confirm our findings, we performed surface plasmon resonance to quantitatively assess the affinities between HLA-F OCs and various KIRs. KIR3DS1 had the highest affinity to HLA-F OCs of the tested KIRs (KD = (25 ± 1) nM), followed by KIR3DL2 (KD = (118 ± 1) nM) and KIR3DL1 (KD = (157 ± 2) nM), while KIR2DS4 and KIR2DL3-Fc did not exhibit any binding even at the highest tested concentrations (Fig. 2a and Table 1). The kinetic data showed that the affinity of KIR3DS1 to HLA-F OCs is mainly driven by a relatively small dissociation rate (kd = (7.0 ± 0.3) × 10−4 s−1) as compared to KIR3DL1 (kd = (6.91 ± 0.09) × 10−3 s−1) and KIR3DL2 (kd = (5.08 ± 0.04) × 10−3 s−1), reflecting a higher stability of the interaction once formed. Together, these data demonstrated that all ‘lineage II KIRs’ (i.e. KIR3DL1, KIR3DS1, and KIR3DL2) can bind HLA-F OCs, although with varying affinities and in a manner that is opposite to that involving binding to classical HLA-I molecules, where inhibitory KIRs normally bind with significantly higher affinity.
Figure 2.
Surface plasmon resonance of KIR binding to HLA-F OCs and functional triggering of KIR3DS1 on reporter cell lines by HLA-F OCs. (a) Each panel represents sensograms of the indicated KIR-Fc constructs at the indicated concentrations flowed over immobilized HLA-F OCs. Legend on left applies to first panel, while legend on right applies to three right panels. (b) Reporter activity of untransduced, KIR3DL1ζ, KIR3DS1hiζ, and KIR3DL2ζ Jurkat cells co-incubated with untreated (black bars) or acid-pulsed (gray bars) cell lines was measured as percentage of CD69hi Jurkat cells. Target cell lines were HLA-Bw4− BCL, 721.221 cells, and Jurkat cells; NT signifies no targets. (c) Reporter activity of untransduced (UT), KIR3DL1ζ, KIR3DS1hiζ, and KIR3DL2ζ Jurkat cells co-incubated with untreated or acid-pulsed HLA-Bw4− BCL was measured as percentage of CD69hi Jurkat cells after controlling for KIRζ expression by flow cytometry; NT signifies no targets. (d) Reporter activity of untransduced (UT), KIR3DL1ζ, and KIR3DS1hiζ Jurkat cells co-incubated with acid-pulsed HLA-Bw4− BCLs in the presence of the indicated anti-KIR or anti-HLA-I antibodies (25 μg/mL each) was measured as percentage of CD69hi Jurkat cells. Blocking antibodies used (clone and specificity) are the following: Z27, anti-KIR3DS1/L1 antibody; DX9, anti-KIR3DL1 antibody; HC10, anti-HLA-I OC antibody; HCA2, anti-HLA-I OC antibody; W6/32, anti-pan-HLA-I complex antibody. Data in a shows three technical replicates and is representative of two independent experiments; b shows pooled data from three independent experiments; c shows three technical replicates; d shows technical duplicates and is representative of three independent experiments; b, c, and d show mean ± s.d.
Table 1.
Kinetic values of KIR binding to HLA-F OCs as determined by surface plasmon resonance. All values represent monomeric interaction kinetic values calculated from the ‘bivalent analyte’ fitting model on BIAevaluation software and are presented as mean ± s.e.m. Asterisk (*) indicates there was no binding detected at highest tested concentration (0.5 μM) of KIR-Fc. Kinetic constants: ka, association rate constant (a.k.a. on-rate); kd, dissociation rate constant (a.k.a. off-rate); KD, equilibrium dissociation constant.
| Constant | ka (M−1s−1) (×104) | kd (s−1) (×10−4) | KD (nM) |
|---|---|---|---|
| KIR3DL1 | 4.40 ± 0.04 | 69.1 ± 0.9 | 157 ± 2 |
| KIR3DS1 | 2.84 ± 0.02 | 7.0 ± 0.3 | 25 ± 1 |
| KIR3DL2 | 4.30 ± 0.02 | 50.8 ± 0.4 | 118 ± 1 |
| KIR2DL3 | –* | –* | –* |
| KIR2DS4 | –* | –* | –* |
HLA-F activates KIR3DS1ζ Jurkat reporter cells
To determine whether cell-expressed KIR3DS1 can bind to and be activated by HLA-F OC-expressing target cells, we developed a reporter cell assay using Jurkat cells, which are HLA-F- and -G-deficient and do not have HLA-Bw4. We performed CRISPR/Cas9-mediated knockout of β2m in Jurkat cells to eliminate surface HLA-I expression (Supplementary Fig. 2a) and prevent potential self-activation. Jurkat cells were then stably transduced with chimeric receptors containing the extracellular and transmembrane domain of the KIR of interest and the cytoplasmic domain of CD3ζ, (herein called ‘KIRζ’) (Supplementary Fig. 2b). For KIR3DS1, the transmembrane domain of KIR3DL1 was used instead to ensure cell surface expression in the absence of its adaptor DAP12, as has been previously shown30; we have called this construct ‘KIR3DS1hiζ’. Surface expression of KIRζ on Jurkat cells was confirmed by staining with the relevant anti-KIR antibodies (Supplementary Fig. 2c), and CD69 expression was used as a reporter cell output, which was validated by anti-KIR antibody-mediated crosslinking (Supplementary Fig. 2d).
Initially, the ability of KIRζ-Jurkat reporter cells to bind to and be triggered by HLA-I-coated cell-sized beads was tested, and this assay showed that KIR3DS1ζ-Jurkat reporter cells avidly bound to and were potently triggered by HLA-F-coated beads, as were KIR3DL2ζ-Jurkat cells and KIR3DL1ζ-Jurkat cells to a much lesser extent (Supplementary Fig. 2e). As expected, no binding or triggering by HLA-F-coated beads was observed for KIR2DL3ζ-expressing or untransduced Jurkat reporter cells. Next, KIRζ-Jurkat reporter cell activity was tested against cell lines that were untreated or acid pulsed to generate HLA-I OCs at the cell-surface, which was confirmed by anti-HLA-I OC antibody staining (Supplementary Fig. 2f). 721.221 cells are a highly-mutated EBV-transformed B-cell line commonly used as NK-cell targets that are deficient in classical HLA-I genes and only express HLA-E and -F (i.e. HLA-A−B−C−E+F+G−). KIRζ-Jurkat reporter activity against 721.221 cells revealed that acid-pulsed 721.221 cells potently stimulated KIR3DS1hiζ-Jurkat cells (Fig. 2b), a finding that occurred independent of other expressed HLA-I allotypes (Supplementary Fig. 3a). KIR3DL2ζ- and KIR3DL1ζ-Jurkat cells, but not KIR2DL3ζ-Jurkat cells, were also triggered by acid-pulsed 721.221 cells, but less potently, especially in the case of KIR3DL1ζ-Jurkat cells, in line with the binding data (Fig. 2b). These data indicated that KIR3DS1 (as well as KIR3DL2 and to a lesser extent KIR3DL1) binds HLA-F OCs independent of classical HLA-I.
In assessing KIRζ-Jurkat reporter activity to other cell lines, we found that acid-pulsed EBV-transformed B cell lines (BCLs) derived from HLA-typed patients also stimulated KIR3DS1hiζ-Jurkat cells, while other cell lines not encoding HLA-F did not (i.e. Jurkat, EL-4, K562, and THP-1 cells) (Fig. 2b and Supplementary Fig. 3b). In a separate experiment where KIRζ surface expression was accounted for, triggering of KIRζ Jurkat reporter cells by acid-pulsed BCLs revealed the highest level of reporter activity by KIR3DS1hiζ, followed by KIR3DL2ζ and then KIR3DL1ζ (Fig. 2c), which correlated to SPR-determined affinities. These data showed that compared to other lineage II KIRs, KIR3DS1 exhibits the most potent functional signaling capacity upon engagement of HLA-F OCs on target cells.
To further confirm the interaction between KIR3DS1 and HLA-F OCs, antibody blockade experiments were performed. Of note, we did not have a specific anti-HLA-F OC blocking antibody at our disposal. Instead, two anti-HLA-I OC antibodies were used: HC10, which binds HLA-B, -C, and -E OCs but has been shown to indirectly down-regulate HLA-F OCs from the cell surface of target cells via endocytosis29, and HCA2, which binds HLA-A and -G OCs but we determined exhibits reactivity to HLA-F OCs (Supplementary Fig. 1c). Accordingly, KIR3DS1hiζ Jurkat reporter cell activity induced by acid-pulsed BCLs could be blocked by anti-KIR3DS1/L1 and both anti-HLA-I OC antibodies, but not by anti-KIR3DL1 or anti-pan-HLA-I complex antibodies (Fig. 2d). In addition, adding soluble KIR3DS1-Fc to block ligands on target cells also abrogated KIR3DS1hiζ Jurkat reporter cell activity induced by acid-pulsed BCLs (Supplementary Fig. 3c). These antibody blockade experiments validated that KIR3DS1 interacts with HLA-F OCs. On the other hand, KIR3DL1ζ Jurkat reporter cell activity triggered by BCLs expressing HLA-Bw4 could be blocked by both anti-KIR3DL1 and anti-pan-HLA-I complex antibodies, but not by either anti-HLA-I open conformer antibody (Supplementary Fig. 3d). This confirms the well-documented interaction between KIR3DL1 and HLA-Bw4 complexes. Of note, the weak reporter cell activity of KIR3DL1ζ-Jurkat cells stimulated by acid-pulsed BCLs without HLA-Bw4 could be blocked by both anti-KIR3DL1 and anti-HLA-I open conformer antibodies, but not the anti-pan-HLA-I complex antibody (Fig. 2d). This further showed a low-level interaction between KIR3DL1 and HLA-F OCs that is functionally much weaker as compared to KIR3DS1, which is remarkable in light of their high homology.
HLA-F triggers polyfunctional response in KIR3DS1+ NK cells
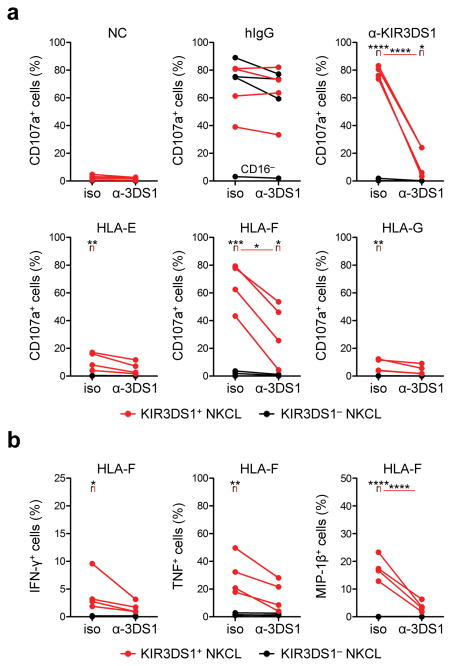
To assess the functional impact of HLA-F OC binding to KIR3DS1 in primary human NK cells, KIR3DS1+ and KIR3DS1− NK-cell clones (NKCLs) were generated via limiting dilution cloning from healthy donor peripheral blood NK cells. NKCLs were confirmed to be CD56+KIR3DL1−LILRB1− to ensure proper functionality, and had variable expression of KIR2Ds (Supplementary Fig. 4a). These NKCLs were seeded into individual well plates coated with human IgG (for CD16 crosslinking), anti-KIR3DS1/L1 antibody (for KIR3DS1 crosslinking), or monomers of HLA-E, -F, or -G. To assess NKCL responses to plate-bound ligands, we measured cytotoxic granule exocytosis (i.e. degranulation), as determined by surface expression of CD107a, and production of antiviral cytokines IFN-γ, TNF, and MIP-1β by intracellular staining. Plate-bound HLA-F strongly and significantly triggered degranulation in KIR3DS1+ NKCLs but not KIR3DS1− NKCLs (Fig. 3a and Supplementary Fig. 4c). The magnitude of this response was comparable to crosslinking of KIR3DS1 and CD16, and was significantly reduced by the addition of soluble anti-KIR3DS1/L1 antibody (Fig. 3a). Plate-bound HLA-E and HLA-G produced a weak response by some but not all NKCLs, and this activation could not be blocked with soluble anti-KIR3DS1/L1 antibody. The observed effects were not due to intrinsic differences in response capacity, as all NKCLs (except for one that was CD16−) were similarly triggered by CD16 crosslinking. In addition, HLA-F induced production of the antiviral cytokines IFN-γ, TNF and MIP-1β in KIR3DS1+ (but not KIR3DS1−) NKCLs, and these responses could be blocked with soluble anti-KIR3DS1/L1 antibody (Fig. 3b and Supplementary Fig. 4c). Interestingly, KIR3DS1+ NKCLs had an intrinsic superior capacity to produce antiviral cytokines when compared to KIR3DS1− NKCLs when encountering the same stimuli (Supplementary Fig. 4d). Altogether, these data demonstrate that HLA-F can trigger a potent activating signal via KIR3DS1 that results in a polyfunctional response by primary NK cells.
Figure 3.
HLA-F OCs trigger degranulation and antiviral cytokine production in primary NK cells via KIR3DS1. (a, b) NK-cell degranulation and cytokine production in KIR3DS1+ (red dots and lines) and KIR3DS1− NKCLs (black dots and lines) seeded onto well plates coated with irrelevant protein (negative control; NC), human IgG (hIgG), anti-KIR3DS1/L1 antibody (anti-KIR3DS1), or monomers of HLA-E, -F, or –G was measured as percentage of CD107a+ cells (in a) and percentages of IFN-γ+, TNF-α+, and MIP-1β+ cells (in b). For each plate-bound ligand condition, antibody-mediated blockade was performed by incubating NKCLs in the presence of mouse IgG1 isotype control antibody (iso) or soluble anti-KIR3DS1/L1 antibody (+α-3DS1) (25 μg/mL each); each line connects a single NKCL in each blocking condition. For each plate-bound ligand and marker in a and b, a one-way ANOVA with Bonferroni multiple comparisons test comparing select columns was performed, and all statistically significant differences are presented (*, **, ***, and **** denotes, p < 0.05, < 0.01, < 0.001, and < 0.0001, respectively). Each graph in a and b show four biological replicated for KIR3DS1+ and KIR3DS1− for each condition, and is representative of two independent experiments.
CD4+ T-cell activation induces KIR3DS1 ligand expression
HLA-F can be expressed on the surface of activated lymphocytes31. To study KIR3DS1 ligand expression in a biologically relevant context, HLA-F expression and KIR3DS1-Fc binding to primary resting CD4+ T cells and CD4+ T cells stimulated with IL-2 and CD3/28 beads was assessed. An immunoblot-compatible anti-HLA-F antibody (validated in Supplementary Fig. 5a) was used to assess protein expression, and resistance to endoglycosidase H (Endo H) digestion served as a measure of cell-surface expression. Endo H is a glycosidase that removes immature N-glycans from glycoproteins that have not undergone Golgi processing; thus, glycoproteins shuttled to the cell surface through the Golgi are rendered Endo H-resistant (i.e. there is no band shift on protein gel electrophoresis). Results showed that while unstimulated CD4+ T cells do not have any detectable levels of HLA-F protein, stimulated CD4+ T cells express HLA-F protein in a predominantly Endo H-resistant form, indicating cell-surface expression (Fig. 4a); similar results were obtained using different stimulation conditions (48 h treatment with 12.5 ng/mL phorbol 12-myristate 13-acetate and 0.335 μM ionomycin; data not shown).
Figure 4.
Activated CD4+ T cells express HLA-F on the cell surface and bind KIR3DS1-Fc. (a) Reducing-denaturing SDS-PAGE and immunoblotting (IB) for HLA-F and β-actin (loading control) was performed on cell lysates of purified CD4+ T cells unstimulated (Unstim) or stimulated (Stim) that underwent Endo H, PNGase F, or mock digestion. Stimulation consisted of high-dose IL-2 and CD3/28 beads for 14 d. (b) Fluorescent in situ hybridization and flow cytometric assessment of HLA-F mRNA levels in unstimulated (Unstim) or stimulated (Stim) CD4+ T cells. Stimulation consisted of 24-h treatment with PMA and ionomycin. (c, d) KIR3DS1-Fc staining of purified CD4+ T cells from four donors unstimulated (Unstim) or stimulated (Stim) with high-dose IL-2 and CD3/28 beads for 6 d. Staining with secondary antibody alone (2° only) was done as a control. Flow histograms from a representative donor are shown in c, and aggregate data are plotted in d. HLA-B types of four donors were the following: B*08:01/B*18:01, B*08:01/B*14:02, B*44:02/B*44:02, and B*27:05/B*57:01. Paired t test was performed for unstimulated versus stimulated CD4+ T cells (* denotes p < 0.001). Data in a are representative of two independent experiments; b is representative of four independent experiments; d shows four donors.
To determine if HLA-F upregulation in activated CD4+ T cells was transcriptionally induced, HLA-F mRNA levels were assessed via flow cytometry-based fluorescent in situ hybridization (FISH) (details in the On-line Methods). HLA-F mRNA-specific probes were used after testing their sensitivity and specificity in HLA-F+ and HLA-F− cell lines (Supplementary Fig. 5b). Activation of CD4+ T cells increased the levels of HLA-F mRNA transcripts (Fig. 4b), indicating that HLA-F is transcriptionally induced in CD4+ T cells upon activation. Furthermore, KIR3DS1-Fc bound significantly to stimulated CD4+ T cells, but not to unstimulated CD4+ T cells (Fig. 4c and Fig. 4d, p < 0.001). These results were consistent across patients with and without HLA-Bw4, demonstrating that KIR3DS1 ligands are expressed on activated CD4+ T cells regardless of their HLA-I genotype, and strongly suggesting KIR3DS1 binding to HLA-F expressed on activated CD4+ T cells.
HIV-1 infection alters KIR3DS1 ligand expression
To assess the influence of HIV-1 infection on expression of KIR3DS1 ligands in CD4+ T cells, HLA-F mRNA levels and KIR3DS1-Fc binding were assessed using activated CD4+ T cells that were uninfected or infected with HIV-1 NL4-3. HLA-F mRNA levels were up-regulated in HIV-1–infected activated CD4+ T cells when compared to uninfected activated CD4+ T cells (Fig. 5c), indicating that HIV-1 infection further stimulated HLA-F mRNA transcription. In contrast, KIR3DS1-Fc binding was significantly decreased upon HIV-1 infection (Fig. 5a and Fig. 5b). The decrease in KIR3DS1-Fc binding was already observed in ‘early’ infected cells (defined as p24loCD4+HLA-I+tetherin+), but was more pronounced in ‘late’ infected cells (defined as p24hiCD4loHLA-Ilotetherinlo), indicating an as-yet-unknown mechanism by which HIV-1 might reduce KIR3DS1 ligand expression. This may be due to direct downregulation of HLA-F protein by HIV-1; however, due to the absence of an available flow cytometry-suitable antibody against HLA-F, it was not possible to directly quantify HLA-F surface expression on HIV-1–infected cells.
Figure 5.
Effect of HIV-1 infection on HLA-F expression, KIR3DS1-Fc binding, and suppression of HIV-1 replication in infected CD4+ T cells by KIR3DS1+ NK cells. (a, b) Staining of KIR3DS1-Fc (25 μg/mL) was measured on CD4+ T cells that were treated with high-dose IL-2 (+IL-2), stimulated with high-dose IL-2 + anti-CD3 + anti-CD28 (Stim), or stimulated and infected with HIV-1. HIV-1-infected CD4+ T cells that were p24loCD4+HLA-I+tetherin+ and p24hiCD4loHLA-Ilotetherinlo were classified as ‘early’ HIV-1 infected (early HIV-1) and ‘late’ HIV-1 infected (late HIV-1), respectively. Staining with secondary antibody alone (2° only) was done as a control. Representative donor staining is shown in a, and aggregate data showing background-subtracted median fluorescence intensity (bsMdFI) is shown in b. (c) HLA-F mRNA levels in CD4+ T cells that were stimulated with high-dose IL-2 and CD3/28 beads (Stim) or stimulated and infected with HIV-1 (HIV-1) were measured by fluorescent in situ hybridization; NC indicates negative control probe. (d, e) Percentage of HIV-1 p24+ CD4+ T cells was measured in wells with HIV-1-infected CD4+ T cell alone or co-cultured with autologous KIR3DS1+ or KIR3DS1− NKCLs for 7 d. Representative flow cytometry plots are presented in e and aggregate results are presented d. For b and d, one-way ANOVA with Tukey multiple comparisons test comparing all columns was performed (* and ** denote p < 0.05 and p < 0.01, respectively). Data in b show five donors; c is representative of two independent experiments.
KIR3DS1+ NK cells suppress HIV-1 replication
To evaluate the antiviral capacity of KIR3DS1+ NK cells, HIV-1–infected autologous CD4+ T cells were co-cultured for seven days with KIR3DS1+ and KIR3DS1− NK-cell clones derived from a KIR3DS1 homozygous donor (for NK-cell receptor phenotypes, see Supplementary Fig. 4b). Intracellular staining for HIV-1 p24 was performed to quantify the percentage of infected cells. Only KIR3DS1+ NK-cell clones were effective at suppressing HIV-1 replication in autologous CD4+ T cells, as seen by significantly less HIV-1 p24+ CD4+ T cells in the presence of KIR3DS1+ NK cells (5.48 % ± 1.60 %), relative to KIR3DS1− NK-cell clones (16.9 % ± 2.32 %; p < 0.01) or no NK cells added (mean 16.0 % ± 2.62 %, p < 0.01) (Fig. 5d and Fig. 5e). Thus, KIR3DS1+ NK cells have superior antiviral capacity towards HIV-1–infected autologous CD4+ T cells, as previously shown32, and HIV-1–mediated downregulation of KIR3DS1 ligands on infected cells is not sufficient to enable complete immune evasion.
DISCUSSION
The expression of the activating NK-cell receptor KIR3DS1 is correlated with the outcome of multiple human diseases; however the precise nature of a ligand that can account for such broad biological effects has remained—although extensively studied—unknown. Here, we identified HLA-F OCs as ligands for KIR3DS1 and showed that HLA-F OCs trigger polyfunctional responses in primary human NK cells through KIR3DS1. It was further demonstrated that KIR3DS1 ligands, in particular HLA-F, are expressed on the surface of activated CD4+ T cells. However, upon HIV-1 infection of activated CD4+ T cells, KIR3DS1 ligand surface expression was downregulated, yet this did not abrogate efficient suppression of viral replication by co-cultured KIR3DS1+ NK cells in vitro. Our findings provide novel and significant insight into the protective effect of KIR3DS1 in HIV-1 infection, and provide an explanation for the widespread influence of KIR3DS1 in human disease.
Differential binding of KIR3DS1 and the other lineage II KIRs1 to HLA-F is remarkable from a structural homology perspective, and is shared with other related receptors. The leukocyte immunoglobulin-like receptor (LILR) family is another group of HLA-I-binding receptors found near the KIR locus in the leukocyte receptor complex (LRC)33. The LILR family has inhibitory members (i.e. LILRBs) that bind HLA-I complexes and activating members (i.e. LILRAs) that bind HLA-I OCs27,34, a property that our data suggests is partially shared with KIRs; two members in particular, LILRB1 and LILRB2, exhibit robust binding to HLA-F35. From this present study, we find that all lineage II KIRs bind HLA-F OCs, with KIR3DS1 exhibiting the highest affinity binding to and the most potent functional signaling capacity upon engagement of HLA-F OCs on target cells. KIR3DL2, which we find also binds to HLA-F OCs as has been previously shown36, shares ~86% extracellular domain sequence identity with KIR3DS1, which readily explains differences in binding affinity. However, it is remarkable that despite >97% extracellular domain sequence identity between KIR3DS1 and KIR3DL1, KIR3DS1 exhibits significantly higher-affinity binding to HLA-F OCs than KIR3DL1, while KIR3DL1 (but not KIR3DS1) bears high-affinity binding to HLA-Bw4 ligands. Structural studies indicate that the few amino acid differences between KIR3DS1 and KIR3DL1 occur at sites critical for HLA-I binding37, which may explain the differences in ligands. From an evolutionary standpoint, these different ligand-binding profiles of KIR3DL1 towards various HLA-Bw4 allotypes and KIR3DS1 towards HLA-F (which is highly conserved and has one predominant *01:01 allele at a frequency >95%38) provide an explanation for the high polymorphicity of KIR3DL1 and the relative monomorphicity of KIR3DS1.
This newly identified KIR3DS1-HLA-F axis has many similarities to the well-known stress-induced NKG2D–MIC-A/B axis, even in the context of HIV-1 infection. Although it is less well-studied, HLA-F bears unique and distinguishing characteristics that separates it from all other HLA-I genes. According to a study examining the genesis and architectural evolution of the MHC locus, HLA-F acted as the ancestral progenitor to today’s HLA-I and MIC-A/B genes39. HLA-F retains features that are similar to MIC-A/B genes, including being peptide-devoid and being able to refold in vitro with and without β2m, indicating its unique ability to be stable as an OC29, in line with data presented here. In agreement with previous studies31,40, our data show that HLA-F OCs are expressed on the surface of activated CD4+ T cells. Also, although only partially assessed in this study with B-cell lines, several studies have indicated that cancers of various tissue origins aberrantly express HLA-F40,41. Additionally, HIV-1 infection of CD4+ T cells increased transcription of HLA-F but reduced KIR3DS1 ligand expression, particularly in late-infected cells, which might suggest the employment of an immune-evasion strategy, potentially through the downregulation of HLA-F by HIV-1 accessory proteins, similar to what has been described for HIV-1 Nef for HLA-A and HLA-B42–44 and NKG2D ligands including MIC-A45, but also HIV-1 Vpu for HLA-G46. Our findings are therefore reminiscent of the up-regulation of the NKG2D ligands MIC-A and MIC-B on the surface of activated T cells, virally infected cells, and transformed cells, which are downregulated in the context of HIV-1 by the actions of the accessory protein Nef45.
HLA-F expression is tightly regulated47, with restricted tissue expression35,40 and is predominantly localized to the endoplasmic reticulum. This indicates that HLA-F expression might serve as a marker for specific kinds of cell stress, such as endoplasmic reticulum stress. Similar to the phenomenon seen for MIC-A48,49, HLA-F can be a target for humoral immune responses, as was shown in one study that reported the presence of anti-HLA-F antibodies in the sera of cancer patients but not healthy controls41. Collectively, these studies underscore a critical role of HLA-F expression in human diseases, and our data demonstrating that KIR3DS1 recognizes HLA-F OCs uncovers a previously unknown mechanism of innate immune surveillance of stressed cells.
While KIR3DS1 binding to HLA-F expounds its widespread influence on human diseases, it does not fully explain the well-known association of KIR3DS1 to HIV-1 disease control described only for HLA-Bw4+ patients. Several studies have confirmed, however, that in the context of HIV-1 infection, presence of KIR3DS1 and KIR3DL1 together associate with decreased viremia, delayed progression to AIDS, and better in-vitro viral inhibition by NK cells in patient bearing HLA-B alleles with a Bw4 motif and an isoleucine at position 807,50. This protective compound genotype can be explained by independent but functionally synergistic effects of KIR3DS1–HLA-F and KIR3DL1–HLA-Bw4 interactions, the latter of which has independently been linked to delayed HIV-1 disease progression21. Of note, a single recent study demonstrated KIR3DS1 binding to HLA-B*57:01 presenting two HIV-1 peptides that arose from an exhaustive screen of various viral peptide databases20. However, it is not clear whether this interaction has real functional consequences, or represents an exceptional case where a peptide can overcome KIR3DS1 mutations that normally abrogate HLA-Bw4 binding. Indeed, KIR3DS1 has been linked to many other human diseases without association to HLA-Bw49,12,15, suggesting that KIR3DS1 exerts disease modulatory effects independent of HLA-Bw4. KIR3DS1–HLA-F interactions between NK cells and pathologically altered target cells would incur the well-known innate function of NK cells to recognize and eliminate target cells expressing “stressed self” ligands. This is supported by our in vitro co-culture assay, which showed that NK cells singly expressing KIR3DS1 are more effective at suppressing HIV-1 replication in autologous CD4+ T cells as compared to KIR3DS1− NK cells. In addition, our data show that KIR3DS1–HLA-F interactions elicit NK-cell production of antiviral and pro-inflammatory cytokines such as IFN-γ, TNF, and MIP-1β, which would have pleiotropic effects on immune responses. Furthermore, KIR3DS1–HLA-F interactions between NK cells and activated CD4+ T cells would also suggest a means of NK cell-mediated adaptive immune regulation. This notion is warranted by previous work in LCMV infection mouse models, which show that NK cells regulate adaptive immunity by killing activated CD4+ T cells, resulting in decreased immunopathology5. Thus, KIR3DS1 recognition of HLA-F OCs expressed on activated immune cells and/or infected target cells provides a mechanistic link between KIR3DS1 and HIV-1 disease progression, and will also have relevance for the pathogenesis of other infectious diseases, autoimmune disorders, and tumor immune-surveillance.
In conclusion, the novel interaction between the activating NK-cell receptor KIR3DS1 and HLA-F OCs described here uncovers a previously unknown mechanism of target cell recognition by NK cells. Further studies will be required to determine how KIR3DS1–HLA-F interactions play a role in NK cell-mediated regulation of immunity and/or elimination of pathologically altered target cells. Each of the clinical diseases KIR3DS1 has been implicated in, including autoimmune disorders10, transplantation outcomes14,15, cancer development/clearance11–13, and viral infections6–9, offers a variety of prospects to exploit KIR3DS1 interactions with HLA-F therapeutically.
ON-LINE METHODS
Cell lines and antibodies
721.221 and Jurkat (clone E6.1; ATCC) cell lines (including transductants) were grown in RPMI-1640 supplemented with 10% fetal bovine serum (Sigma-Aldrich), 2 mM L-glutamine (Gibco), 100 U/mL penicillin (Gibco), and 100 U/mL streptomycin (Gibco) at 37°C/5% CO2. EBV-transformed B-cell lines (BCLs) were generated from peripheral blood mononuclear cells from donors bearing specific HLA genotypes; BCLs were also grown in the same media and conditions as 721.221 and Jurkat cells. All antibodies used for flow cytometry staining, blocking assays, and immunoblotting are tabulated in Supplementary Table 1.
KIR-Fc binding to HLA-I-coated beads
Classical HLA-I-coated beads used were LABScreen single HLA-I beads (One Lambda). This bead set contains 97 different classical HLA-I allotypes individually coated onto fluorescently barcoded beads. Classical HLA-I allotypes were grouped into the following categories: (i) HLA-A allotypes without a Bw4 motif (HLA-A), (ii) HLA-A and -B allotypes with a Bw4 motif and an isoleucine at position 80 (HLA-Bw4-I80), (iii) HLA-B allotypes with a Bw4 motif and a threonine at position 80 (HLA-Bw4-T80), (iv) HLA-B allotypes with a Bw6 motif (HLA-Bw6), (v) HLA-C alleles with a serine at position 77 and an asparagine at position 80 (HLA-C1), and (vi) HLA-C alleles with an asparagine at position 77 and a lysine at position 80 (HLA-C2). Acid pulsing of HLA-I-coated beads was performed by resuspending beads in 50 μL of 300 mM glycine (pH 2.4), incubating at room temperature for exactly 2 min, and washing three times with 1 mL of HBSS. Untreated and acid-pulsed beads were stained according to the manufacturer’s instructions with KIR-Fc constructs (R&D Systems) diluted in PBS to 200 μg/mL. Binding to beads was measured on a Bio-Plex 3D Suspension Array system using Luminex xMAP technology (Bio-Rad). To generate non-classical HLA-I-coated beads, biotinylated monomers of HLA-E+β2m+VMAPRTLVL, HLA-F+β2m, and HLA-G+β2m+KGPPAALTL were purchased from Immune Monitoring Lab at Fred Hutchinson Cancer Research Center, Seattle, WA, and loaded onto streptavidin-coated beads (Life Technologies). Acid pulsing was performed as before. Staining of KIR-Fc constructs (25 μg/mL) was performed for 45 min at 4°C while shaking. Beads were then washed and stained with goat anti-human IgG(Fc) F(ab’)2 antibody for 30 min at 4°C while shaking (for antibody details, see Supplementary Table 1). Beads were subsequently washed and fixed with 4% paraformaldehyde in PBS (Affymetrix) before flow cytometric analysis.
Lentiviral transduction/transfection
Jurkat cells stably expressing genes of interest were generated via lentiviral transduction. Gene constructs were designed accordingly and ordered from GeneArt (Life Technologies). Constructs were cloned into a lentiviral transfer vector containing an SFFV promoter and IRES-driven puromycin resistance. This backbone vector was generated by cloning the SFFV promoter from pAPM51 into pLVX-EF1α-IRES-Puro (Clontech). HEK293T cells (ATCC) were transfected with a VSV-G envelope vector (pHEF-VSVG, obtained from NIH AIDS Reagent Program), HIV-1 gag-pol packaging vector (psPAX2, obtained from NIH AIDS Reagent Program), and the transfer vector of interest. Lentivirus-containing supernatants were harvested 3 d after transfection and used to transduce Jurkat cells, which were subsequently selected in 1 μg/mL puromycin and sorted for gene expression by fluorescence-activated cell sorting (FACS). 721.221 HLA transductants were previously generated by retroviral transduction or generated via lentiviral transduction for this study, with the exception of HLA-G-expressing 721.221 cells. β2m-knockout (β2m-KO) Jurkat cells were generated KIRζ Jurkat reporter cell lines. For this, Jurkat cells were stably transduced with S. pyogenes Cas9 (published in 52; Addgene plasmid # 52962) and selected in 5 μg/mL blasticidin S (Gibco). Jurkat-Cas9 cells were electroporated with a β2m-targeting gRNA CRISPR vector (published in 53) and sorted for loss of HLA expression. Jurkat-β2m-KO cells were subsequently transduced with KIRζ chimeric constructs. KIRζ expression was confirmed by staining with anti-KIR3DS1/L1, anti-KIR3DL1, anti-KIR3DL2, and anti-KIR2DL2/L3 antibodies.
KIRζ Jurkat reporter cell assay
KIRζ-Jurkat reporter cells were incubated with target cells at a reporter-target cell ratio of 1:10 at 37°C/5% CO2 for 2–2.5 h for acid-pulse experiments or for 8 h for experiments where beads were used or acid pulsing was not a condition. For antibody blockade experiments, blocking antibodies used were pre-incubated with relevant cells (reporters or targets) for 30 min at 4°C before co-incubation of reporter and target cells; the antibodies remained during the co-incubation. After the co-incubation, cells were stained with anti-CD3 and anti-CD69 antibodies (and in certain experiments with the corresponding anti-KIR antibodies) and CD69 expression of reporter cells relative to negative and positive controls was assessed and used as a measure of reporter activity.
Primary human CD4+ T-cell isolation, stimulation, infection, and staining with KIR-Fc and HLA-F mRNA probes
CD4+ T cells were isolated from donor PBMCs by indirect magnetic labeling and negative selection, using the CD4+ T cell isolation kit II (Miltenyi) according to the manufacturer’s protocol. Stimulations were done differently depending on the experiment. Stimulations for immunoblot analyses were done with CD3/28 beads (Life Technologies) at a bead-to-cell ratio of 2:1 and culturing in supplemented RPMI with high-dose IL-2 (100 U/mL) for the indicated amounts of time. Stimulations for flow cytometry-based fluorescent in situ hybridization were done with phorbol 12-myristate 13-acetate (12.5 ng/mL) and ionocymin (0.335 μM) (Cell Stimulation Cocktail used at 0.25X; eBioscience) for the indicated amounts of time. Stimulations for KIR-Fc staining and HIV-1 infection were done for the indicated amounts of time in supplemented RPMI with high-dose IL-2 (100 U/mL) on non-tissue culture-treated flat-bottom 48-well plates previously pre-coated with anti-CD3 (OKT3) and anti-CD28 antibodies each at 10 μg/mL. HIV-1 infection was done by resuspending ~1 × 106 activated CD4+ T cells in 1 mL of supplemented RPMI containing 100 U/mL of IL-2 and adding 1 × 105 TCID50 of replication-competent HIV-1 NL4-3. Cell were immediately spinfected (centrifuging for 2 h at 1,500 × g at 37°C) and incubated at 37°C/5% CO2 for 72 h to allow for viral replication, at which time cells were stained and analyzed by flow cytometry. For KIR-Fc staining of cells, cells were first stained with LIVE/DEAD® Fixable Blue Dead Cell Staining Kit (Life Technologies) following manufacturer’s instructions, and then stained with 25 μg/mL of KIR3DS1-Fc for 45 min at 4°C while shaking, after which the cells were washed and stained with anti-CD3, anti-CD4, anti-pan-HLA-I complex, anti-tetherin, and goat anti-human IgG(Fc) F(ab’)2 antibodies for 30 min at 4°C while shaking. Cells were subsequently washed and subjected to fixation and permeabilization with the FIX & PERM ® Cell Fixation and Cell Permeabilization Kit (Life Technologies) to stain with anti-HIV-1 p24 antibody. Cells were subsequently washed and resuspended in PBS, except in assays where fixation and permeabilizationn was not done, in which case cells were fixed with 4% paraformaldehyde in PBS (Affymetrix) before flow cytometric analysis. Fluorescent in situ hybdridization was done with the PrimeFlow™ RNA Assay from Affymetrix with custom designed HLA-F mRNA specific probes following manufacturer’s intructions. Flow cytometric analyses of samples were performed on a BD LSRFortessa.
Immunoblot analysis
2 × 106 Cells were lysed in 100 μL of lysis buffer (20 mM Tris·HCl, 100 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, pH 8.0, all reagents from Sigma-Aldrich) containing 1X Halt™ protease and phosphatase inhibitor cocktail (Life Technologies). Nuclei were pelleted supernatants containing cytoplasmic and plasma membrane proteins were collected. Lysates were treated with Endo Hf (New England Biolabs), PNGase F (New England Biolabs), or mock treated according to manufacturer’s instructions. Reaction products were denatured and reduced, and run on NuPAGE Novex 4–12% Bis-Tris protein gels (Life Technologies). Gel products were transferred over to a PVDF membrane, and dual immunoblotting was done with mouse anti-β-actin and rabbit anti-HLA-F primary antibodies and IRDye 800CW goat anti-mouse IgG(H+L) and IRDye 680RD goat anti-rabbit IgG(H+L) secondary antibodies. The blots were visualized using an Odyssey imaging system (LI-COR) and images were analyzed using Image Studio 3.1 software (LI-COR).
Surface Plasmon Resonance (SPR)
SPR measurements were conducted in HBS-EP buffer using a Biacore 3000 system (Biacore AB). To assess binding of various KIR-Fc constructs to HLA-F open conformers, biotinylated HLA-F monomers were immobilized onto a SA (streptavidin) sensor chip (GE Healthcare) to approximately 1000 response units (RU). A blank flow cell with no immobilized ligand was used as a reference flow cell. Injections of 60 μL of KIR-Fc constructs diluted in PBS to the indicated concentrations were performed at a flow rate of 20 μL/min, with a subsequent 10 min run of buffer to allow sufficient dissociation. Regeneration after each injection was done with two pulses of 100 μL of 10 mM glycine·HCl, pH 2.5, at a flow rate of 100 μL/min. Raw sensograms were corrected by double referencing (subtracting from the reference flow cell response and from PBS injection response). All experiments were done at standard temperature (25°C).
Generation of primary NK-cell clones
Primary human NK cells isolated from peripheral blood mononuclear cells (PBMCs) of healthy human donors were subcloned by limiting dilution in two manners. For the HIV-1 replication inhibition assay, NK cells were subcloned in the presence of feeders and maintained in NK-cell cloning medium consisting of supplement RPMI additionally supplemented with 5% human serum (Sigma-Aldrich), 1X MEM-NEAA (Gibco), 1X sodium pyruvate (Gibco), 100 μg/mL kanamycin, 450 U/mL IL-2 (AIDS Reagent Program, NIH) using a protocol adapted from a previously reported method54. Briefly, NK cells were isolated from peripheral blood mononuclear cells (PBMCs) from a KIR3DS1+/+(homozygous) donor via magnetic negative selection (NK-cell isolation kit from Miltenyi), added to a mix of irradiated feeders consisting of freshly isolated allogeneic (PBMCs) combined with log-phase-growth RPMI 8866 cells (Sigma-Aldrich) at a 10:1 ratio in cloning medium containing 1 μg/mL phytohaemagglutinin (PHA; Fisher) and mixed thoroughly before plating at 100 μL/well (1 NK cell/well) in 96-well plates and incubated for 14 days at 37°C/5% CO2. After 14 days, wells that had outgrowth of cells were transferred to 48-well plates and maintained in NK-cell medium with frequent media exchange (approximately every 3 days). For plate-bound ligand assays, NK cells were subcloned in the presence of irradiated K562 cells expressing mbIL-15 and CD137L (kind gift from Dario Campana, published in 55) and modified NK-cell cloning media consisting of RPMI supplemented with 15% fetal bovine serum (Sigma-Aldrich), 5% human serum (Sigma-Aldrich), 2 mM L-glutamine (Gibco), 1X MEM-NEAA (Gibco), 1X sodium pyruvate (Gibco), 100 μg/mL Primocin™ (Invivogen), 500 U/mL IL-2 (AIDS Reagent Program, NIH), with the addition of the following four cytokines to supplant the need for PBMC feeders: 5 ng/mL IL-15 (Peprotech), 10 ng/mL IL-12 (Peprotech), 40 ng/mL IL-18 (Peprotech), and 20 ng/mL IL-21 (Peprotech). NK-cell clones that grew out were continually cultured in modified NK-cell cloning media without additional cytokines. Cells were stained with the following antibodies for flow cytometric phenotyping: anti-CD3, anti-CD56, anti-CD16, anti-NKG2A, anti-LILRB1, anti-KIR3DS1/L1, anti-KIR2D1/S1, and anti-KIR2DL2/L3. Only NK-cell clones that were CD3−CD56+LILRB1− were used for subsequent assays to ensure proper functionality.
NK-cell degranulation and intracellular cytokine staining assay
Non-tissue culture-treated flat-bottom 96-well plates were coated overnight at 4°C with 50 μL the following ligands diluted in PBS: 1 mg/mL human γ-globulins (Thermo Scientific), 1 μg/mL of anti-KIR3DS1/L1, 10 μg/mL of HLA-E, -F, or -G monomer, or 0.5% bovine serum albumin (Miltenyi). ~1 × 104 NK cells in 100 μL of supplemented RPMI containing 1.5 μL of anti-CD107a antibody and 5 μg/mL brefeldin A (Biolegend) and in the presence of anti-KIR3DS1/L1 antibody or isotype control (mIgG1) antibodies (25 μg/mL each) were seeded onto ligand-coated plate wells. After a 5-h incubation at 37°C/5% CO2, cells were first stained with LIVE/DEAD® Fixable Blue Dead Cell Staining Kit (Life Technologies) following manufacturer’s instructions, and then stained with anti-CD56 and anti-CD16 antibodies for 15 min at 4°C. Cells were then fixed with BD Cytofix/Cytoperm solution (BD Biosciences) and permeabilized with BD Perm/Wash solution (BD Biosciences), after which intracellular cytokine staining was carried out using the following antibodies: anti-IFN-γ, anti-TNF-α, and anti-MIP-1β. After washing, flow cytometric analysis was performed on a BD LSRFortessa.
NK-cell co-incubation with HIV-1–infected autologous CD4+ T cells
PBMCs from the same donor from which NK-cell clones were generated were thawed and CD4+ T cells were enriched via magnetic negative selection with the EasySep™ Human CD4+ T Cell Enrichment Kit (StemCell). CD4+ T cells were cultured overnight in supplemented RPMI containing 50 U/mL of IL-2 and 1 μg/mL PHA. The next day, cells were washed and resuspended at 1 × 106 cells/mL in supplemented RPMI containing 50 U/mL of IL-2. HIV-1 JR-CSF was added to a final concentration of 1 × 104 TCID50/mL, and cells were incubated for 4 h at 37°C/5% CO2. Afterwards, infected CD4+ T cells were washed, resuspended in NK cell medium, and plated in a round-bottom 96-well plate at 5 × 104 cells/well. NK-cell clones from the same donor were added to individuals wells at 1.25 × 105 cells/well (NK-to-CD4+ T cell ratio = 2.5:1). Cells were co-cultured for 7 days, after which surface staining with anti-CD3 and anti-CD4 and intracellular staining with anti-HIV-1 p24 was performed for flow cytometric assessment.
Human Samples and Viruses
Frozen peripheral blood mononuclear cells (PBMCs) from healthy donors, which gave informed consent, were used in this study in accordance to protocols approved by Partners Human Research Committee and Institutional Review Board of Massachusetts General Hospital. HLA/KIR genotypes of selected human samples were determined prior to this study by high-resolution HLA typing performed at the HLA-typing laboratory of the National Cancer Institute, National Institutes of Health, and Sanger Sequencing-based KIR genotyping performed by Mary Carrington’s laboratory. Human donor samples were either completely anonymous or chosen based solely on KIR/HLA genotype. HIV-1 NL4-3 and HIV-1 JR-CSF were purchased from the Virology Core of the Ragon Institute of MGH, MIT, and Harvard, and are quality controlled for infectivity and titered on stimulated human PBMCs by standard endpoint TCID50 assay.
Data Acquisition and Analysis
Flow cytometry data were analyzed using FlowJo software version 7.6 (Tree Star) and statistical analyses were performed using GraphPad Prism 6 (GraphPad Software). KIR-Fc binding and CD69 reporter cell expression values are shown as mean values with error bars representing one standard deviation (SD). SPR data was analyzed using BIAevaluation Software (GE Healthcare); given the dimeric nature of the KIR-Fc analyte, the bivalent analyte model was used to obtain proper fit of kinetic curves.
Supplementary Material
Acknowledgments
The authors gratefully thank C. Körner and A. Crespo for scientific advice and helpful discussions. We would also like to thank the following reagent donors: C. Brander (Ragon Institute of MGH, MIT, and Harvard, Cambridge, MA) for 721.221 HLA transductants; J. Strominger (Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA) for 721.221-HLA-G cells; F. Zhang (Broad Institute of MIT and Harvard, Cambridge, MA) for lentiCas9-Blast plasmid; L. Ferreira (Department of Stem Cell and Regenerative Biology and Department of Molecular and Cellular Biology, Harvard University, Cambridge, MA) and T. Meissner (Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA) for β2m-targeting sgRNA plasmid. The authors appreciate the support and assistance from the Ragon Institute Flow Cytometry Core and Virology Core. This study was supported by the National Institute of Health (NIH) (R01-AI067031-08, P01-AI104715), the National Institute of General Medical Sciences (T32GM007753), and the Ragon Institute of MGH, MIT and Harvard. W. F. Garcia-Beltran was supported by the NIH (F31AI116366). This project has been funded in whole or in part with federal funds from the Frederick National Laboratory for Cancer Research, under Contract No. HHSN261200800001E. The content is solely the responsibility of the authors and does not necessarily represent the official views or policies of the National Institute of General Medical Sciences, the National Institutes of Health, or the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported in part by the Intramural Research Program of the NIH, Frederick National Lab, Center for Cancer Research. This research was supported in part by the Program Area Antiviral Targets and Strategies of the Heinrich-Pette Institute, Hamburg, Germany, a Leibniz Institute for Experimental Virology, and by the German Center for Infection Research (DZIF).
Footnotes
AUTHOR CONTRIBUTIONS
W.F. Garcia-Beltran, A.H., G.M., and A.W.C., Y.P., C.R.S., T.K., and H.D. performed the experiments; M.R. performed mass spectrometry analysis not presented in manuscript; P.A.L.M. and T.P. provided resources and reagents; J.D.M. designed Jurkat reporter cell assay; S.J. developed NK-cell cloning protocol; M.A., M.C., S.J., and G.A. supervised the research. W.F. Garcia-Beltran analyzed the data and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Parham P, Norman PJ, Abi-Rached L, Guethlein LA. Variable NK cell receptors exemplified by human KIR3DL1/S1. J Immunol. 2011;187:11–19. doi: 10.4049/jimmunol.0902332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vivier E, Ugolini S. Natural killer cells: From basic research to treatments. Front Immunol. 2011;2:2–5. doi: 10.3389/fimmu.2011.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajalingam R. Human diversity of killer cell immunoglobulin-like receptors and disease. Korean J Hematol. 2011;46:216. doi: 10.5045/kjh.2011.46.4.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Min-Oo G, Kamimura Y, Hendricks DW, Nabekura T, Lanier LL. Natural killer cells: Walking three paths down memory lane. Trends Immunol. 2013;34:251–258. doi: 10.1016/j.it.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waggoner SN, Cornberg M, Selin LK, Welsh RM. Natural killer cells act as rheostats modulating antiviral T cells. Nature. 2011:2–7. doi: 10.1038/nature10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin MP, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 7.Jiang Y, et al. KIR3DS1/L1 and HLA-Bw4-80I are associated with HIV disease progression among HIV typical progressors and long-term nonprogressors. BMC Infect Dis. 2013;13:405. doi: 10.1186/1471-2334-13-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rivero-Juarez A, et al. Natural Killer KIR3DS1 Is Closely Associated with HCV Viral Clearance and Sustained Virological Response in HIV/HCV Patients. PLoS One. 2013;8:8–13. doi: 10.1371/journal.pone.0061992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trydzenskaya H, et al. The genetic predisposition of natural killer cell to BK virus-associated nephropathy in renal transplant patients. Kidney Int. 2013;84:359–65. doi: 10.1038/ki.2013.59. [DOI] [PubMed] [Google Scholar]
- 10.Díaz-Peña R, et al. Association of the KIR3DS1*013 and KIR3DL1*004 alleles with susceptibility to ankylosing Spondylitis. Arthritis Rheum. 2010;62:1000–1006. doi: 10.1002/art.27332. [DOI] [PubMed] [Google Scholar]
- 11.López-Vázquez A, et al. Protective effect of the HLA-Bw4I80 epitope and the killer cell immunoglobulin-like receptor 3DS1 gene against the development of hepatocellular carcinoma in patients with hepatitis C virus infection. J Infect Dis. 2005;192:162–165. doi: 10.1086/430351. [DOI] [PubMed] [Google Scholar]
- 12.Besson C, et al. Association of killer cell immunoglobulin-like receptor genes with Hogkin’s lymphoma in a familial study. PLoS One. 2007;2:1–10. doi: 10.1371/journal.pone.0000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrington M, et al. Hierarchy of resistance to cervical neoplasia mediated by combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci. J Exp Med. 2005;201:1069–1075. doi: 10.1084/jem.20042158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gagne K, et al. Donor KIR3DL1/3DS1 gene and recipient Bw4 KIR ligand as prognostic markers for outcome in unrelated hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2009;15:1366–1375. doi: 10.1016/j.bbmt.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 15.Venstrom JM, et al. Donor activating KIR3DS1 is associated with decreased acute GVHD in unrelated allogeneic hematopoietic stem cell transplantation Brief report Donor activating KIR3DS1 is associated with decreased acute GVHD in unrelated allogeneic hematopoietic stem cell. Blood. 2010;115:3162–3165. doi: 10.1182/blood-2009-08-236943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carr WH, et al. Cutting Edge: KIR3DS1, a gene implicated in resistance to progression to AIDS, encodes a DAP12-associated receptor expressed on NK cells that triggers NK cell activation. J Immunol. 2007;178:647–651. doi: 10.4049/jimmunol.178.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cella M, Longo a, Ferrara GB, Strominger JL, Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med. 1994;180:1235–1242. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Connor GM, et al. Functional Polymorphism of the KIR3DL1/S1 Receptor on Human NK Cells. J Immunol. 2010;178:235–241. doi: 10.4049/jimmunol.178.1.235. [DOI] [PubMed] [Google Scholar]
- 19.Gillespie GMa, et al. Lack of KIR3DS1 binding to MHC class I Bw4 tetramers in complex with CD8+ T cell epitopes. AIDS Res Hum Retroviruses. 2007;23:451–455. doi: 10.1089/aid.2006.0165. [DOI] [PubMed] [Google Scholar]
- 20.O’Connor GM, et al. Peptide-dependent recognition of HLA-B*57:01 by KIR3DS1. J Virol. 2015 doi: 10.1128/JVI.03586-14. JVI.03586–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin MP, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbour JD, et al. Synergy or independence? Deciphering the interaction of HLA class I and NK cell KIR alleles in early HIV-1 disease progression. PLoS Pathog. 2007;3:1540. doi: 10.1371/journal.ppat.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long BR, et al. Conferral of enhanced natural killer cell function by KIR3DS1 in early human immunodeficiency virus type 1 infection. J Virol. 2008;82:4785–4792. doi: 10.1128/JVI.02449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardiner CM, et al. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. 2001;166:2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- 25.Thananchai H, et al. Cutting Edge: Allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol. 2007;178:33–37. doi: 10.4049/jimmunol.178.1.33. [DOI] [PubMed] [Google Scholar]
- 26.Sharma D, et al. Dimorphic motifs in D0 and D1+D2 domains of killer cell Ig-like receptor 3DL1 combine to form receptors with high, moderate, and no avidity for the complex of a peptide derived from HIV and HLA-A*2402. J Immunol. 2009;183:4569–4582. doi: 10.4049/jimmunol.0901734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arosa Fa, Santos SG, Powis SJ. Open conformers: the hidden face of MHC-I molecules. Trends Immunol. 2007;28:115–123. doi: 10.1016/j.it.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Wainwright SD, Biro Pa, Holmes CH. HLA-F is a predominantly empty, intracellular, TAP-associated MHC class Ib protein with a restricted expression pattern. J Immunol. 2000;164:319–328. doi: 10.4049/jimmunol.164.1.319. [DOI] [PubMed] [Google Scholar]
- 29.Goodridge JP, Burian A, Lee N, Geraghty DE. HLA-F complex without peptide binds to MHC class I protein in the open conformer form. J Immunol. 2010;184:6199–6208. doi: 10.4049/jimmunol.1000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trundley A, Frebel H, Jones D, Chang C, Trowsdale J. Allelic expression patterns of KIR3DS1 and 3DL1 using the Z27 and DX9 antibodies. Eur J Immunol. 2007;37:780–787. doi: 10.1002/eji.200636773. [DOI] [PubMed] [Google Scholar]
- 31.Lee N, Ishitani A, Geraghty DE. HLA-F is a surface marker on activated lymphocytes. Eur J Immunol. 2010;40:2308–2318. doi: 10.1002/eji.201040348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alter G, et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204:3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen RL, Raine T, Haude a, Trowsdale J, Wilson MJ. Leukocyte receptor complex-encoded immunomodulatory receptors show differing specificity for alternative HLA-B27 structures. J Immunol. 2001;167:5543–5547. doi: 10.4049/jimmunol.167.10.5543. [DOI] [PubMed] [Google Scholar]
- 34.Jones DC, et al. HLA class I allelic sequence and conformation regulate leukocyte Ig-like receptor binding. J Immunol. 2011;186:2990–2997. doi: 10.4049/jimmunol.1003078. [DOI] [PubMed] [Google Scholar]
- 35.Lepin EJM, et al. Functional characterization of HLA-F and binding of HLA-F tetramers to ILT2 and ILT4 receptors. Eur J Immunol. 2000;30:3552–3561. doi: 10.1002/1521-4141(200012)30:12<3552::AID-IMMU3552>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 36.Goodridge JP, Burian A, Lee N, Geraghty DE. HLA-F and MHC class I open conformers are ligands for NK cell Ig-like receptors. J Immunol. 2013;191:3553–62. doi: 10.4049/jimmunol.1300081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vivian JP, et al. Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature. 2011;479:401–405. doi: 10.1038/nature10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan FH, Liu XX, Tian W. Characterization of HLA-F polymorphism in four distinct populations in Mainland China. Int J Immunogenet. 2013;40:369–376. doi: 10.1111/iji.12047. [DOI] [PubMed] [Google Scholar]
- 39.Shiina T, et al. Molecular dynamics of MHC genesis unraveled by sequence analysis of the 1,796,938-bp HLA class I region. Proc Natl Acad Sci. 1999;96:13282–13287. doi: 10.1073/pnas.96.23.13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee N, Geraghty DE. HLA-F surface expression on B cell and monocyte cell lines is partially independent from tapasin and completely independent from TAP. J Immunol. 2003;171:5264–5271. doi: 10.4049/jimmunol.171.10.5264. [DOI] [PubMed] [Google Scholar]
- 41.Noguchi K, et al. Detection of anti-HLA-F antibodies in sera from cancer patients. Anticancer Res. 2004;42:3387–3392. [PubMed] [Google Scholar]
- 42.Le Gall S, et al. Nef Interacts with the μ Subunit of Clathrin Adaptor Complexes and Reveals a Cryptic Sorting Signal in MHC I Molecules. Immunity. 1998;8:483–495. doi: 10.1016/s1074-7613(00)80553-1. [DOI] [PubMed] [Google Scholar]
- 43.Collins KL, Chen BK, Kalams Sa, Walker BD, Baltimore D. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature. 1998;391:397–401. doi: 10.1038/34929. [DOI] [PubMed] [Google Scholar]
- 44.Cohen GB, et al. The Selective Downregulation of Class I Major Histocompatibility Complex Proteins by HIV-1 Protects HIV-Infected Cells from NK Cells. Immunity. 1999;10:661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 45.Cerboni C, et al. Human immunodeficiency virus 1 Nef protein downmodulates the ligands of the activating receptor NKG2D and inhibits natural killer cell-mediated cytotoxicity. J Gen Virol. 2007;88:242–50. doi: 10.1099/vir.0.82125-0. [DOI] [PubMed] [Google Scholar]
- 46.Derrien M, et al. Human immunodeficiency virus 1 downregulates cell surface expression of the non-classical major histocompatibility class I molecule HLA-G1. J Gen Virol. 2004;85:1945–1954. doi: 10.1099/vir.0.79867-0. [DOI] [PubMed] [Google Scholar]
- 47.Boyle LH, Gillingham AK, Munro S, Trowsdale J. Selective export of HLA-F by its cytoplasmic tail. J Immunol. 2006;176:6464–6472. doi: 10.4049/jimmunol.176.11.6464. [DOI] [PubMed] [Google Scholar]
- 48.Jinushi M, Hodi FS, Dranoff G. Therapy-induced antibodies to MHC class I chain-related protein A antagonize immune suppression and stimulate antitumor cytotoxicity. Proc Natl Acad Sci. 2006;103:9190–9195. doi: 10.1073/pnas.0603503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jinushi M, et al. MHC class I chain-related protein A antibodies and shedding are associated with the progression of multiple myeloma. Proc Natl Acad Sci. 2008;105:1285–1290. doi: 10.1073/pnas.0711293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pelak K, et al. Copy number variation of KIR genes influences HIV-1 control. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pertel T, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mandal PK, et al. Efficient Ablation of Genes in Human Hematopoietic Stem and Effector Cells using CRISPR/Cas9. Cell Stem Cell. 2014;15:643–652. doi: 10.1016/j.stem.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cella M, Colonna M. In: Methods in Molecular Biology: Natural Killer Cell Protocols: Cellular and Molecular Methods. Campbell KS, Colonna M, editors. Vol. 121. Humana Press; 1999. pp. 1–4. [DOI] [PubMed] [Google Scholar]
- 55.Imai C, Iwamoto SCD. Genetic modification of primary natural killer cells overcomes inhibitory signals and induces specific killing of leukemic cells. Leukemia. 2005;106:376–383. doi: 10.1182/blood-2004-12-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.