Abstract
This study aimed to determine if an association exists between circulating microRNA (miRNA) levels and disease progression in chronic hepatitis C (CHC); whether plasma or extracellular vesicles (EVs) were optimal for miRNA measurement and their correlation with hepatic miRNA expression; and the mechanistic plausibility in this association. We studied 130 CHC patients prospectively followed over decades. A comprehensive miRNA profile in plasma using microarray with 2,578 probe sets showed 323 miRNAs differentially expressed between healthy individuals and CHC patients, but only 6 that distinguished patients with mild versus severe chronic hepatitis. Eventually, let-7a/7c/7d-5p and miR-122-5p were identified as candidate predictors of disease progression. Cross-sectional analyses at the time of initial liver biopsy showed that reduced levels of let-7a/7c/7d-5p (let-7s) in plasma were correlated with advanced histological hepatic fibrosis stage and other fibrotic markers, whereas miR-122-5p levels in plasma were positively correlated with inflammatory activity, but not fibrosis. Measuring let-7s levels in EVs was not superior to intact plasma for discriminating significant hepatic fibrosis. Longitudinal analyses in 60 patients with paired liver biopsies showed that let-7s levels in plasma markedly declined over time in parallel with fibrosis progression. However, circulating let-7s levels did not parallel those in liver.
Conclusion
Of all miRNAs screened, the let-7 family showed the best correlation with hepatic fibrosis in CHC. A single determination of let-7s levels in plasma did not have superior predictive value for significant hepatic fibrosis compared to that of fibrosis-4 index, but the rate of let-7s decline in paired longitudinal samples correlated well with fibrosis progression. Pathway analysis suggested that low levels of let-7 may influence hepatic fibrogenesis through activation of TGF-β signaling in hepatic stellate cells.
Keywords: HCV, microRNA, extracellular vesicle, exosome, miR-122
Introduction
Chronic hepatitis C virus (HCV) infection is at significant risk for progressive hepatic fibrosis, subsequent liver cirrhosis (LC) and hepatocellular carcinoma (HCC). The histologic pattern of chronic hepatitis C (CHC) and the duration over which it develops into LC or HCC differ greatly among individuals. Recently developed interferon-free oral regimens combining direct-acting antiviral agents have achieved remarkably high rates of HCV eradication even in patients with LC.(1) However, the risk of developing HCC will not completely disappear even after the elimination of HCV, and the advanced state of hepatic fibrosis is its major risk factor.(2) Therefore, evaluating the state of disease progression in CHC is critical, and identifying a predictive biomarker for it will be helpful for implementing personalized treatment and surveillance of disease progression and HCC.
MicroRNAs (miRNAs) comprise a class of small noncoding RNAs, on average only 22 nucleotides in length, which are post-transcriptional gene regulators. Specifically, they negatively regulate gene expression by base-pairing to partially complementary sites typically in the 3’ untranslated regions (UTR) of target mRNA.(3) MicroRNAs are involved in various biological phenomena such as cell development, differentiation, proliferation, apoptosis and metabolism.(3) In addition, they play roles in the pathogenesis of inflammation, fibrogenesis and carcinogenesis in liver diseases.(4) Circulating miRNAs are found in cell-free serum, plasma and other body fluids in a highly stable form, including in association with Argonaute2, exosomes or high-density lipoprotein,(5, 6) by which they are protected from RNase, extreme temperatures, extreme pHs, or freeze-thaw cycle.(7, 8) Therefore, expression profiling of circulating miRNAs holds promise as a novel noninvasive marker for disease progression and severity. Furthermore, miRNAs in extracellular vesicles (EVs), including exosomes, can be incorporated into other cells and may alter gene expression in the recipient cells,(9) suggesting that they have potential as a mechanism of cell to cell communication.
Previous studies identified that several circulating miRNAs were associated with chronic HCV infection, hepatic necroinflammatory activity or fibrosis progression in CHC. Serum levels of miR-155, miR-125b, miR-146a, miR-134, miR-320c and miR-483-5p have been shown to be elevated in CHC patients compared to healthy individuals.(10, 11) In addition, several studies have shown that circulating miRNA-122 and miR-21 levels were correlated with elevated aminotransferases and pathological necroinflammatory activities in livers of CHC patients.(12-15) Additional studies have correlated either up-or down-regulation of circulating miRNAs such as miR-20a, miR-29a, miR-34a, miR-133a, miR-513-3p and miR-571 with hepatic fibrosis progression.(16-20) Murakami et al. investigated miRNA expression profile in exosome-rich fractionated serum of 64 CHC patients using microarray and quantitative real-time polymerase chain reaction (qRT-PCR). They demonstrated that several miRNA expression levels were correlated with the histological grade of hepatic inflammation and fibrosis, and the expression pattern of these miRNAs was useful for discriminating the histological stage.(21)
However, despite these numerous studies, no consistent association between circulating miRNA levels and disease progression has been identified. Comparison among studies has been difficult because of differences in sample collection, sample size, methodology, data normalization and analysis, and because diverse etiologies of liver disease were included within and between studies. Importantly, all these reported studies were cross-sectional. To address these uncertainties, we performed circulating miRNA expression analysis comprehensively, cross-sectionally and longitudinally to identify circulating miRNAs associated with disease progression in the natural course of chronic HCV infection using a prospectively followed and well-characterized HCV-infected blood donor cohort.
Patients and Methods
Study Population and Design
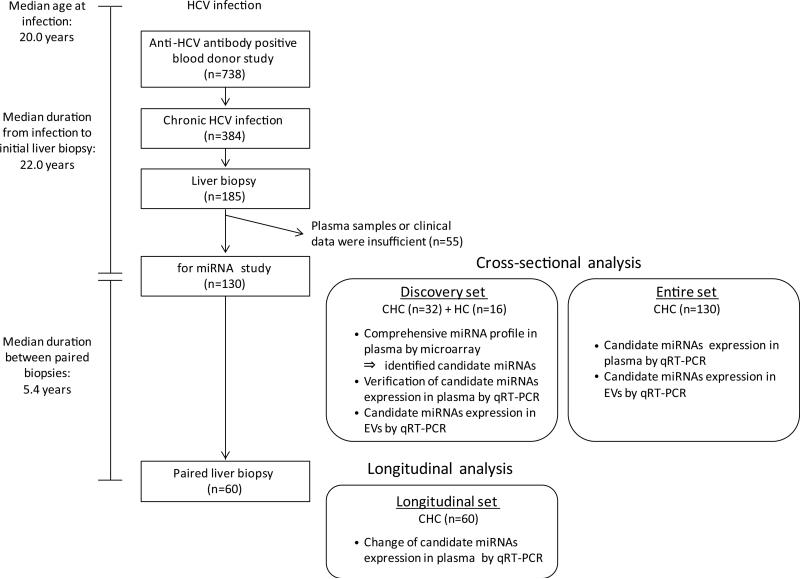
Details of the anti-HCV antibody positive blood donor cohort are described in Supporting Material and Methods and our previous reports.(22, 23) The study design is shown in Figure 1. In the present miRNA analysis, we enrolled 130 CHC patients from this cohort; the clinical characteristics at the initial liver biopsy are shown in Table 1. Sixty patients underwent paired liver biopsies during a median of 5.4 years. No patients received treatment for their HCV infection before their initial liver biopsy or between the paired biopsies. Additionally, plasma samples from 16 healthy blood donors were utilized as healthy controls (HC). No participants were infected with hepatitis B virus or human immunodeficiency virus, nor had other liver diseases such as autoimmune hepatitis, non-alcoholic steatohepatitis or primary biliary cirrhosis. Ethics committees of the American Red Cross and the National Institutes of Health (NIH) approved the study protocol in accordance with the Declaration of Helsinki and the study has been reviewed annually by an NIH Institutional Review Board (NIH Protocol 91-CC-0017). Written informed consent was obtained from each study subject.
Fig. 1.
Study design for circulating microRNA expression analysis in patients with chronic hepatitis C. We enrolled 130 CHC patients from the anti-HCV antibody positive blood donor cohort. Initially, we analyzed miRNA expression patterns in plasma in the discovery set consisting of 16 healthy controls, 16 patients with mild fibrosis and 16 with significant fibrosis. Then, we verified the microarray results by qRT-PCR using the same samples, and selected candidate miRNAs for further study. Next, we analyzed the association between the candidate miRNA levels in plasma or EVs and clinical features in the entire set that included the 32 patients in the discovery set. Finally, we analyzed changes in the candidate miRNA levels in plasma between the dates of paired liver biopsies in the longitudinal set. HCV, hepatitis C virus; miRNA, microRNA; CHC, chronic hepatitis C; HC, healthy control; qRT-PCR, quantitative real-time polymerase chain reaction; EVs, extracellular vesicles.
Table 1.
Clinical characteristics of 130 chronic hepatitis C patients at initial liver biopsy stratified by Ishak fibrosis score
| Ishak fibrosis score |
||||
|---|---|---|---|---|
| 0 (n = 43) | 1 (n = 39) | 2 (n = 24) | 3 - 6 (n = 24)* | |
| Gender, male / female | 18 / 25 | 20 / 19 | 16 / 8 | 13 / 11 |
| Age at initial biopsy, years | 38 (35 - 42) | 42 (35 - 48) | 42 (38 - 47) | 44 (40 - 50) |
| Age at infection, years† | 18 (14 - 20) | 21 (18 - 23) | 21 (19 - 25) | 23 (20 - 25) |
| Duration of infection, years† | 22 (19 - 27) | 23 (16 - 28) | 19 (17 - 27) | 21 (14 - 25) |
| Race, Caucasian / African American/ Others | 38 / 4 / 1 | 30 / 9 / 0 | 23 / 0 / 1 | 20 / 4 / 0 |
| Platelet count, ×109/L | 241 (205 - 275) | 210 (197 - 248) | 245 (198 - 264) | 179 (155 - 227) |
| AST, IU/L | 31 (26 - 38) | 35 (31 - 45) | 43 (31 - 61) | 57 (43 - 79) |
| ALT, IU/L | 38 (31 - 57) | 53 (39 - 68) | 75 (48 - 94) | 74 (53 - 114) |
| ALP, IU/L | 78 (55 - 98) | 67 (60 - 78) | 74 (66 - 93) | 76 (66 - 100) |
| y-GTP, IU/mL | 27 (22 - 61) | 33 (23 - 76) | 54 (27 - 86) | 50 (33 - 104) |
| Total bilirubin, mg/dL | 0.5 (0.5 - 0.6) | 0.6 (0.4 - 0.9) | 0.6 (0.6 - 0.8) | 0.7 (0.5 - 0.8) |
| Albumin, g/dL | 4.3 (4.1 - 4.5) | 4.2 (4.0 - 4.4) | 4.4 (4.1 - 4.5) | 4.3 (3.9 - 4.5) |
| AFP, ng/mL | 2.4 (1.9 - 3.6) | 2.9 (2.0 - 5.4) | 3.1 (2.5 - 5.3) | 4.7 (2.7 - 9.9) |
| APRI | 0.38 (0.28 - 0.52) | 0.50 (0.38 - 0.64) | 0.53 (0.38 - 0.73) | 0.92 (0.64 - 1.36) |
| FIB-4 | 0.76 (0.63 - 1.06) | 0.97 (0.75 - 1.17) | 0.98 (0.78 - 1.19) | 1.61 (1.19 - 2.40) |
| HCV RNA, log copies/mL‡ | 6.76 (6.08 - 7.21) | 6.52 (6.35 - 6.92) | 6.82 (6.23 - 7.19) | 6.27 (6.03 - 6.70) |
| HAI score, ≤7 / ≥8 | 29 / 14 | 21 / 18 | 11 / 13 | 6 / 18 |
| HCV genotype, 1 / 2 / 3 / 1+2 / N.D. | 32 / 6 / 1 / 1 / 3 | 31 / 5 / 1 / 2 / 0 | 18 / 6 / 0 / 0/ 0 | 18 / 3 / 2 / 0 / 1 |
| SNP genotype | ||||
| rs12979860: IFNL4, CC / CT / TT | 10 / 23 / 10 | 14 / 19 / 6 | 8 / 11 / 5 | 6 / 15 / 3 |
| rs738409: PNPLA3, CC / CG / GG | 23 / 20 / 0 | 19 /18 / 2 | 12 / 10 / 2 | 16 / 8 / 0 |
| rs4374383: MERTK, AA / AG / GG | 3 / 23 / 17 | 13 / 17 / 9 | 3 / 10 / 11 | 2 / 14 / 8 |
| rs9380516: TULP1, CC / CT / TT | 28 / 11 / 4 | 22 / 17 / 0 | 19 / 5 / 0 | 18 / 6 / 0 |
| rs16851720: RNF7, AA / AC / CC / N.D. | 21 / 12 / 0 / 10 | 21 / 6 / 1 / 11 | 8 / 6 / 2 / 8 | 5 / 10 / 0 / 9 |
| rs910049: HLA class II, GG / GA / AA / N.D. | 20 / 16 / 2 / 5 | 15 / 15 / 4 / 5 | 13 / 4 / 3 /4 | 14 / 7 / 0 / 3 |
| rs3135363: HLA class II, TT / TC / CC | 28 /13/2 | 23 / 13/3 | 10 / 10 / 4 | 15 / 7 /2 |
Data are expressed as number for categorical data or the median (first-third quartiles) for non-categorical data.
Ishak 3 (n=13); 4 (n=7); 5 (n=2); 6 (n=2).
Data were available in 90 patients: Ishak 0 (n=23); 1 (n=29); 2 (n=20); 3 (n=18), respectively.
HCV RNA loads were measured at least once in 109 patients, however these data were obtained at different periods from the initial liver biopsy in most cases. Therefore we collected the data of average HCV RNA load in each patient. AST, aspartate aminotransferase; ALT, alanine aminotransferase; ALP, alkaline phosphatase; γ-GTP, γ-glutamyl transpeptidase; AFP, alpha-fetoprotein; APRI, AST-to-platelet ratio index; FIB-4, fibrosis-4; HAI, histologic activity index; N.D., not determined; SNP, single nucleotide polymorphism.
Sampling Plasma and Liver Tissue, and Preparation for RNA
Peripheral blood was collected from each participant in tubes containing ethylenediaminetetraacetic acid as an anticoagulant. The tubes were centrifuged at 1,100 g for 10 minutes at room temperature. After plasma separation, samples were stored at −80°C until use. Liver specimens were obtained by needle biopsy, fixed in formalin and embedded in paraffin. Total RNAs including miRNAs in plasma, EVs and whole liver tissues were purified with miRNeasy Serum/Plasma kit, exoRNeasy Serum/Plasma Midi Kit and miRNeasy FFPE Kit (Qiagen, Hilden, Germany) respectively, following the manufacturer's instructions with minor modification.
Microarray Analysis
We performed miRNA expression profiling in plasma samples using miRNA microarray: Affymetrix GeneChip miRNA 4.0 Array (Affymetrix, Santa Clara, CA), which contains 2,578 mature human miRNA probe sets. Total RNA was poly(A) tailed and then directly ligated to a fluorescent dendrimer using the FlashTag Biotin HSR RNA Labeling Kit (Affymetrix). Standard protocols were used for hybridization, staining, washing and scanning of the arrays. All samples passed the quality control assessment using chip-specific quality control probes. Raw microarray data (CEL files) were imported into Partek Genomics Suite (Partek Inc., St. Louis, MO), and probe set summaries were computed using Robust Multi-Array algorithm which consists of background correction, quantile normalization across all the chips, log2 transformation and median polish.(24) The relationship between the exclusive miRNAs and disease progression were also visualized by principal component analysis (PCA) and hierarchical clustering in which dissimilarity was measured by the Euclidean distance and the average linkage method is used for the clustering. The complete miRNA microarray data have been deposited in the Gene Expression Omnibus (GSE74872) and is publicly available.
qRT-PCR for miRNA analysis
Quantitative miRNA levels were determined using qRT-PCR with the Applied Biosystems 7900 HT Sequence Detection System (Applied Biosystems, Foster City, CA) and TaqMan MicroRNA Assay (Applied Biosystems). Cycle threshold (Ct) values were calculated using SDS Software v2.3 (Applied Biosystems). Expression levels of miRNAs were normalized to those of the following reference genes: spike-in Caenorhabditis elegans (cel)-miR-39, endogenous miR-16-5p, or the average levels of endogenous miR-92a-3p and miR-486-5p for analyses using plasma samples; spike-in cel-miR-39 for EVs; and endogenous RNU6B for liver tissues. The expression levels were determined by the 2−ΔCt method in which ΔCt was calculated as follows: ΔCt = Ct (miRNA of interest) – Ct (reference gene).
Further information about methods is provided in Supporting Material and Methods.
Results
miRNA Profile in Plasma of Healthy Individuals and CHC Patients Using Microarray
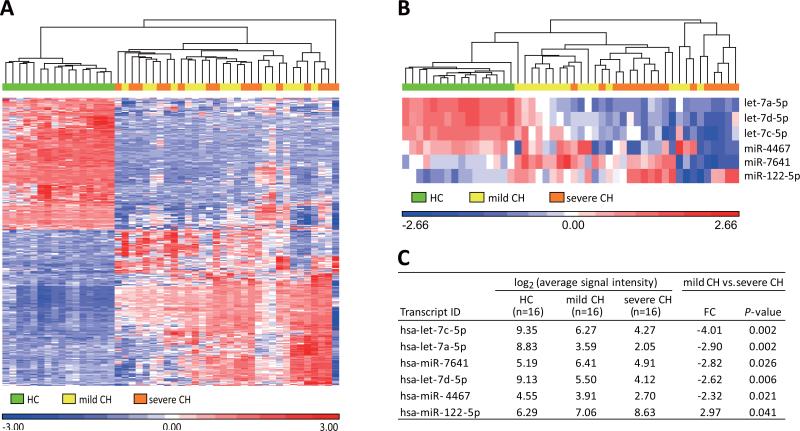
The clinical characteristics of the subjects in the discovery set are shown in Supporting Table 1. Gender and age were matched. The Ishak fibrosis scores were 0 for all patients in mild chronic hepatitis (CH), whereas they were 3-6 in severe CH. Alanine aminotransferase (ALT) levels, histologic activity index (HAI) scores, aspartate aminotransferase (AST)-to-platelet ratio index (APRI) and fibrosis-4 (FIB-4) index were significantly higher, and platelet counts were significantly lower in severe CH compared to mild CH. Principal component analysis (PCA) based on the result of the normalized signal intensities of probes for human miRNAs, showed that miRNA profiles were markedly different between HC and CHC patients (mild CH + severe CH), whereas no discernable differences in miRNA profiles were observed when cases with mild CH were compared to those with severe CH (Supporting Fig. 1). Next, we compared the normalized signal intensities between HC and CHC patients, showing that 323 miRNAs were differentially expressed (176 up-regulated and 147 down-regulated) in CHC compared with HC, based on the following conditions: fold change (FC) < −2 or >2; P-value < 0.05 by the one-way analysis of variance (ANOVA); Q-value < 0.05 by the false discovery rate (Supporting Table 2). The hierarchical clustering based on the result of these 323 miRNAs showed that the miRNA profiles were remarkably different between HC and CHC; however those with mild CH or severe CH were not clustered under these conditions (Fig. 2A). Then we compared the normalized signal intensities between mild CH and severe CH. Contrary to our expectations, there were only a few miRNAs that showed significant differences of expression levels between them: let-7a-5p, let-7c-5p, let-7d-5p, miR-4467 and miR-7641 levels were down-regulated, and miR-122-5p levels were up-regulated in severe CH compared to mild CH (FC < −2 or > 2; P-value < 0.05 by the one-way ANOVA) (Fig. 2B and 2C).
Fig. 2.
MicroRNA expression profiling in plasma using microarray in the discovery set. (A) The hierarchical clustering based on the normalized signal intensities of 323 miRNAs which were differentially expressed (176 up-regulated and 147 down-regulated) in chronic hepatitis C patients (mild CH + severe CH) compared with healthy controls (HC). Up-regulated microRNAs are represented in red, and down-regulated ones are in blue. (B) The hierarchical clustering based on the normalized signal intensities of 6 miRNAs that were differentially expressed between mild CH and severe CH, and (C) the expression levels of these miRNAs. Fold changes (FC) and P-values by the one-way analysis of variance were calculated by comparisons between mild CH and severe CH. CH, chronic hepatitis.
Verification of miRNA Expression Levels by qRT-PCR
We re-extracted total RNA from the same plasma samples as the discovery set and verified the microarray results by qRT-PCR. There is no consensus about an optimal normalization strategy for analysis of circulating miRNAs by qRT-PCR. In previous studies on CHC, exogenous spike-in RNAs(10, 13, 16-20) or endogenous miRNAs such as miR-16(15, 21) have been commonly used as a reference. According to our results by microarray and the previous studies, we selected exogenous spike-in cel-miR-39, endogenous miR-16-5p, or the average level of endogenous miR-92a-3p and miR-486-5p as a reference for normalization (Supporting Fig. 2), and then compared the correlation of the expression levels of several miRNAs between microarray and qRT-PCR. Overall, the correlations were the strongest when we used spike-in cel-miR-39 as a reference (Supporting Fig. 3A-C): for let-7a-5p (r = 0.858, P < 0.001), let-7c-5p (r = 0.787, P < 0.001), let-7d-5p (r = 0.848, P < 0.001) and miR-122-5p (r = 0.628, P < 0.001), whereas only a moderate correlation was seen for miR-7641 (r = 0.463, P < 0.001) and there was no correlation for miR-4467 (Supporting Fig. 3A). Based on the analyses of the discovery set, we selected four miRNAs: let-7a-5p, let-7c-5p, let-7d-5p and miR-122-5p as potential candidates for predicting disease severity, and examined their expression levels using spike-in cel-miR-39 as a reference for normalization in subsequent analyses by qRT-PCR.
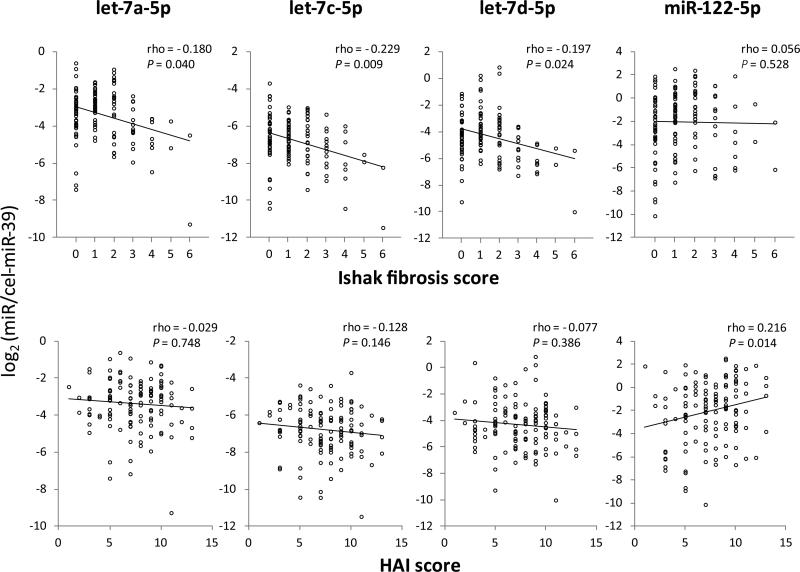
Correlations of the Candidate miRNA Levels in Plasma with Clinical Features in the Entire Set at Initial Liver Biopsy
We analyzed the association between the candidate miRNA levels in plasma and clinical features in the entire set that included the 32 patients of the discovery set. The levels of let-7a-5p, let-7c-5p and let-7d-5p were inversely correlated with Ishak fibrosis scores (rho = −0.180, −0.229, −0.197 and P = 0.040, 0.009, 0.024, respectively) (Fig. 3), and also inversely correlated with FIB-4 (r = −0.383, −0.368, −0.285, respectively and P < 0.001 for all let-7s) and APRI (r = −0.345, −0.355, −0.240 and P < 0.001, < 0.001, = 0.007, respectively), whereas positively correlated with platelet count (r = 0.318, 0.240, 0.192 and P < 0.001, = 0.007, = 0.031, respectively) (Table 2). Thus, let-7s (let-7a-5p, let-7c-5p and let-7d-5p) levels in plasma were associated with hepatic fibrosis severity. Meanwhile, miR-122-5p levels were positively correlated with HAI scores (rho = 0.216, P = 0.014) (Fig. 3) and ALT (r = 0.283, P = 0.001) (Table 2), suggesting that miR-122-5p levels in plasma were associated with hepatic inflammatory activity, but not with the degree of fibrosis. The levels of let-7s had weak inverse correlations with age (Table 2), but not with gender (data not shown). The levels of let-7s and miR-122-5p showed no correlations with genotypes of the single nucleotide polymorphisms previously reported to be associated with hepatic fibrosis progression or inflammatory activity in CHC: rs12979860 (IFNL4), rs738409 (PNPLA3), rs4374383 (MERTK), rs9380516 (TULP1), rs16851720 (RNF7), rs910049 and rs3135363 (HLA class II)(25-28) (data not shown).
Fig. 3.
Correlations of let-7s and miR-122 expression levels in plasma with histological findings at initial liver biopsy using Spearman's correlation coefficient (rho). HAI, histologic activity index.
Table 2.
Correlations of microRNA expression levels in plasma with clinical parameters in chronic hepatitis C patients
| let-7a-5p |
let-7c-5p |
let-7d-5p |
miR-122-5p |
|||||
|---|---|---|---|---|---|---|---|---|
| r | P-value | r | P-value | r | P-value | r | P-value | |
| Age | −0.218 | 0.013 | −0.188 | 0.032 | −0.223 | 0.011 | −0.161 | N.S. |
| AST | −0.176 | 0.046 | −0.236 | 0.007 | −0.092 | N.S. | 0.11 | N.S. |
| ALT | −0.023 | N.S. | −0.094 | N.S. | 0.012 | N.S. | 0.283 | 0.001 |
| ALP | −0.176 | 0.046 | −0.102 | N.S. | −0.235 | 0.007 | −0.005 | N.S. |
| γ-GTP | −0.017 | N.S. | 0.048 | N.S. | 0.006 | N.S. | 0.018 | N.S. |
| Total bilirubin | −0.045 | N.S. | −0.011 | N.S. | 0.012 | N.S. | 0.023 | N.S. |
| Albumin | 0.156 | N.S. | 0.051 | N.S. | 0.170 | N.S. | 0.111 | N.S. |
| AFP | −0.160 | N.S. | −0.126 | N.S. | −0.132 | N.S. | −0.034 | N.S. |
| Platelet count | 0.318 | < 0.001 | 0.240 | 0.007 | 0.192 | 0.031 | 0.106 | N.S. |
| APRI | −0.345 | < 0.001 | −0.355 | < 0.001 | −0.240 | 0.007 | −0.053 | N.S. |
| FIB-4 | −0.383 | < 0.001 | −0.368 | < 0.001 | −0.285 | < 0.001 | −0.196 | 0.027 |
| HCV RNA | −0.061 | N.S. | −0.003 | N.S. | −0.050 | N.S. | 0.017 | N.S. |
N.S., not significant; AST, aspartate aminotransferase; ALT, alanine aminotransferase; ALP, alkaline phosphatase; γ-GTP, γ-glutamyl transpeptidase; AFP, alpha-fetoprotein; APRI, AST-to-platelet ratio index; FIB-4, fibrosis-4. Correlations were evaluated by Pearson's correlation coefficient (r).
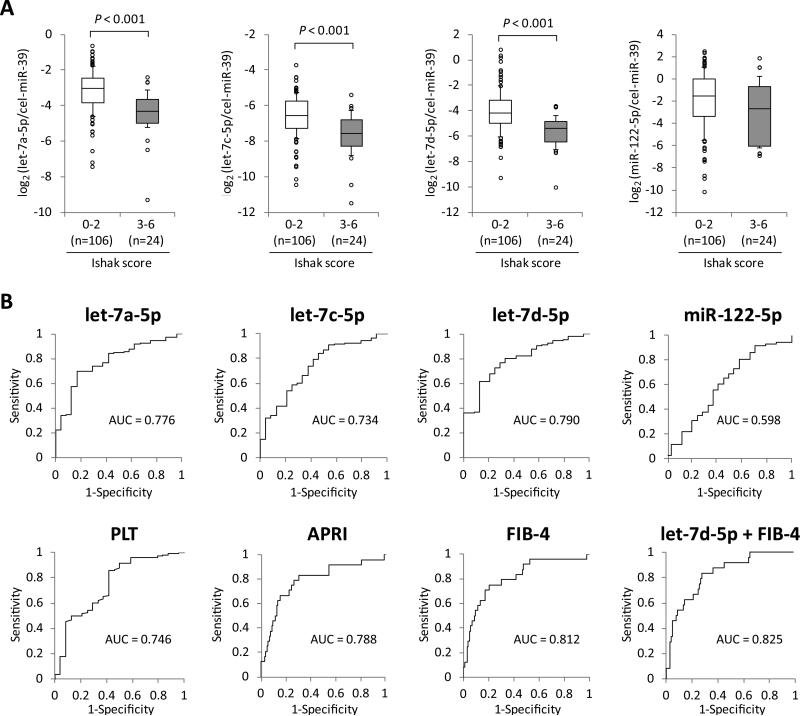
Discrimination of Significant Hepatic Fibrosis by let-7s Expression Levels in Plasma
We divided the 130 CHC patients into two groups: mild fibrosis with Ishak score 0-2 (n = 106) and significant fibrosis with Ishak score 3-6 (n = 24). The clinical characteristics of the two groups are shown in Supporting Table 3. The expression levels of let-7a-5p, let-7c-5p and let-7d-5p were significantly lower in patients with significant fibrosis than in those with mild fibrosis (P < 0.001 for all let-7s) (Fig. 4A). The receiver operating characteristic (ROC) curve analysis for discriminating significant fibrosis indicated that the area under the curve (AUC) levels of let-7a-5p, let-7c-5p and let-7d-5p (AUC = 0.776, 0734 and 0.790, respectively) were comparable to those of platelet count and APRI (AUC = 0.746 and 0.788, respectively) (Fig. 4B). Meanwhile, several other factors were associated with significant fibrosis by univariate analysis as shown in Supporting Table 3. As described in Supporting Material and Methods, APRI is calculated by AST and platelet count, whereas FIB-4 is done by age, AST, ALT and platelet count. Considering the correlations between these factors, we analyzed factors associated with significant fibrosis in logistic regression models including the following variables: age, APRI and let-7s levels or FIB-4 and let-7s levels. The levels of let-7s were independently associated with significant fibrosis in both analyses (Supporting Table 4). We then calculated the AUC levels when each of the let-7s was used in combination with either APRI or FIB-4. The AUC value was most enhanced when let-7d-5p and FIB-4 were used in combination (AUC = 0.825) (Fig. 4B and Supporting Table 5).
Fig. 4.
Discrimination of significant hepatic fibrosis by let-7s expression levels in plasma. (A) Expression levels of the candidate microRNAs in plasma between chronic hepatitis C patients with or without significant fibrosis (Ishak score 3-6 at initial liver biopsy). Boxes represent the interquartile range of the data. The lines across the boxes and the numbers indicate the median values. The hash marks above and below the boxes indicate the 90th and 10th percentiles for each group, respectively. P-values were calculated by Mann-Whitney U-test. (B) The receiver operating characteristic curve analyses were carried out and the area under the curves (AUC) were calculated to evaluate capabilities of the candidate miRNA levels in plasma and other hepatic fibrotic markers for discriminating significant hepatic fibrosis. PLT, platelet count; APRI, aspartate aminotransferase-to-platelet ratio index; FIB-4, fibrosis-4.
Longitudinal miRNA Expression Analysis in Patients with Paired Liver Biopsies
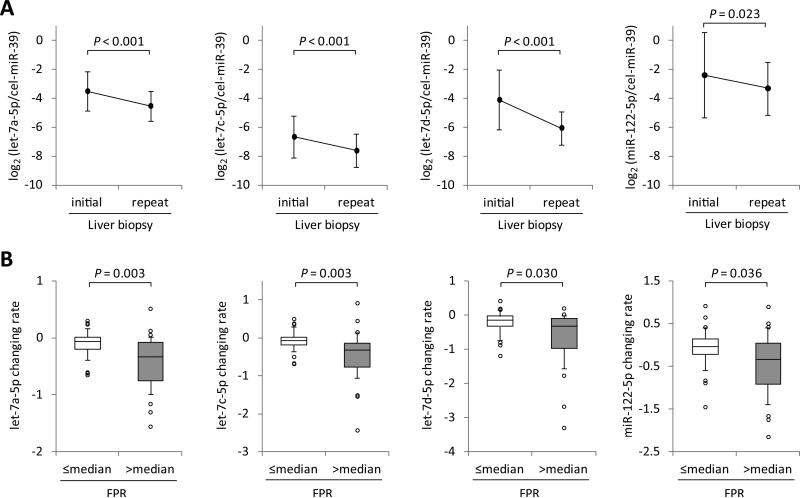
The clinical characteristics of 60 CHC patients at the initial liver biopsy, who underwent a repeat biopsy, are shown in Supporting Table 6. Over the interval between the paired biopsies, 22 patients had fibrosis that increased by at least 1 Ishak stage (17 increased by 1 stage; two by 2 stages; two by 3 stages; one by 4 stages), whereas 38 had no change. We investigated the changes in expression levels of let-7a-5p, let-7c-5p, let-7d-5p and miR-122-5p in plasma between the paired biopsies, showing that the levels of let-7a-5p, let-7c-5p and let-7d-5p markedly declined over time (P < 0.001 for all let-7s), whereas miR-122-5p levels gradually declined (P = 0.023) (Fig. 5A). Next, we calculated fibrosis progression rates (FPR) of the 60 patients between the paired biopsies, and divided the population into two groups according to the median FPR: the slowly progressive fibrosis group was defined as those with ≤ median FPR and the rapidly progressive fibrosis group as those with > median FPR, and compared the changing rates of miRNA levels between the two groups (Supporting Fig. 4). Although these miRNA levels declined over the interval between biopsies in most patients, the declines were significantly greater in the rapidly progressive fibrosis group compared to the slowly progressive fibrosis group (P = 0.003, 0.003, 0.030 and 0.036, for let-7a-5p, let-7c-5p, let-7d-5p and miR5 122-5p respectively) (Fig. 5B). Meanwhile, the levels of miR-16-5p, which are considered to be expressed abundantly and stably in plasma/serum, did not change between the paired biopsies, and their changing rates were not different between the slowly progressive fibrosis group and the rapidly progressive fibrosis group (Supporting Fig. 5), which implying that the decline of let-7s and miR-122-5p did not simply reflect the total levels of miRNAs in plasma.
Fig. 5.
Association between changes in let-7s and miR-122 levels in plasma and hepatic fibrosis progression in longitudinal analysis. (A) Changes in let-7s and miR-122 expression levels between paired liver biopsies (n = 60). Data represent mean ± standard deviation. P-values were calculated by the paired t-test. (B) We divided the 60 patients into two groups according to the median fibrosis progression rates (FPR) between the paired biopsies, and compared the changing rates of let-7s and miR-122 levels between the two groups. Boxes represent the interquartile range of the data. The lines across the boxes and the numbers indicate the median values. The hash marks above and below the boxes indicate the 90th and 10th percentiles for each group, respectively. P-values were calculated by Mann-Whitney U-test.
Expression Levels of the Candidate miRNAs in EVs
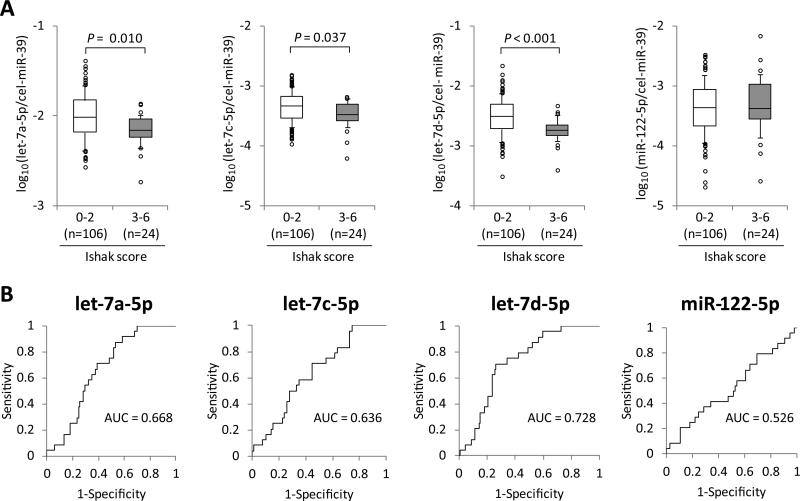
Since Arroyo et al. previously showed that circulating miR-122 was detected in the protein-associated fractions, whereas let-7a is predominant in EVs in healthy individuals,(29) we expected that let-7s levels in EVs might reflect hepatic fibrosis progression in CHC more clearly than those in total plasma. Therefore, we extracted total RNA from inside or outside EVs in plasma of the discovery set, and analyzed the levels of the candidate miRNAs in each component (Supporting Fig. 6). Levels of let-7a-5p and let-7d-5p inside EVs were higher than that of miR-122-5p, whereas levels of let-7s outside EVs were lower than that of miR-122-5p (Supporting Fig. 7A); hence the ratio of inside to outside EVs for let-7s were higher compared to that for miR-122-5p. Levels of miR-122-5p in plasma were correlated with those inside EVs and outside EVs almost equally, while levels of let-7s in plasma were more strongly correlated with those inside EVs than those outside EVs (Supporting Fig. 7B). Next, we analyzed the association between the candidate miRNA levels in EVs and clinical features in the entire set. The expression levels of let-7a-5p, let-7c-5p and let-7d-5p were significantly lower in patients with significant hepatic fibrosis than in those with mild fibrosis (P = 0.010, = 0.037, < 0.001, respectively) (Fig. 6A). Although the levels of let-7a-5p, let-7c-5p and let-7d-5p in both EVs and plasma were inversely correlated with the severity of hepatic fibrosis, ROC curve analysis for discriminating significant fibrosis showed that the AUC values of them in EVs (AUC = 0.668, 0.636 and 0.728 for let-7a-5p, let-7c-5p and let-7d-5p, respectively) (Fig. 6B) were lower than those in plasma (Fig. 4B). Hence, measurement of let-7s levels in EVs had no advantage over direct measurement in plasma.
Fig. 6.
Discrimination of significant hepatic fibrosis by let-7s expression levels in extracellular vesicles. (A) Expression levels of the candidate microRNAs in extracellular vesicles (EVs) between chronic hepatitis C patients with or without significant fibrosis (Ishak score 3-6 at initial liver biopsy). Boxes represent the interquartile range of the data. The lines across the boxes and the numbers indicate the median values. The hash marks above and below the boxes indicate the 90th and 10th percentiles for each group, respectively. P-values were calculated by Mann-Whitney U-test. (B) The receiver operating characteristic curve analyses were carried out and the area under the curves (AUC) were calculated to evaluate capabilities of the candidate miRNA levels in EVs for discriminating significant hepatic fibrosis.
Correlations of let-7s Levels between Liver Tissue and Plasma or EVs
Formalin-fixed paraffin-embedded specimens of liver tissues were available for only a subset of patients: only 6 of 43 cases where a specimen was available at the initial biopsy had a paired specimen at the repeat biopsy, and additional 37 specimens were available only at the repeat biopsy. Thus, the number of available specimens was limited. Therefore we combined the samples at the initial and the repeat biopsy, and analyzed the expression levels of let-7s and miR-122-5p in liver tissues. The clinical characteristics of these patients are shown in Supporting Table 7. Our results indicated no correlations in their expression levels between liver and plasma, but demonstrated inverse correlations between liver and EVs (Supporting Fig. 8A). Meanwhile, we found that there were no associations between their expression in liver and hepatic fibrosis severity (Supporting Fig. 8B).
Pathway Analysis of Target Genes of let-7
We extracted 1,072 target genes of the let-7 family by TargetScan, and then performed pathway analysis by importing these genes into Ingenuity Pathway Analysis (Qiagen). Of the 92 canonical pathways on which these target genes were mapped, the top was “Hepatic Fibrosis / Hepatic Stellate Cell (HSC) Activation” (Supporting Table 8).
Discussion
Our cohort consisted of prospectively followed CHC patients who were unaware of their HCV infection at the time of blood donation and had not received anti-HCV therapy prior to the initial or repeat liver biopsy analyzed in this report. Therefore, we believe this cohort provides a valid model for assessing long-term disease progression in the natural course of chronic HCV infection. In this cohort of patients with CHC, we observed that circulating let-7s expression levels in plasma and EVs, as measured by comprehensive microarray and qRT-PCR, were inversely correlated with the severity of hepatic fibrosis. In addition, longitudinal analysis showed that let-7s levels in plasma continued to decline during the interval between paired liver biopsies in parallel with fibrosis progression. To our knowledge, this is the first demonstration of changes in circulating miRNA expression over time during the natural course of chronic HCV infection. In addition, we validated that circulating miR-122-5p levels in plasma were correlated with elevated ALT levels and pathological necroinflammatory activity,(12-14) and decreased in parallel with fibrosis progression.(13)
The distribution of circulating miRNAs in vesicle-rich or protein-rich fractions in plasma/serum has been controversial. Gallo et al. suggested that the majority of miRNAs in plasma/serum are concentrated in exosomes.(30) In contrast, Arroyo et al. demonstrated that up to 90% of circulating miRNAs are associated with proteins, and showed that different miRNAs were enriched in specific extracellular compartments, e.g., circulating miR-122 was detected in the protein-rich fractions, whereas let-7a was predominant in the vesicle-rich fractions in healthy individuals.(23) Intriguingly, Bala et al. indicated that the distributions of miR-122 and miR-155 in the exosome-rich or the protein-rich fractions were different according to the underlying pathological condition as demonstrated in mice with alcoholic and inflammatory liver diseases versus drug induced liver injury.(31) Together, these results suggest that the blood compartment or fraction most suitable for measuring miRNA expression levels depends on the underlying disease, and the particular miRNA being measured. Murakami et al. showed that the levels of several miRNAs, including let-7a, in exosome-rich fractionated serum in CHC patients were correlated with the histological grade of hepatic fibrosis using microarray.(18) This is consistent with our results, but we found that measuring let-7s in EVs was not superior to direct measurement in plasma. In the present study, when purifying EVs, including exosomes, binding of vesicles to the membrane of the exoRNeasy kit was not selective for a specific sub-population or size range of EVs. This restriction might account in part for the observation that let-7s levels in EVs did not reflect hepatic fibrosis progression more clearly than those in total plasma. Future study is required to analyze the expression levels of these miRNAs in specific sub-populations of EVs.
The cellular source of circulating miRNAs and the mechanism of regulation of miRNA expression are not well known. It is presumed that circulating let-7s derive from various cell sources, since members of the let-7 family are abundantly expressed in most cell types. Murakami et al. analyzed the correlation of miRNA expression levels between liver tissue and exosome-rich fractionated serum in 60 CHC patients using microarray data, and showed that the levels of almost all the miRNAs including let-7 were not comparable in serum and liver.(21) We showed no correlations in let-7s expression levels between whole liver tissue and plasma, but weak inverse correlations between liver and EVs (Supporting Fig. 8A). Although it is not completely clear why we unexpectedly observed these inverse correlations, it is clear that the expression levels of circulating let-7s in plasma and EVs were not parallel with those in liver. Diehl, et al showed that the profiles of miRNAs in microvesicles were found to be markedly different from those in their maternal cells, indicating an active mechanism of selective packaging of miRNAs from cells into microvesicles.(32) These results suggest that it would be difficult to identify the specific source of circulating let-7s, responsible for the differences of let-7s expression in plasma/serum and EVs in CHC patients using current technology. Primary miRNAs are, like protein-coding mRNAs, transcribed by polymerase II, suggesting that miRNA expression is regulated principally at the level of transcription. Indeed, transcriptional regulation has been demonstrated for several miRNA genes.(33) However, the mature let-7 family members are reported to be regulated, at least in part, post-transcriptionally.(34-36) The present study does not allow us to distinguish whether the decreased levels of let-7a-5p, let-7c-5p and let-7d-5p, as well as other let-7 family members, in the plasma of CHC patients (Supporting Table 2) represent transcriptional or post-transcriptional suppression in hepatic or other cell types.
The biological functions of circulating miRNAs in HCV infection are being actively explored. Previous studies suggest that circulating miRNA in EVs, including exosomes, might provide a means of communication between neighboring cells and influence gene expression on target cells.(9) The let-7 family is highly conserved and abundantly expressed across most cells and animal species. The let-7 miRNAs regulate cell proliferation and differentiation, and their reduced expression has been implicated in epithelial to mesenchymal transition and enhanced cell migration/invasion.(37, 38) Moreover, they inhibit the expression of multiple oncogenes, including RAS, MYC and HMGA2,(39, 40) and their expression levels decrease in several cancers including HCC.(41, 42) In HCV infection, let-7b inhibits HCV replication by directly binding to 5'UTR and NS5B coding regions of the HCV genome,(43) and by targeting host factor insulin-like growth factor 2 mRNA-binding protein 1.(44) Interestingly, our pathway analysis showed that the estimated target genes of let-7 were most closely related to the pathway of “Hepatic Fibrosis / HSC Activation” (Supporting Table 8). In particular, an effect on transforming growth factor-beta (TGF-β) signaling by let-7 is a plausible mechanism for enhanced hepatic fibrogenesis. Chronic HCV infection up-regulates TGF-β signaling and subsequently activates HSCs, resulting in excessive accumulation of extracellular matrix proteins including collagen in the liver.(45) Let-7 family members are predicted to target TGFBR1, SMAD2, COL1A1 and COL1A2 in the TGF- β signaling pathway (Supporting Fig. 9), and thereby are thought to negatively regulate fibrosis progression. Indeed, a previous study showed that let-7 targeted TGFBR1 and regulated TGF-β signaling in liver development.(46) Moreover, let-7 has been shown to be involved in renal profibrotic processes by regulating TGFBR1 or COL1A2.(47-49) Another study indicated that let-7a regulated the expression of type I collagen in skin fibroblasts by targeting its 3'UTR in systemic and localized scleroderma. Interestingly, this study also showed that serum let-7a levels were significantly decreased in these patients.(50) Our pathway analysis along with previous findings(46-50) lead us to speculate that decreased levels of let-7 may activate hepatic profibrotic processes associated with TGF-β signaling pathway in HSCs of CHC patients. As a result, secretion of let-7 in EVs from HSCs might change according to HSCs activity and/or hepatic fibrosis severity. An alternative possibility is that EVs including let-7 might incorporate into HSCs and influence the activation of TGF-β signaling. Future studies need to clarify 1) the expression of let-7 in different cell subsets in the liver, especially HSCs and hepatocytes; 2) the let-7 levels secreted from these cells; 3) the function of let-7 in HSCs and hepatocytes. These studies might suggest to the possibility of enhancing let-7 levels as a therapeutic modality.
There are several limitations in the present study. First, the number of patients, especially those with significant hepatic fibrosis was limited. Second, the fact that the ability of let-7s in plasma to discriminate significant hepatic fibrosis was not superior to that of FIB-4, with only a marginal additional effect of let-7s in addition to FIB-4. Notably, measuring FIB-4 and APRI is easier than examining circulating let-7 expression levels. Thus, measuring circulating let-7 levels may not be a useful clinical tool to evaluate hepatic fibrosis. However, our longitudinal analysis implies that calculating the rate of change in plasma let-7 levels measured in serial samples during follow up might predict future hepatic fibrosis progression. Validation studies in independent cohorts including patients with more advanced hepatic fibrosis are clearly needed, which might enhance the utility of circulating let-7 as a hepatic fibrotic marker. Even in the absence of clinical utility, this study has identified let-7s, more than other miRNAs, as mechanistically important in hepatic fibrosis progression in CHC.
In conclusion, the present study provides evidence that circulating let-7 may serve as a surrogate marker for hepatic fibrosis progression in CHC, and suggests a plausible mechanism for this effect. Further studies are also necessary to elucidate the intracellular and cell-to-cell functions of let-7 in hepatic fibrogenesis and the specific mechanisms through which let-7 exert these effects.
Supplementary Material
Acknowledgments
Financial support: This research was supported by the Intramural Research Program of the Warren G. Magnuson Clinical Center and the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. Kentaro Matsuura was supported by the Uehara Memorial Foundation of Japan.
Abbreviations
- HCV
hepatitis C virus
- LC
liver cirrhosis
- HCC
hepatocellular carcinoma
- CHC
chronic hepatitis C
- miRNA
microRNA
- UTR
untranslated regions
- EVs
extracellular vesicles
- qRT-PCR
quantitative real-time polymerase chain reaction
- HC
healthy control
- PCA
principal component analysis
- Ct
Cycle threshold
- CH
chronic hepatitis
- ALT
alanine aminotransferase
- HAI
histologic activity index
- AST
aspartate aminotransferase
- APRI
AST-to-platelet ratio index
- FIB-4
fibrosis-4
- FC
fold change
- ANOVA
analysis of variance
- ROC
receiver operating characteristic
- AUC
area under the curve
- FPR
fibrosis progression rates
- HSC
hepatic stellate cell
- TGF-β
transforming growth factor-beta
Footnotes
Conflicts of interests: The all authors have no conflict of interest.
References
- 1.Reddy KR, Bourliere M, Sulkowski M, Omata M, Zeuzem S, Feld JJ, Lawitz E, et al. Ledipasvir and sofosbuvir in patients with genotype 1 hepatitis C virus infection and compensated cirrhosis: An integrated safety and efficacy analysis. Hepatology. 2015;62:79–86. doi: 10.1002/hep.27826. [DOI] [PubMed] [Google Scholar]
- 2.Chang KC, Hung CH, Lu SN, Wang JH, Lee CM, Chen CH, Yen MF, et al. A novel predictive score for hepatocellular carcinoma development in patients with chronic hepatitis C after sustained response to pegylated interferon and ribavirin combination therapy. J Antimicrob Chemother. 2012;67:2766–2772. doi: 10.1093/jac/dks269. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Wang XW, Heegaard NH, Orum H. MicroRNAs in liver disease. Gastroenterology. 2012;142:1431–1443. doi: 10.1053/j.gastro.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemoinne S, Thabut D, Housset C, Moreau R, Valla D, Boulanger CM, Rautou PE. The emerging roles of microvesicles in liver diseases. Nat Rev Gastroenterol Hepatol. 2014;11:350–361. doi: 10.1038/nrgastro.2014.7. [DOI] [PubMed] [Google Scholar]
- 6.Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11:145–156. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 10.Bala S, Tilahun Y, Taha O, Alao H, Kodys K, Catalano D, Szabo G. Increased microRNA-155 expression in the serum and peripheral monocytes in chronic HCV infection. J Transl Med. 2012;10:151. doi: 10.1186/1479-5876-10-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shwetha S, Gouthamchandra K, Chandra M, Ravishankar B, Khaja MN, Das S. Circulating miRNA profile in HCV infected serum: novel insight into pathogenesis. Sci Rep. 2013;3:1555. doi: 10.1038/srep01555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bihrer V, Friedrich-Rust M, Kronenberger B, Forestier N, Haupenthal J, Shi Y, Peveling-Oberhag J, et al. Serum miR-122 as a biomarker of necroinflammation in patients with chronic hepatitis C virus infection. Am J Gastroenterol. 2011;106:1663–1669. doi: 10.1038/ajg.2011.161. [DOI] [PubMed] [Google Scholar]
- 13.Trebicka J, Anadol E, Elfimova N, Strack I, Roggendorf M, Viazov S, Wedemeyer I, et al. Hepatic and serum levels of miR-122 after chronic HCV-induced fibrosis. J Hepatol. 2013;58:234–239. doi: 10.1016/j.jhep.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 14.van der Meer AJ, Farid WR, Sonneveld MJ, de Ruiter PE, Boonstra A, van Vuuren AJ, Verheij J, et al. Sensitive detection of hepatocellular injury in chronic hepatitis C patients with circulating hepatocyte-derived microRNA-122. J Viral Hepat. 2013;20:158–166. doi: 10.1111/jvh.12001. [DOI] [PubMed] [Google Scholar]
- 15.Bihrer V, Waidmann O, Friedrich-Rust M, Forestier N, Susser S, Haupenthal J, Welker M, et al. Serum microRNA-21 as marker for necroinflammation in hepatitis C patients with and without hepatocellular carcinoma. PLoS One. 2011;6:e26971. doi: 10.1371/journal.pone.0026971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shrivastava S, Petrone J, Steele R, Lauer GM, Di Bisceglie AM, Ray RB. Up-regulation of circulating miR-20a is correlated with hepatitis C virus-mediated liver disease progression. Hepatology. 2013;58:863–871. doi: 10.1002/hep.26296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, Janssen J, et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 18.Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6:e23937. doi: 10.1371/journal.pone.0023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roderburg C, Luedde M, Vargas Cardenas D, Vucur M, Mollnow T, Zimmermann HW, Koch A, et al. miR-133a mediates TGF-beta-dependent derepression of collagen synthesis in hepatic stellate cells during liver fibrosis. J Hepatol. 2013;58:736–742. doi: 10.1016/j.jhep.2012.11.022. [DOI] [PubMed] [Google Scholar]
- 20.Roderburg C, Mollnow T, Bongaerts B, Elfimova N, Vargas Cardenas D, Berger K, Zimmermann H, et al. Micro-RNA profiling in human serum reveals compartment-specific roles of miR-571 and miR-652 in liver cirrhosis. PLoS One. 2012;7:e32999. doi: 10.1371/journal.pone.0032999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murakami Y, Toyoda H, Tanahashi T, Tanaka J, Kumada T, Yoshioka Y, Kosaka N, et al. Comprehensive miRNA expression analysis in peripheral blood can diagnose liver disease. PLoS One. 2012;7:e48366. doi: 10.1371/journal.pone.0048366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conry-Cantilena C, VanRaden M, Gibble J, Melpolder J, Shakil AO, Viladomiu L, Cheung L, et al. Routes of infection, viremia, and liver disease in blood donors found to have hepatitis C virus infection. N Engl J Med. 1996;334:1691–1696. doi: 10.1056/NEJM199606273342602. [DOI] [PubMed] [Google Scholar]
- 23.Allison RD, Conry-Cantilena C, Koziol D, Schechterly C, Ness P, Gibble J, Kleiner DE, et al. A 25-year study of the clinical and histologic outcomes of hepatitis C virus infection and its modes of transmission in a cohort of initially asymptomatic blood donors. J Infect Dis. 2012;206:654–661. doi: 10.1093/infdis/jis410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 25.Noureddin M, Wright EC, Alter HJ, Clark S, Thomas E, Chen R, Zhao X, et al. Association of IL28B genotype with fibrosis progression and clinical outcomes in patients with chronic hepatitis C: a longitudinal analysis. Hepatology. 2013;58:1548–1557. doi: 10.1002/hep.26506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trepo E, Pradat P, Potthoff A, Momozawa Y, Quertinmont E, Gustot T, Lemmers A, et al. Impact of patatin-like phospholipase-3 (rs738409 C>G) polymorphism on fibrosis progression and steatosis in chronic hepatitis C. Hepatology. 2011;54:60–69. doi: 10.1002/hep.24350. [DOI] [PubMed] [Google Scholar]
- 27.Patin E, Kutalik Z, Guergnon J, Bibert S, Nalpas B, Jouanguy E, Munteanu M, et al. Genome-wide association study identifies variants associated with progression of liver fibrosis from HCV infection. Gastroenterology. 2012;143:1244–1252. e1241–1212. doi: 10.1053/j.gastro.2012.07.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Urabe Y, Ochi H, Kato N, Kumar V, Takahashi A, Muroyama R, Hosono N, et al. A genome-wide association study of HCV-induced liver cirrhosis in the Japanese population identifies novel susceptibility loci at the MHC region. J Hepatol. 2013;58:875–882. doi: 10.1016/j.jhep.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 29.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7:e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bala S, Petrasek J, Mundkur S, Catalano D, Levin I, Ward J, Alao H, et al. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology. 2012;56:1946–1957. doi: 10.1002/hep.25873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diehl P, Fricke A, Sander L, Stamm J, Bassler N, Htun N, Ziemann M, et al. Microparticles: major transport vehicles for distinct microRNAs in circulation. Cardiovasc Res. 2012;93:633–644. doi: 10.1093/cvr/cvs007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 34.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 37.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 38.Chang CJ, Hsu CC, Chang CH, Tsai LL, Chang YC, Lu SW, Yu CH, et al. Let-7d functions as novel regulator of epithelial-mesenchymal transition and chemoresistant property in oral cancer. Oncol Rep. 2011;26:1003–1010. doi: 10.3892/or.2011.1360. [DOI] [PubMed] [Google Scholar]
- 39.Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 40.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lan FF, Wang H, Chen YC, Chan CY, Ng SS, Li K, Xie D, et al. Hsa-let-7g inhibits proliferation of hepatocellular carcinoma cells by downregulation of c-Myc and upregulation of p16(INK4A). Int J Cancer. 2011;128:319–331. doi: 10.1002/ijc.25336. [DOI] [PubMed] [Google Scholar]
- 43.Cheng JC, Yeh YJ, Tseng CP, Hsu SD, Chang YL, Sakamoto N, Huang HD. Let-7b is a novel regulator of hepatitis C virus replication. Cell Mol Life Sci. 2012;69:2621–2633. doi: 10.1007/s00018-012-0940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng M, Si Y, Niu Y, Liu X, Li X, Zhao J, Jin Q, et al. High-throughput profiling of alpha interferon- and interleukin-28B-regulated microRNAs and identification of let-7s with anti-hepatitis C virus activity by targeting IGF2BP1. J Virol. 2013;87:9707–9718. doi: 10.1128/JVI.00802-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuppan D, Krebs A, Bauer M, Hahn EG. Hepatitis C and liver fibrosis. Cell Death Differ. 2003;10(Suppl 1):S59–67. doi: 10.1038/sj.cdd.4401163. [DOI] [PubMed] [Google Scholar]
- 46.Tzur G, Israel A, Levy A, Benjamin H, Meiri E, Shufaro Y, Meir K, et al. Comprehensive gene and microRNA expression profiling reveals a role for microRNAs in human liver development. PLoS One. 2009;4:e7511m. doi: 10.1371/journal.pone.0007511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brennan EP, Nolan KA, Borgeson E, Gough OS, McEvoy CM, Docherty NG, Higgins DF, et al. Lipoxins attenuate renal fibrosis by inducing let-7c and suppressing TGFbetaR1. J Am Soc Nephrol. 2013;24:627–637. doi: 10.1681/ASN.2012060550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang B, Jha JC, Hagiwara S, McClelland AD, Jandeleit-Dahm K, Thomas MC, Cooper ME, et al. Transforming growth factor-beta1-mediated renal fibrosis is dependent on the regulation of transforming growth factor receptor 1 expression by let-7b. Kidney Int. 2014;85:352–361. doi: 10.1038/ki.2013.372. [DOI] [PubMed] [Google Scholar]
- 49.Park JT, Kato M, Lanting L, Castro N, Nam BY, Wang M, Kang SW, et al. Repression of let-7 by transforming growth factor-beta1-induced Lin28 upregulates collagen expression in glomerular mesangial cells under diabetic conditions. Am J Physiol Renal Physiol. 2014;307:F1390–1403. doi: 10.1152/ajprenal.00458.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Makino K, Jinnin M, Hirano A, Yamane K, Eto M, Kusano T, Honda N, et al. The downregulation of microRNA let-7a contributes to the excessive expression of type I collagen in systemic and localized scleroderma. J Immunol. 2013;190:3905–3915. doi: 10.4049/jimmunol.1200822. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.