Summary
TM4SF20 (transmembrane 4 L6 family 20) is a polytopic membrane protein that inhibits proteolytic processing of CREB3L1 (cAMP response element-binding protein 3-like 1), a membrane-bound transcription factor that blocks cell division and activates collagen synthesis. Here we report that ceramide stimulates CREB3L1 cleavage by inverting the orientation of TM4SF20 in membranes. In the absence of ceramide, the N-terminus of the 1st transmembrane helix of TM4SF20 is inserted into the endoplasmic reticulum (ER) lumen. This translocation requires TRAM2 (translocating chain-associated membrane protein 2), a membrane protein containing a putative ceramide-interacting domain. In the presence of ceramide, the N-terminus of the 1st transmembrane domain of TM4SF20 is exposed to cytosol. Consequently, the membrane topology of TM4SF20 is inverted, and this form of TM4SF20 stimulates CREB3L1 cleavage. In the presence of ceramide, translocation of TM4SF20 is TRAM2-independent. We designate this mechanism causing regulated inversion of the membrane topology as “regulated alternative translocation”.
Introduction
The topology of integral membrane proteins, which depicts the orientation of the membrane-spanning segments within the membrane, is crucial for the function of these proteins. In mammalian cells, the topology of polytopic membrane proteins is largely determined by the orientation of the first transmembrane helix, which is inserted into membranes of the endoplasmic reticulum (ER) through three different mechanisms (Lodish et al., 2007) (Zimmermann et al., 2011). Type I insertion refers to proteins that contain a cleavable ER-targeting signal peptide N-terminal to the first transmembrane helix (Lodish et al., 2007). The signal peptide directs the hydrophilic sequence N-terminal to the transmembrane helix through the ER translocon into the ER lumen before the signal peptide is cleaved from the mature protein by signal peptidase (Zimmermann et al., 2011). The N-terminus of the first transmembrane helix of the mature protein is embedded into the ER lumen following this type of insertion. Type II and Type III insertions refer to proteins that do not contain the signal peptide. These insertions are both initiated by interaction of the ER translocon with the first transmembrane helix, but differ in direction of the insertion: While type II insertion directs the N-terminus of the transmembrane helix to cytosol, type III insertion directs the N-terminus of the first transmembrane helix to the ER lumen (Lodish et al., 2007). Currently, it is assumed that membrane proteins always adopt the same topology (Lodish et al., 2007).
This assumption is challenged by our exploration of the topology of a transmembrane protein called TM4SF20 (transmembrane 4 L6 family member 20). We previously reported that TM4SF20 inhibited proteolytic activation of CREB3L1 (cAMP response element binding protein 3-like 1), a transcription factor synthesized as a membrane-bound precursor (Chen et al., 2014). Stimulation of cells with either transforming-growth factor β (TGF-β) or ceramide causes proteolytic activation of CREB3L1 (Chen et al., 2014; Denard et al., 2012) through a process of regulated intramembrane proteolysis (RIP) (Brown et al., 2000). This proteolytic cleavage releases the N-terminal domain of CREB3L1 from membranes, allowing it to enter the nucleus where it activates transcription of genes that inhibit cell cycle progression and stimulate assembly of collagen-containing extracellular matrix (Denard et al., 2011; Murakami et al., 2009). TGF-β stimulates the cleavage of CREB3L1 by inhibiting the expression of TM4SF20 mRNA (Chen et al., 2014).
In the current study, we show that ceramide activates CREB3L1 cleavage by a mechanism different from that of TGF-β: Rather than inhibiting TM4SF20 expression, ceramide inverts the membrane orientation of the protein. This inversion mechanism creates a form of TM4SF20 that stimulates the cleavage of CREB3L1.
Results
Ceramide alters membrane topology of TM4SF20
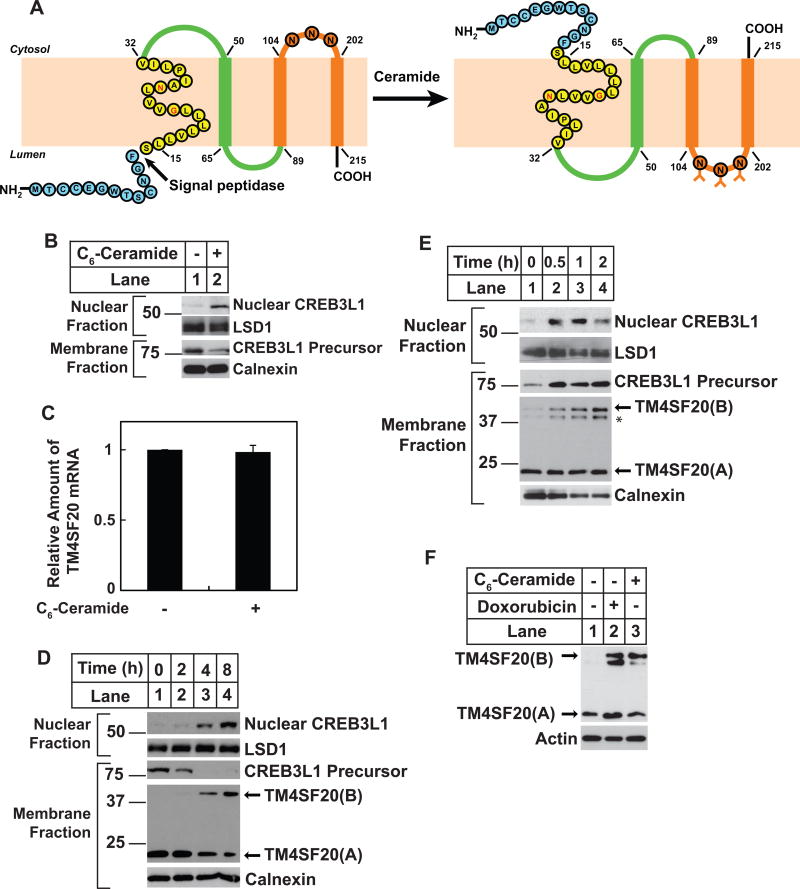
For orientation purposes, Figure 1A shows the two forms of TM4SF20 revealed in this study. In the absence of ceramide, the N-terminus of the first transmembrane helix of TM4SF20 is inserted into the ER lumen so that the preceding peptide is cleaved off by signal peptidase. In this form of TM4SF20, which is designated as TM4SF20(A), the N-terminus of the mature protein lies in the lumen. The B form of TM4SF20 is induced by ceramide. In this form, the N-terminus of the first transmembrane domain faces the cytosol. As a result, the N-terminal sequence of the protein is not removed by signal peptidase. The protein, which we designate as TM4SF20(B), adopts a membrane topology opposite to that of TM4SF20(A). Exposed loops that are cytosolic in TM4SF20(A) are luminal in TM4SF20(B) and vice versa.
Figure 1. Ceramide induces appearance of TM4SF20(B).
(A) Proposed model illustrating ceramide-induced RAT of TM4SF20. The N-terminal loop and the 1st transmembrane helix are highlighted in blue and yellow, respectively. The two polar residues G22 and N26 in the first transmembrane helix are marked in red. The three N-linked glycosylation sites in loop 3 are indicated. N80 in loop 2 is not glycosylated because of its close proximity to the transmembrane helix. The A and B forms of TM4SF20 are designated as TM4SF20(A) and TM4SF20(B), respectively.
(B and C) On day 0, A549 cells were seeded at 4×105 cells per 60-mm dish. On day 1, cells were treated with or without 6 μM C6-ceramide for 4 h. (B) Cells were harvested and separated into nuclear and membrane fractions, and analyzed by immunoblot analysis with indicated antibodies. Immunoblot analysis with antibodies against calnexin and lysine-specific demethylase 1 (LSD1) served as loading controls for membrane and nuclear fractions, respectively. (C) The amount of TM4SF20 mRNA was quantified through RT-QPCR with the value in untreated cells set to 1. Results are reported as mean ± S.E.M. of three independent experiments.
(D) On day 0, A549/pTM4SF20 cells were seeded at 4×105 cells per 60-mm dish. On day 1, cells were treated with 6 μM C6-ceramide for the indicated time. Cells were then harvested for the analysis of RIP of CREB3L1 as described in B. Immunoblot with anti-Myc was used to detect the stably transfected TM4SF20 tagged with the Myc epitope at the C-terminus of the protein.
(E) On day 0, A549/pTM4SF20 cells were seeded at 4×105 cells per 60-mm dish. On day 1, cells were treated with 0.24 units/ml of sphingomyelinase for the indicated time. Cells were then harvested for immunoblot analysis as described in D. Asterisk denotes a band derived from TM4SF20 the appearance of which is not always reproducible (e.g. Figure 1D) and is not always induced by ceramide treatment (e.g. Figure 3A). This band is thus not further pursued in this study.
(F) On day 0, A549/pTM4SF20 cells were seeded at 4×105 cells per 60-mm dish. On day 1, cells were treated with 500 nM doxorubicin for 24 h, or 6 μM C6-ceramide for 8 h followed by immunoblot analysis with the indicated antibodies.
See also Figure S1
We previously reported that TGF-β stimulated the cleavage of CREB3L1 through inhibiting the expression of TM4SF20 in A549 cells, a line of human lung cancer cells (Chen et al., 2014). Since ceramide also induces cleavage of CREB3L1 (Denard et al., 2012), we expected that ceramide would also reduce the amount of TM4SF20. As shown in Figure 1B, treatment of the cells with C6-ceramide, a cell permeable analogue of ceramide, caused cleavage of CREB3L1 as indicated by appearance of the cleaved nuclear CREB3L1 in immunoblots with an antibody against the N-terminal domain of the protein. However, unlike TGF-β, ceramide did not lower the amount of TM4SF20 mRNA (Figure 1C). Further evidence that ceramide stimulates cleavage of CREB3L1 through a mechanism different from that of TGF-β came from analysis of the cleavage in A549/pTM4SF20 cells. In these cells, expression of stably transfected TM4SF20 tagged with the Myc epitope is driven by a constitutive promoter so cleavage of CREB3L1 was not stimulated by TGF-β (Chen et al., 2014). However, C6-ceramide still induced CREB3L1 cleavage in these cells (Figure 1D, 1st panel lanes 3 and 4). Instead of reducing the amount of TM4SF20, ceramide treatment produced a new form of TM4SF20 with a higher molecular mass. In untreated cells, TM4SF20 migrated on immunoblot as a single band close to its predicted molecular mass (~23 kDa) (TM4SF20(A), Figure 1D, 4th panel lane 1); After ceramide treatment, a new form of TM4SF20 migrating at ~45 kDa appears (TM4SF20(B), Figure 1D, 4th panel lanes 3 and 4). The appearance of TM4SF20(B) correlated with generation of the nuclear form of CREB3L1 (Figure 1D).
Although C6-ceramide is not a natural product, it is converted in cells to naturally-existing ceramide through the ceramide salvage pathway that replaces the 6-carbon acyl chain in C6-ceramide with long chain fatty acids (Denard et al., 2012; Kitatani et al., 2008). To confirm that TM4SF20(B) is induced by naturally-existing ceramide, we treated the cells with reagents that stimulate endogenous production of ceramide. For this purpose, we treated the cells with a bacterial sphingomyelinase that hydrolyzes sphingomyelin to produce ceramide. This treatment generated TM4SF20(B) and induced cleavage of CREB3L1 (Figure 1E). We also treated the cells with doxorubicin, which stimulates cleavage of CREB3L1 by enhancing de-novo synthesis of ceramide (Denard et al., 2012). This treatment also stimulated production of TM4SF20(B) (Figure 1F). Lipid measurement through mass spectroscopy analysis indicated that ceramide was the only sphingolipid increased by at least two folds in all three treatments (i.e. C6-ceramide, sphingomyelinase and doxorubicin) that produced TM4SF20(B) (Figure S1).
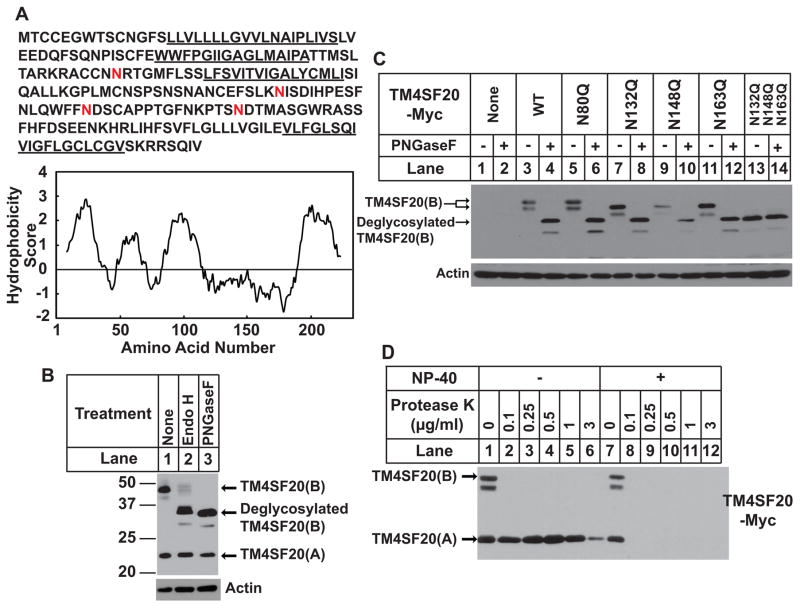
We eventually determined that TM4SF20(B) exhibited a membrane topology opposite to that of TM4SF20(A) (Figure 1A). The first line of evidence supporting this conclusion came from analysis of N-linked glycosylation. TM4SF20 belongs to a family of proteins containing four transmembrane domains (Wright et al., 2000). Hydropathy analysis shows four hydrophobic peaks corresponding to the four transmembrane helices (Figure 2A). There are four consensus N-linked glycosylation sites in TM4SF20: One (N80) is in loop 2, and the other three (N132, 148, 163) lie in loop 3 (Figures 1A and 2A). To monitor glycosylation status of TM4SF20, we treated lysate of A549/pTM4SF20 cells with endo H and PNGase F, two endoglycosidases. This treatment reduced the apparent molecular weight of TM4SF20(B) but not that of TM4SF20(A) (Figure 2B). We then performed site-directed mutagenesis to determine the glycosylation sites of TM4SF20(B). When each of the three asparagines located in loop 3 was changed to glutamine the apparent molecular weight of TM4SF20(B) was reduced (Figure 2C, lanes 3, 7, 9 and 11). Simultaneous replacement of all three asparagines reduced the apparent molecular weight of TM4SF20(B) to that of the wild type protein treated with PNGase F, which removes almost all of the N-linked sugar chains (Figure 2C, lanes 4 and 13). Treatment with the endoglycosidase did not further reduce the apparent molecular weight of the mutant protein lacking the three asparagines (Figure 2C, lanes 13 and 14). These results demonstrate that N132, N148 and N163 are the only N-linked glycosylation sites in TM4SF20(B). In contrast to these sites located in loop 3, N80 located in loop 2 is not glycosylated in TM4SF20(B) as the N80Q mutation did not alter the migration of the protein (Figure 2C, lanes 3 and 5). This site is also not glycosylated in TM4SF20(A), presumably because it is too close to the predicted transmembrane helix (Nilsson and von Heijne, 1993). We noticed that the deglycosylation treatment did not reduce the apparent molecular weight of TM4SF20(B) completely to that of TM4SF20(A) (Figure 2B), suggesting that TM4SF20(B) may contain additional post translational modifications. Ceramide appears to alter the glycosylation pattern of TM4SF20 specifically, as C6-ceramide treatment did not change that of Grp97, an ER luminal protein, or NPC1, a transmembrane protein (Figures S2A and B).
Figure 2. Ceramide alters the membrane topology of TM4SF20.
(A) The amino acid sequence and hydropathy plot of TM4SF20. The putative membrane-spanning sequence is underlined. Potential N-linked glycosylation sites are highlighted in red. The residue-specific hydropathy index was calculated over a window of 18 residues by the method of Kyte and Doolittle.
(B) On day 0, A549/pTM4SF20 cells were seeded at 4×105 cells per 60-mm dish. On day 1, after incubation with 6 μM C6-ceramide for 8 h, cells were harvested and cell lysate were incubated in the absence and presence of the indicated endoglycosidase, subjected to SDS/PAGE followed by immunoblot analysis.
(C) On day 0, A549 cells were seeded at 4×105 cells per 60-mm dish. On day 1, cells were transfected with 0.1 μg of wild type or the mutant pCMV-TM4SF20-Myc as indicated. On day 2, cells were treated with 6 μM C6-ceramide for 8 h. Cell lysates were incubated in the absence or presence of PNGase F followed by immunoblot analysis.
(D) A549/pTM4SF20 cells were seeded and treated as described in C. Membrane vesicles were subjected to protease protection assay as described in Experimental Procedure followed by immunoblot analysis with anti-Myc detecting the epitope tagged at the C-terminus of TM4SF20.
See also Figure S2
Since N-linked glycosylation only occurs in the lumen of the ER (Breitling and Aebi, 2013), the results shown above suggest that loop 3 of TM4SF20(B) is located in the ER lumen, whereas the same loop in TM4SF20(A) is exposed to the cytosol. Loop 3 is separated from the C-terminal end of the protein by a single transmembrane helix. If loop 3 is inverted, then the C-terminal end of the protein should be in the ER lumen and cytosol for TM4SF20(A) and TM4SF20(B), respectively. To test this hypothesis, we isolated sealed membrane vesicles from A549/pTM4SF20 cells in which the Myc epitope is located at the C-terminus of TM4SF20, and incubated them with proteinase K in the absence or presence of the detergent NP-40 that solubilizes microsome membranes. As shown in Figure 2D, the Myc epitope at the C-terminus of TM4SF20(A) but not TM4SF20(B) was protected from protease digestion by microsomes. These results support the conclusion that the membrane topology of TM4SF20(B) is different from that of TM4SF20(A).
Ceramide prevents the N-terminal sequence of TM4SF20 from cleavage by signal peptidase through regulated alternative translocation
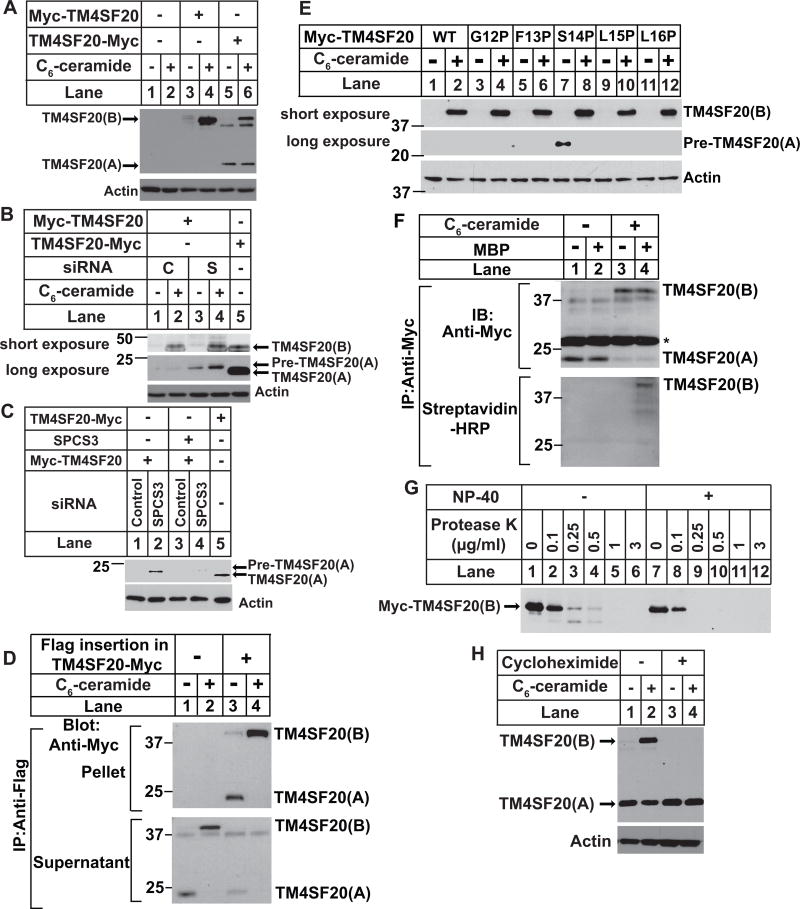
The second line of evidence supporting the model shown in Figure 1A comes from the different accessibility of the N-terminal sequence of the two forms of TM4SF20 to cleavage catalyzed by signal peptidase, a reaction that occurs only in the ER lumen (Zimmermann et al., 2011). We made this observation by performing immunoblot analysis of TM4SF20 tagged at the N-terminus with the Myc epitope. In cells treated with C6-ceramide, we detected the Myc epitope at the N-terminus of TM4SF20(B) (Figure 3A, lane 4). We did not detect the Myc epitope at the N-terminus of TM4SF20(A) (Figure 3A, lanes 3 and 4). In contrast, when we transfected the cells with a plasmid encoding TM4SF20 tagged with the myc epitope at C-terminus of the protein, we detected both forms of TM4SF20 (Figure 3A, lanes 5 and 6). These observations suggest that the N-terminal sequence of TM4SF20(A) together with the Myc epitope but not that of TM4SF20(B) may be co-translationally removed from the mature protein by signal peptidase. To further test the hypothesis, we transfected cells with a siRNA targeting signal peptidase complex subunit 3 (SPCS3), a component of signal peptidase (Meyer and Hartmann, 1997), and a plasmid encoding TM4SF20 tagged with the Myc epitope at the N-terminus. The siRNA knocked down SPCS3 mRNA by more than 90% (Figure S3A), causing the N-terminal Myc tag to remain associated with TM4SF20(A) (Figure 3B). This protein had an apparent molecular weight slightly higher than that of TM4SF20(A) present in cells transfected with a plasmid encoding C-terminally tagged TM4SF20 (Figure 3B). To confirm that the retention of the N-terminal myc tag in TM4SF20(A) was caused by knockdown of SPCS3, we transfected the cells with a cDNA encoding SPCS3 that contained synonymous mutations at the region targeted by the siRNA. Restoration of SPCS3 expression (Figure S3B) caused removal of the Myc tag from TM4SF20(A) (Figure 3C). We named this form of TM4SF20 as TM4SF20(A) precursor (pre-TM4SF20(A)) that contains the N-terminal sequence normally cleaved off by signal peptidase.
Figure 3. Ceramide induces RAT of TM4SF20.
(A) On day 0, A549 cells were seeded at 4×105 cells per 60-mm dish. On day 1, cells were transfected with 0.1 μg of plasmid encoding TM4SF20 tagged with the Myc epitope at C- (TM4SF20-Myc) or N-terminus of the protein (Myc-TM4SF20). On day 2, cells were treated with or without 6 μM C6-ceramide for 8 h. Cell lysates were subjected to immunoblot analysis with anti-Myc to detect TM4SF20.
(B and C) On day 0, A549 cells were seeded at 1×105 cells per 60-mm dish. On day 1, the cells were transfected with a control siRNA (C) or that targeting SPCS3 (S). On day 3, the cells were transfected with 0.1 μg of plasmids encoding N or C-terminally tagged TM4SF20 and SPC3 containing synonymous mutations at the region targeted by the siRNA as described in A. On day 4, some cells were treated with 6 μM C6-ceramide for 8 h as indicated (B) while others were not treated with the lipid (C). Cell lysates were subjected to immunoblot analysis with the indicated antibody.
(D) On day 0, A549 cells were seeded at 5×105 cells per 60-mm dish. On Day 1, cells were transfected as described in A with 0.1 μg of indicated plasmids encoding C-terminally Myc-tagged TM4SF20 with or without the Flag epitope inserted N-terminal to the first transmembrane helix. After incubation for 8 h, cells were treated with 5 μM C6-ceramide. On day 2, after 16 h of the treatment, cells were harvested and lysates of the cells were subject to immunoprecipitation with anti-Flag. Supernatant and pellet fractions of the precipitation were loaded at 1:1 ratio and analyzed by immunoblot analysis with anti-Myc.
(E) A549 cells were seeded, transfected with the indicated plasmids encoding the proline scanning mutants of TM4SF20, and treated with C6-ceramide as described in (D). Cell lysates were subjected to immunoblot analysis with anti-Myc to detect TM4SF20.
(F) A549 cells were seeded on day 0 and transfected on day 1 as described in (A) with pCMV-TM4SF20-Myc(C47, 77, 78, 101, 116, 126, 151, 217, 219S) in which cysteine residues are only presented in peptide N-terminal to the putative signal peptidase cleavage site. After 6 h, the cells were treated with 5 μM C6-ceramide as indicated for 16 h. On day 2, cysteines residues from the cell lysate were labeled with maleimide-biotin (MBP) as indicated. Following isolation of TM4SF20 through immunoprecipitation with anti-Myc, the presence of cysteine residues in TM4SF20 was detected by immunoblot with streptavidin-HRP as described in Experimental Procedure. Asterisk denotes light chain of anti-Myc.
(G) A549 cells were seeded on day 0 and transfected on day 1 as described in (A) with Myc-TM4SF20. On day 2, cells were treated with 6 μM C6-ceramide for 8 h. Membrane vesicles were subjected to protease protection assay as described in Figure 2D.
(H) On day 0, A549/pTM4SF20 cells were seeded at 4×105 cells per 60-mm dish. On day 1, cells were treated with 50 μM cycloheximide or 4 μM C6-ceramide as indicated for 5 h. Cell lysates were subjected to immunoblot analysis with anti-Myc to detect TM4SF20.
See also Figure S3
Calculation of the difference in molecular weight between pre-TM4SF20(A) and TM4SF20(A) suggested that the N-terminal 13–15 amino acids located N-terminal to the first transmembrane helix may be cleaved off by signal peptidase. To test this hypothesis, we inserted the Flag epitope immediately before the first transmembrane helix between F13 and S14 of C-terminally Myc-tagged TM4SF20. Following immunoprecipitation with anti-Flag, we detected in immunoprecipitates both A and B forms of TM4SF20 with the Flag insertion by immunoblot with anti-Myc (Figure 3D, lanes 3 and 4). This immunoprecipitation is Flag-specific as neither form of wildtype TM4SF20 without the Flag insertion was immunoprecipitated (Fig. 3D, lanes 1 and 2). Since the Flag epitope is still present in TM4SF20(A) detected by the Myc epitope tagged at the C-terminus of the protein, signal peptidase is unlikely to cleave at a position C-terminal to the Flag insertion site.
To identify the signal peptidase cleavage site more precisely, we took advantage of previous observations that proline is not tolerated at the P1′ position (i.e., the residue at the C-terminus of the cleaved peptide bond) of the substrates for signal peptidase (Choo and Ranganathan, 2008; Nilsson and Von Heijne, 1992). We made mutations altering each individual amino acid from positions 12 to 16 to proline in N-terminally Myc-tagged TM4SF20. None of the mutation we made affected ceramide-induced production of TM4SF20(B) (Figure 3E, upper panel). Pre-TM4SF20(A) was only detectable in TM4SF20(S14P) in the immunoblot detecting the Myc epitope tagged at the N-terminus of the protein (Figure 3E, middle panel, lane 7). Thus, it appeared that the signal peptide cleaved TM4SF20(A) between F13 and S14.
To rule out the possibility that the results shown above are caused by artifacts of the Myc epitope tagged at the N-terminus of the protein, we also analyzed cleavage of the N-terminal sequence of TM4SF20 that is not N-terminally tagged. For this purpose, we determined the presence of the N-terminal sequence of TM4SF20 by detecting cysteine residues within the sequence. TM4SF20 contains 12 cysteine residues, with 3 of them located in the sequence N-terminal to the putative signal peptidase cleavage site. We mutated all 9 cysteines C-terminal to the cleavage site to serines so that all cysteine residues are located in the putative signal sequence in the mutant protein. If the N-terminal sequence is cleaved off by signal peptidase in TM4SF20(A) but not TM4SF20(B), then only TM4SF20(B) but not TM4SF20(A) should contain any cysteine residues in the mutant protein. To test this hypothesis, we transfected cells with a plasmid encoding the mutant protein tagged with Myc epitope at the C-terminus, incubated the cell lysate with or without maleimide-biotin that specifically labels cysteine residues with biotin, immunoprecipitated TM4SF20 with anti-Myc, and analyzed the immunoprecipitates through immunoblot analysis. Similar to the wildtype protein, immunoblot with anti-Myc detected the A form of TM4SF20 containing the cysteine mutations in untreated cells and B form of the protein in ceramide-treated cells (Figure 3F, upper panel). In cell lysate treated with maleimide-biotin, TM4SF20(B) was visible in the streptavidin-HRP blot that specifically detects biotin-labeled cysteine residues in cells treated with ceramide (Figure 3F, lower panel, lane 4). In contrast, TM4SF20(A) in the untreated cells was not detectable in the same blot (Figure 3F, lower panel, lane 2). These results demonstrate that cysteine residues located in the N-terminal 13 amino acid residues are presented in TM4SF20(B) but not in TM4SF20(A).
The reason that the N-terminal sequence is not cleaved off from TM4SF20(B) may be that this sequence resides in cytosol so that it is not accessible to signal peptidase located in the ER lumen (Zimmermann et al., 2011). To test this hypothesis, we performed a protease protection assay using C6-ceramide-treated cells transfected with a plasmid encoding N-terminally Myc-tagged TM4SF20. The Myc epitope at the N-terminus of TM4SF20(B) was not protected from protease digestion by microsomes (Figure 3G). These results are consistent with the hypothesis that the N-terminus of TM4SF20(B) is exposed to cytosol.
We also investigated whether ceramide altered membrane topology of endogenous TM4SF20. Since we were unable to generate an antibody to detect the protein, we knocked in a Flag epitope at the N-terminus of the protein through the CRISPR/CAS9 approach. Similar to the transfected protein shown in Figure 3A, the Flag epitope tagged at the N-terminus of the endogenously expressed TM4SF20 can only be detected in TM4SF20(B) when cells were treated with ceramide (Figure S3C).
We then delineated the mechanism through which ceramide induces appearance of TM4SF20(B). We determined that the disappearance of TM4SF20(B) in cells that were not treated with ceramide was not owing to rapid degradation because addition of MG132, a proteasome inhibitor, did not increase the amount of TM4SF20(B) in these cells (Figure S3D). Ceramide is also unlikely to flip the topology of previously synthesized TM4SF20 as it is difficult to explain how the N-terminal sequence cleaved off from TM4SF20(A) synthesized in the absence of ceramide reappears in TM4SF20(B) generated after the ceramide treatment. To further determine whether ceramide stimulates transformation of previously synthesized TM4SF20(A) into TM4SF20(B), we performed a pulse-chase analysis. For this purpose, we transfected cells with a plasmid encoding C-terminally Myc-tagged TM4SF20, pulse labeled the cells with [35S]methionine and cysteine in the absence of ceramide, chased the cells in the presence or absence of C6-ceramide, and immunoprecipitated TM4SF20 with anti-Myc. If TM4SF20(A) can be transformed into TM4SF20(B), then we should observe radiolabeled TM4SF20(B) in immunoprecipitates of ceramide-treated cells. While radiolabeled TM4SF20(A) was visible in the immunoprecipitates regardless of the ceramide treatment, we were unable to detect such a band specific for TM4SF20(B) in that of ceramide-treated cells (Figure S3E). Thus, TM4SF20(B) is not derived from previously synthesized TM4SF20(A).
Since ceramide does not flip the topology of TM4SF20 that has been synthesized before the lipid treatment, it is likely that the newly synthesized protein after the lipid treatment adopts an inversed topology to generate TM4SF20(B). If this is the case, then treatment with cycloheximide, an inhibitor of protein synthesis, should block ceramide-induced production of TM4SF20(B). As predicted, ceramide-induced appearance of TM4SF20(B) was completely blocked by co-treatment with cycloheximide (Figure 3H, lanes 2 and 4). Cycloheximide treatment did not reduce the amount of TM4SF20(A) in cells treated with C6-ceramide (Figure 3H, lanes 2 and 4), presumably because the protein was synthesized before the lipid treatment. These results suggest that ceramide alters the direction through which transmembrane helices are translocated into ER membrane during translation of TM4SF20. We thus designate this mechanism causing regulated inversion of the membrane topology as ‘regulated alternative translocation’ (RAT).
RAT of TM4SF20 depends on polar residues located in the first transmembrane helix
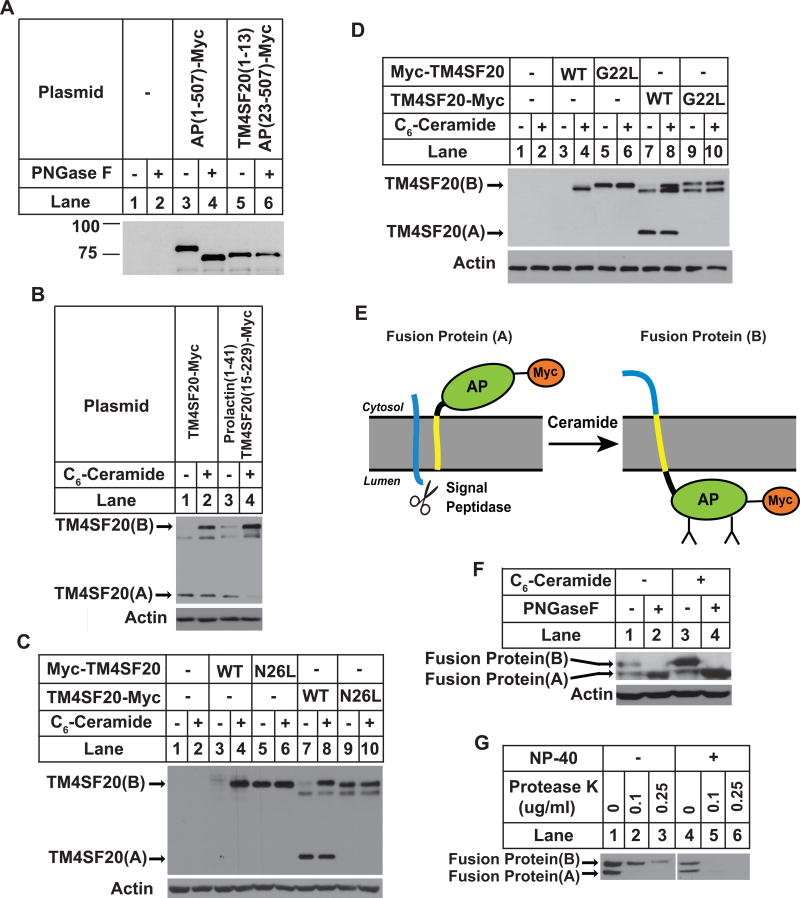
Since the N-terminal sequence of TM4SF20(A) but not TM4SF20(B) is cleaved off the mature protein by signal peptidase, we speculate that this sequence may function as a signal peptide in TM4SF20(A) but not in TM4SF20(B). However, this sequence appears to be shorter and less hydrophobic than a typical signal sequence and is not predicted to be a signal peptide according to currently established criteria (Petersen et al., 2011). To determine whether this sequence may function as a signal peptide, we replaced the signal peptide of alkaline phosphatase (AP), a secreted glycosylated protein (Sakai et al., 1998), with the N-terminal 13 amino acid residues of TM4SF20. Unlike wild type AP that is N-glycosylated, the fusion protein is not N-glycosylated as treatment with PNGase F did not reduce the apparent molecular weight of the protein (Figure 4A). These results suggest that the N-terminal sequence of TM4SF20 cannot substitute for the signal peptide of AP to direct the protein into ER lumen where N-linked glycosylation occurs.
Figure 4. The first transmembrane helix of TM4SF20 is critical for RAT of the protein.
(A) A549 cells were seeded, transfected with a plasmid encoding either wildtype AP or mutant AP in which the signal peptide is replaced by the N-terminal sequence of TM4SF20, and analyzed by immunoblot analysis following PNGase F treatment as described in Figure 2C.
(B–D) A549 cells were seeded, treated, transfected with a plasmid encoding TM4SF20-Myc in which the N-terminal sequence was replaced by the signal peptide of prolactin (B) and that contains the indicated point mutation (C and D), and analyzed as described in Figure 3A.
(E) A model predicting ceramide-induced RAT of TM4SF20-AP fusion protein. The putative signal sequence and the first transmembrane helix of TM4SF20 are highlighted in blue and yellow, respectively.
(F and G) A549 cells were set up at 4×105 cells per 60-mm dish. On day 1 they were transfected with 0.5 μg of the plasmid encoding the fusion protein. On day 2 the cells were treated with 3 μM C6-ceramide for 8 h as indicated. (F) Cell lysates were subjected to PNGaseF treatment followed by immunoblot analysis with anti-Myc to detect the transfected fusion protein. (G) Membrane vesicles of ceramide-treated cells were subjected to protease protection assay as described in Figure 2D.
To further determine the role of the N-terminal sequence of TM4SF20 in RAT of the protein, we replaced the sequence with the signal peptide of prolactin. Surprisingly, ceramide still stimulated RAT of the fusion protein (Figure 4B). Thus, ceramide-induced RAT of TM4SF20 is independent of the presence of a functional signal peptide at the N-terminus of the protein. We then focused our attention on the first transmembrane helix of TM4SF20. This helix contains hydrophobic residues typical for a transmembrane domain except for two polar residues, namely G22 and N26 (Figure 1A). To determine the importance of these residues, we mutated each of them to leucine. As expected, immunoblot with the Myc epitope tagged at the N-terminus of wild type TM4SF20 only detected TM4SF20(B) in cells treated with ceramide (Figure 4C, lanes 3–4). In contrast, N-terminally Myc-tagged TM4SF20(N26L)(B) was readily visible regardless of C6-ceramide treatment (Figure 4C, lanes 5–6). Unlike the wild type C-terminally Myc-tagged TM4SF20 that existed as the A form in the absence of ceramide and existed as both the A and B forms in cells treated with C6-ceramide (Figure 4C, lanes 7–8), the C-terminally tagged TM4SF20(N26L) only existed as the B form and the A form was undetectable regardless of the ceramide treatment (Figure 4C, lanes 9–10). Exactly the same result was observed for TM4SF20(G22L) (Figure 4D). These results suggest that G22 and N26 within the first transmembrane helix are required for generation of TM4SF20(A) but not for TM4SF20(B) by allowing the insertion of the N-terminus of the first transmembrane helix into the ER lumen.
To determine whether the first transmembrane helix of TM4SF20 is sufficient to induce RAT, we fused the N-terminal 36 amino acid residues encompassing the N-terminal loop and the first transmembrane helix of TM4SF20 to AP that has its N-terminal signal peptide deleted, and tagged the fusion protein with the Myc epitope at the C-terminus. Since the N-terminal signal sequence is deleted from AP, its membrane orientation should be determined by the N-terminal sequence derived from TM4SF20. A model shown in Figure 4E predicts the localization of AP based on the hypothesis that the N-terminal sequence of the fusion protein derived from TM4SF20 is capable of inducing RAT in response to ceramide: The AP portion of the fusion protein is expected to be in cytosol in the absence of ceramide. As a result, the fusion protein should not be glycosylated, and the C-terminal Myc epitope is not expected to be protected by microsomes (A form of the fusion protein); If ceramide treatment leads to RAT of the fusion protein, then the AP portion of the fusion protein is expected to be in the ER lumen so the fusion protein should be glycosylated, and the C-terminal tag should be protected from protease digestion by microsomes as well (B form of the fusion protein). In the absence of ceramide, the fusion protein migrated as two bands in immunoblot analysis detecting the C-terminally tagged Myc epitope (Figure 4F, lane 1). The higher band was glycosylated, as treatment with PNGase F reduced its apparent molecular weight (Figure 4F, lanes 1 and 2). The C-terminal Myc tag in the band was also protected from protease digestion by intact microsomes (Figure 4G). Thus, the higher band represented the B form of the fusion protein. The lower band was not glycosylated, as PNGase F treatment did not further reduce its apparent molecular weight (Figure 4F, lanes 1 and 2). This band was also not protected from protease digestion by microsomes (Figure 4G). The lower band was thus the A form of the protein. The observation that C6-ceramide treatment only increased the amount of the B but not the A form of the protein suggested that ceramide also induced RAT of the fusion protein as predicted by the model shown in Figure 4E (Figure 4F, lanes 1 and 3). These results suggest that the first transmembrane helix of TM4SF20 is not only required but also sufficient to induce RAT. However, unlike TM4SF20 that only existed as the A form in the absence of ceramide, the fusion protein existed as both A and B forms under this condition (Figure 4F, lane 1). Thus, it appears that amino acid residues C-terminal to the first transmembrane helix may also be involved for efficient production of TM4SF20(A) in the absence of ceramide.
Translocating chain-associated membrane protein 2 (TRAM2) is required for ER translocation of TM4SF20 in the absence but not presence of ceramide
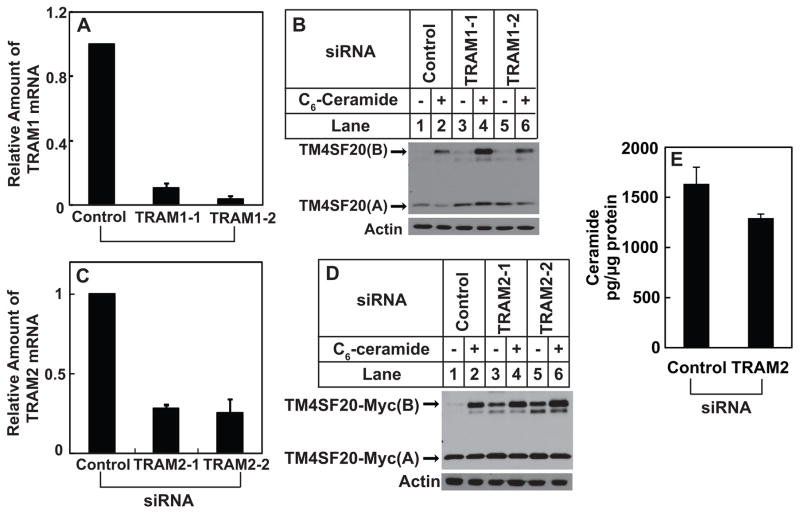
The next puzzle to solve is how ceramide induces RAT of TM4SF20 by altering the direction through which the first transmembrane helix is inserted into the ER. Since transmembrane helix is inserted into ER membranes through ER translocon, we looked for translocon-associated proteins that could be regulated by ceramide. This analysis led us to translocating chain-associated membrane protein 1 (TRAM1), a translocon-associated protein (Görlich et al., 1992; Görlich and Rapoport, 1993) that also contains a TLC domain, which is shared by ceramide synthase and is postulated to bind ceramide or related sphingolipids (Winter and Ponting, 2002). We thus wondered whether TRAM1 was required for insertion of the N-terminus of the first transmembrane helix of TM4SF20 into ER lumen, and whether ceramide triggered RAT of the protein by inactivating TRAM1. However, this hypothesis does not appear to be correct as knockdown of TRAM1 by more than 90% with two different siRNA did not affect ceramide-induced RAT of TM4SF20 (Figures 5A and B).
Figure 5. Ceramide induces RAT of TM4SF20 through TRAM2.
(A–D) On day 0, A549/pTM4SF20 cells were seeded at 1×105 cells per 60-mm dish. On day 1, the cells were transfected with indicated siRNAs. On day 3, some of the cells were harvested for quantification of indicated mRNA through RT-QPCR with the value in cells transfected with the control siRNA set to 1 (A and C). Some of the cells were harvested for quantification of sphingolipids (E). The rest of the cells were treated with 6 μM C6-ceramide as indicated for 8 h. Cell lysates were subjected to immunoblot analysis of TM4SF20 with anti-Myc (B and D). (A, C and E) Results are reported as mean ± S.E.M. of three independent experiments.
Database searches revealed that in addition to TRAM1, mammalian cells expressed two TRAM1 homologues, namely TRAM1L1 and TRAM2, the function of which in protein translocation has never been characterized. Since A549 cells only express TRAM2 but not TRAM1L1, we determined the requirement of this protein for RAT of TM4SF20. Knockdown of TRAM2 by ~70% with two different siRNA (Figure 5C) produced TM4SF20(B) even in the absence of the treatment with exogenous ceramide (Figure 5D). Under this condition the endogenous production of ceramide was not increased (Figure 5E). These results suggest that TRAM2 is required for insertion of the N-terminus of the first transmembrane helix into the ER lumen, as knockdown of the protein promoted generation of TM4SF20(B) in which the N-terminus of the first transmembrane helix faces cytosol.
TM4SF20(B) stimulates CREB3L1 cleavage
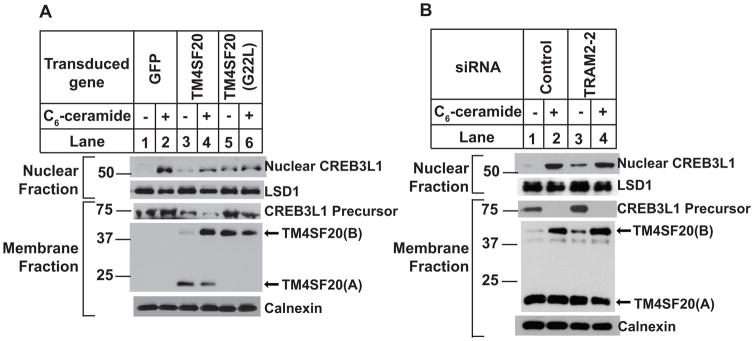
The results in Figure 1 showed that the appearance of TM4SF20(B) correlated with generation of the nuclear form of CREB3L1. If TM4SF20(B) indeed triggers CREB3L1 cleavage, then treatments producing TM4SF20(B) other than those causing accumulation of ceramide should trigger CREB3L1 cleavage as well. We thus determined whether overexpression of TM4SF20(G22L) that exists as TM4SF20(B) regardless of ceramide treatment (Figure 4D) would cause cleavage of CREB3L1 even in the absence of ceramide. For this purpose, we infected A549 cells with lentivirus encoding wild type or the G22L mutant of TM4SF20. While infection with the control virus encoding the green fluorescent protein or that encoding the wild type TM4SF20 did not affect ceramide-induced cleavage of CREB3L1 (Figure 6A, 1st panel, lanes 1–4), infection with the virus encoding TM4SF20(G22L) resulted in cleavage of CREB3L1 even in the absence of ceramide (Figure 6A, 1st panel lanes 5).
Figure 6. TM4SF20(B) activates CREB3L1 cleavage.
(A) A549 cells were seeded on day 0 at 1.5×106 cells per 100-mm dish. On day 1, cells were infected with lentivirus encoding the indicated proteins. On day 2, 24 h later, the cells were switched into fresh medium containing 2 μg/ml puromycin for selection of the virus-infected cells. On day 4, cells were treated with or without 6 μM C6-ceramide for 8 h. Cells were then harvested and analyzed as described in Figure 1D.
(B) A549/pTM4SF20 cells were set up and treated as described in Figure 5A. Cells were also fractionated into nucleus and membranes for analysis of RIP of CREB3L1 as described in Figure 1B.
Another way to test the hypothesis is to knockdown TRAM2, which also caused RAT of TM4SF20 regardless of ceramide treatment (Figure 5D). In the absence of ceramide treatment, TM4SF20(B) and the nuclear form of CREB3L1 were barely detectable in cells transfected with a control siRNA, but both proteins became readily detectable in those transfected with a siRNA targeting TRAM2 (Figure 6B, lanes 1 and 3).
Discussion
The findings in the current study support the model presented in Figure 1A. According to this model, RAT of TM4SF20 is caused by different directions through which the first transmembrane helix of the protein is inserted into ER membranes. Since the N-terminus of TM4SF20(A) produced in the absence of ceramide is cleaved off from the mature protein by signal peptidase, it is tempting to conclude that the first transmembrane helix adopts the Type I insertion (Lodish et al., 2007) under this circumstance. However, we have demonstrated that the N-terminal sequence does not function as a signal peptide to direct the N-terminus of the first transmembrane helix into ER lumen. Thus, it appears that the cleavage catalyzed by the signal peptidase is not a result of the function of the N-terminal peptide as a signal sequence but rather a consequence of the accessibility of the peptide to the protease in the ER lumen. This type of cleavage catalyzed by signal peptidase has been reported in proteolytic processing of hepatitis C virus protein in which the protease cleaves the viral polyprotein precursor at multiple sites in the ER lumen distal to the N-terminal signal sequence (Hijikata et al., 1991). In contrast to the N-terminal peptide, the first transmembrane helix itself plays a critical role for its insertion in such orientation. Thus, this insertion may be more appropriately categorized as the Type III insertion (Lodish et al., 2007). Insertion of the first transmembrane helix through this orientation appears to require TRAM2, and depends on residues G22 and N26 in the first transmembrane domain of TM4SF20. It is possible that these two residues are critical for the nascent polypeptide chain of TM4SF20 to interact with TRAM2, and ceramide may block this interaction by interacting with the TLC domain of TRAM2. In the absence of TRAM2-mediated translocation, the first transmembrane helix of TM4SF20 is inserted into the ER membrane through the Type II orientation. Consistent with this scenario, knockdown of TRAM2 or mutations in G22 and N26 triggered RAT of TM4SF20 even in the absence of ceramide.
TRAM2 is a homologue of TRAM1 that has been shown to be an ER translocon-associated protein that facilitates translocation of certain signal peptides that are less hydrophobic than a typical signal sequence (Voigt et al., 1996). Unlike TRAM1, the function of TRAM2 in protein translocation through ER membranes has never been demonstrated previously. The current study suggests that TRAM2 may be involved in RAT of TM4SF20. While we do not have direct biochemical evidence, we suspect that TRAM2 is the ceramide sensor for RAT of TM4SF20, as the protein contains a TLC domain, which is also present in ceramide synthase and is postulated to bind ceramide or ceramide-derived lipids (Winter and Ponting, 2002). Exactly which species of ceramide acts as a ligand for TRAM2 remains unclear. We also cannot rule out the possibility that a metabolite of ceramide is the ligand that inactivates TRAM2. It is also possible that other lipids can interact with TRAM2 to stimulate RAT of transmembrane proteins. These questions may only be addressed by measuring the direct binding of ceramide or other lipids to purified TRAM2. Such an assay is technically challenging, owing to the difficulty of finding a detergent suitable for the binding between a protein with multiple transmembrane domains and hydrophobic ligands.
Exactly how TM4SF20 regulates CREB3L1 cleavage remains unclear. Previous studies have identified Scap and Insig proteins as key regulators for RIP of sterol regulatory element binding proteins (SREBPs), the prototype of transcription factors activated through RIP (Brown and Goldstein, 2009; Ye and DeBose-Boyd, 2011). Scap binds to SREBPs (Sakai et al., 1997). In the absence of cholesterol, Scap escorts SREBPs from the ER to Golgi complex where SREBPs are cleaved by the Golgi-localized proteases that liberate the N-terminal domain of SREBPs from membranes, allowing it to activate transcription of all genes required for cholesterol synthesis and uptake (Brown and Goldstein, 2009; Horton et al., 2003; Ye and DeBose-Boyd, 2011). Excess cholesterol triggers binding of Insig proteins to Scap in order to retain the Scap/SREBPs complex in the ER, thereby preventing proteolytic activation of SREBPs by separating the proteins from the Golgi-localized proteases (Brown and Goldstein, 2009; Yang et al., 2002; Ye and DeBose-Boyd, 2011). In analogy to the RIP of SREBPs, TM4SF20(A) and TM4SF20(B) may perform the function similar to that of Insig proteins and Scap to inhibit and activate the RIP of CREB3L1, respectively.
What is remarkable in ceramide-induced RIP of CREB3L1 is that both the inhibitor (i.e. TM4SF20(A)) and the activator (TM4SF20(B)) of CREB3L1 cleavage is encoded by the same mRNA. In the absence of ceramide, TM4SF20(A) is the translational product that inhibits CREB3L1 cleavage. Upon ceramide treatment, instead of producing TM4SF20(A), TM4SF20(B) becomes the translational product that activates CREB3L1 cleavage. This regulatory system allows ceramide to increase expression of the activator and at the same time prevent further synthesis of the inhibitor of the RIP reaction, thereby efficiently triggering proteolytic activation of CREB3L1.
The most important finding in the current study is the identification of RAT as a mechanism to regulate the function of a transmembrane protein by reversing its membrane topology. The topology of several bacteria membrane proteins has been reported to be altered by changing the level of phosphatidylethanolamine in membranes, but this bacterial mechanism appears to be different from the current mammalian example of RAT: In the bacterial system, reversal of the topology occurs to proteins already synthesized before the lipid manipulation (Bogdanov and Dowhan, 2012; Levy, 1996; Von Heijne, 2006), whereas in the mammalian system reversal occurs only to newly synthesized proteins made after the lipid treatment. A few mammalian transmembrane proteins have been reported to adopt dual topology (Dunlop et al., 1995; Sebag and Hinkle, 2009), but the underlying mechanism has not been identified. It will be interesting to determine whether RAT is also responsible for these proteins to acquire different topology.
While TM4SF20 is the first mammalian protein observed to undergo RAT, we believe that this is not the only protein that undergoes this type of regulation. Any one of the three TRAM proteins, namely TRAM1, TRAM1L1 and TRAM2, may facilitate RAT of other transmembrane proteins. Remarkably, all three TRAM proteins contain a TLC domain, making it possible for ceramide to be a global regulator of RAT. Thus, identification of other proteins subjected to ceramide-regulated RAT will provide new insights into the mechanism through which ceramide affects cell physiology.
Experimental Procedures
Materials
We obtained doxorubicin, sphingomyelinase, N-hexanoyl-D-erythro-sphingosine (C6-ceramide), cycloheximide, rabbit anti-actin and mouse anti-HA from Sigma; rabbit anti-LSD1 from Cell Signaling Technology; mouse anti-calnexin from Enzo Life Sciences; Hybridoma cells producing IgG-9E10, a mouse monoclonal antibody against Myc tag, were obtained from the American Type Culture Collection. A rabbit polyclonal antibody against human CREB3L1 was generated as previously described (Denard et al., 2011). A mouse monoclonal antibody against human CREB3L1 (10H1) was generated by immunizing mice with polypeptides corresponding to amino acid residues 7–41 of CREB3L1.
Immunoblot
Cells were harvested and separated into nuclear and membrane fractions as described (Sakai et al., 1996), and analyzed by SDS-PAGE followed by immunoblot analysis with the indicated antibodies (1:4000 dilution for anti-calnexin and mouse monoclonal anti-CREB3L1 10H1, 1:2000 dilution for anti-Myc, anti-HA, rabbit polyclonal anti-CREB3L and anti-actin, and 1:1000 dilution for the remaining antibodies). Bound antibodies were visualized with a peroxidase-conjugated secondary antibody using the SuperSignal ECL-HRP substrate system (Pierce). Except for figure 1A in which immunoblot of CREB3L1 was carried out by the rabbit polyclonal antibody, all other experiments utilized 10H1 to detect CREB3L1.
Protease protection assay
Protease protection assay was performed as previously described (Feramisco et al., 2004) with minor modifications: Aliquots of membranes containing 50 μg protein were treated with various amount of proteinase K in the absence or presence of 1% NP-40 in a total volume of 50 μl for 1 h at 37°C. The reactions were stopped by addition of 2 μl of phenylmethylsulfonyl fluoride at a final concentration of 5 mM. The samples were then mixed with 5× SDS loading buffer, boiled for 5 min, and analyzed by immunoblot analysis.
Detection of cysteine residues in TM4SF20
Cells pulled from five 60-mm dishes were lysed in buffer A containing 1% NP-40. Cell lysates were incubated with 350 μM MBP (Life Technologies) at room temperature for 1 h protected from light for biotinylation of cysteine residues. Following immunoprecipitation with anti-Myc, the immunoprecipitates were subjected to immunoblot analysis with High Sensitivity Streptavidin-HRP (ThermoScientific) to detect biotinylated cysteine residues according to instruction of the manufacturer.
Supplementary Material
Acknowledgments
We thank Drs. Brown and Goldstein for their helpful comments and constant supports; Lisa Beatty, Ijeoma Dukes, Nimisha Jacob and Lauren Valsin for help with tissue culture; Jeff Cormier for RT-QPCR; and Nancy Heard for graphic illustration. This work was supported by grants from the National Institutes of Health (GM-116106, AI-70116 and HL-20948).
Footnotes
Supplemental information containing four figures and supplemental experimental procedures and references can be found with this article online at __________.
Author Contribution
Q.C., B.D., C.L. and S.H. designed and performed experiments, analyzed data, and drafted the manuscript; J.S.Y. performed experiments; J.Y. supervised the entire study, designed experiments, analyzed data and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bogdanov M, Dowhan W. Lipid-dependent generation of dual topology for a membrane protein. J Biol Chem. 2012;287:37939–37948. doi: 10.1074/jbc.M112.404103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling J, Aebi M. N-linked protein glycosylation in the endoplasmic reticulum. Cold Spring Harb Perspect Biol. 2013;5:10. doi: 10.1101/cshperspect.a013359. 1101/cshperspect.a013359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M, Ye J, Rawson R, Goldstein J. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. Cholesterol feedback: from Schoenheimer’s bottle to Scap’s MELADL. J Lipid Res. 2009;50:S15–S27. doi: 10.1194/jlr.R800054-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Lee CE, Denard B, Ye J. Sustained induction of collagen synthesis by TGF-β requires regulated intramembrane proteolysis of CREB3L1. PLoS ONE. 2014;9:e108528. doi: 10.1371/journal.pone.0108528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denard B, Lee C, Ye J. Doxorubicin blocks proliferation of cancer cells through proteolytic activation of CREB3L1. eLife Sciences. 2012;1:10. doi: 10.7554/eLife.00090. 7554/eLife.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denard B, Seemann J, Chen Q, Gay A, Huang H, Chen Y, Ye J. The membrane-bound transcription factor CREB3L1 is activated in response to virus infection to inhibit proliferation of virus-infected cells. Cell Host & Microbe. 2011;10:65–74. doi: 10.1016/j.chom.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop J, Jones PC, Finbow ME. Membrane insertion and assembly of ductin: a polytopic channel with dual orientations. EMBO J. 1995;14:3609–3616. doi: 10.1002/j.1460-2075.1995.tb00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feramisco JD, Goldstein JL, Brown MS. Membrane topology of human Insig-1, a protein regulator of lipid synthesis. J Biol Chem. 2004;279:8487–8496. doi: 10.1074/jbc.M312623200. [DOI] [PubMed] [Google Scholar]
- Görlich D, Hartmann E, Prehn S, Rapoport TA. A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature. 1992;357:47–52. doi: 10.1038/357047a0. [DOI] [PubMed] [Google Scholar]
- Görlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci USA. 1991;88:5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitatani K, Idkowiak-Baldys J, Hannun YA. The sphingolipid salvage pathway in ceramide metabolism and signaling. Cell Signal. 2008;20:1010–1018. doi: 10.1016/j.cellsig.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D. Membrane proteins which exhibit multiple topological orientations. Essays Biochem. 1996;31:49–60. [PubMed] [Google Scholar]
- Lodish H, Berk A, Kaiser C, Krieger M, Scott M, Bretscher A, Ploegh H, Matsudaira P. Molecular Cell Biology. 6 W. H. Freeman; 2007. [Google Scholar]
- Murakami T, Saito A, Hino S-i, Kondo S, Kanemoto S, Chihara K, Sekiya H, Tsumagari K, Ochiai K, Yoshinaga K, et al. Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat Cell Biol. 2009;11:1205–1211. doi: 10.1038/ncb1963. [DOI] [PubMed] [Google Scholar]
- Nilsson IM, von Heijne G. Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J Biol Chem. 1993;268:5798–5801. [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Meth. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Sakai J, Duncan EA, Rawson RB, Hua X, Brown MS, Goldstein JL. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 1996;85:1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- Sakai J, Nohturfft A, Cheng D, Ho YK, Brown MS, Goldstein JL. Identification of complexes between the COOH-terminal domains of sterol regulatory element-binding proteins (SREBPs) and SREBP cleavage-activating protein. J Biol Chem. 1997;272:20213–20221. doi: 10.1074/jbc.272.32.20213. [DOI] [PubMed] [Google Scholar]
- Sakai J, Rawson RB, Espenshade PJ, Cheng D, Seegmiller AC, Goldstein JL, Brown MS. Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol Cell. 1998;2:505–514. doi: 10.1016/s1097-2765(00)80150-1. [DOI] [PubMed] [Google Scholar]
- Sebag JA, Hinkle PM. Regions of melanocortin 2 (MC2) receptor accessory protein necessary for dual topology and MC2 receptor rrafficking and signaling. J Biol Chem. 2009;284:610–618. doi: 10.1074/jbc.M804413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt S, Jungnickel B, Hartmann E, Rapoport TA. Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J Cell Biol. 1996;134:25–35. doi: 10.1083/jcb.134.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Heijne G. Membrane-protein topology. Nat Rev Mol Cell Biol. 2006;7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- Winter E, Ponting CP. TRAM, LAG1 and CLN8: members of a novel family of lipid-sensing domains? Trends Biochem Sci. 2002;27:381–383. doi: 10.1016/s0968-0004(02)02154-0. [DOI] [PubMed] [Google Scholar]
- Wright MD, Rudy GB, Ni J. The L6 membrane proteins—A new four-transmembrane superfamily. Protein Sci. 2000;9:1594–1600. doi: 10.1110/ps.9.8.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, Goldstein JL, Brown MS. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- Ye J, DeBose-Boyd RA. Regulation of cholesterol and fatty acid synthesis. Cold Spring Harb Perspect Biol. 2011;3:10. doi: 10.1101/cshperspect.a004754. 1101/cshperspect.a004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R, Eyrisch S, Ahmad M, Helms V. Protein translocation across the ER membrane. Biochim Biophys Acta. 2011;1808:912–924. doi: 10.1016/j.bbamem.2010.06.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.