Abstract
BACKGROUND
Prostate cancer (PCa) is clinically and biologically heterogeneous, making it difficult to predict at detection whether it will take an indolent or aggressive disease course. Cell cycle-regulated genes may be more highly expressed in actively dividing cells, with transcript levels reflecting tumor growth rate. Here we evaluated expression of cell cycle genes in relation to PCa outcomes in a population-based cohort.
METHODS
Gene expression data were generated from tumor tissues obtained at radical prostatectomy for 383 population-based patients (12.3-years average follow-up). The overall mean and individual transcript levels of 30 selected cell cycle genes was compared between patients with no evidence of recurrence (73%) and those who recurred (27%) or died (7%) from PCa.
RESULTS
The multivariate adjusted hazard ratio (HR) for a change from the 25th to 75th percentile of mean gene expression level (range 8.02–10.05) was 1.25 (95% CI 0.96–1.63; P = 0.10) for PCa recurrence risk, and did not vary substantially by Gleason score, TMPRSS2-ERG fusion status, or family history of PCa. For lethal PCa, the HR for a change (25th to 75th percentile) in mean gene expression level was 2.04 (95% CI 1.26–3.31; P = 0.004), adjusted for clinicopathological variables. The ROC curve for mean gene expression level alone (AUC = 0.740) did not perform as well as clinicopathological variables alone (AUC = 0.803) for predicting lethal PCa, and the addition of gene expression to clinicopathological variables did not substantially improve prediction (AUC = 0.827; P = 0.18). Higher TK1 expression was strongly associated with both recurrent (P = 6.7×10−5) and lethal (P = 6.4×10−6) PCa.
CONCLUSIONS
Mean expression level for 30 selected cell cycle-regulated genes was unrelated to recurrence risk, but was associated with a two-fold increase in risk of lethal PCa. However, gene expression had less discriminatory accuracy than clinical variables alone for predicting lethal events. Transcript levels for several genes in the panel were significantly overexpressed in lethal vs. non-recurrent PCa.
Keywords: cell cycle-regulated genes, gene expression, patient outcomes, population-based cohort, prostate cancer
INTRODUCTION
Prostate cancer (PCa) is highly prevalent, and it is second only to lung cancer as a leading cause of cancer-related death among American men [1]. In 2015, an estimated 220,000 PCa cases will be diagnosed and more than 27,000 men will die from the disease. PCa is characterized by substantial biological heterogeneity, which makes it clinically challenging to predict tumor aggressiveness and to identify which patients need treatment. Therefore, additional prognostic biomarkers that are independent of the current standard clinicopathological measures of diagnostic prostate-specific antigen (PSA) level, tumor stage, and Gleason score are urgently needed to more accurately predict individual patient outcomes.
The dysregulation of cell cycle-regulated genes is a common feature shared among different cancer types that has been associated with cancer aggressiveness and poor patient outcomes [2–6]. Cell cycle-regulated genes are those with expression levels that peak at specific times in the cell cycle and oscillate with a periodicity corresponding to cell cycle duration [7]. These genes are crucial for normal growth and development as they are involved in processes necessary for cell duplication including DNA replication and repair, spindle assembly, and the accurate segregation of daughter chromosomes. The observation that increased expression of cell cycle genes is often associated with proliferative tumors may be due in part to the presence of large numbers of cycling cells in these tissues. However, many of these genes are also likely directly involved in tumorigenesis (e.g., STK and PLK, with peak expression levels at cell cycle phase G2/M, that exhibit transforming activity in cell lines) [8]. Other cell cycle-regulated genes have decreased expression level in proliferative tumors that is associated with metastasis and poor prognosis. Included in this group are genes with peak expression levels at the M/G1 phase of the cell cycle that are involved in cell adhesion and actin cytoskeleton regulation, such as VCL, CNN2, SMTN, and AFAP [7,9,10].
Molecular signatures of tumor aggressiveness based on the expression profile of cell cycle-regulated genes were initially developed to sort breast cancer [2,3,11–13] and lymphoma [5] patients into good versus poor prognostic subgroups. Subsequently, several studies (Suppl. Table 1) of PCa patients evaluated a panel of 31 cell cycle-regulated genes to assess expression levels in relation to prognosis. Cuzick et al. [14,15] studied a cohort of men treated with radical prostatectomy (RP) at a U.S. facility and a conservatively managed PCa cohort from the U.K. and reported that the average expression level of the 31 genes was an independent prognostic marker for PCa recurrence and mortality. In a more recent study, Cooperberg et al. [16] analyzed a series of RP patients from an academic medical center and also reported that the gene expression information improved the ability to stratify PCa patients into lower versus higher risk groups for biochemical recurrence. To further evaluate the relationship between tumor tissue RNA expression profiles of genes in the cell cycle pathway (selected based on Cuzick et al, [14,15]) and prognosis, we studied a population-based cohort of PCa patients who had undergone RP. We evaluated the expression profile of 30 genes (Suppl. Table 2) in relation to outcomes and its predictive accuracy for predictive accuracy for classifying patients at risk for PCa recurrence and lethality. In addition, we tested for differences in expression level of each gene according to patient outcome groups.
MATERIALS AND METHODS
Patients
The present study included 383 men of European American ancestry who had undergone RP as primary treatment for clinically localized PCa and were previously enrolled in population-based studies of PCa in King County, Washington [17,18]. The first study ascertained cases under age 65 years who were diagnosed between 1993 and 1996, and for the second study men were under age 75 and were diagnosed between 2002 and 2005. The study was approved by the Fred Hutchinson Cancer Research Center Institutional Review Board, and all participants signed informed consent statements. Patient information including demographic factors, family cancer history, pathological tumor stage, Gleason score, PSA level at diagnosis, and tumor TMPRSS2-ERG fusion status was available. Vital status and underlying cause of death were obtained from the Seattle-Puget Sound SEER cancer registry and coded according to SEER guidelines [19]. Follow-up surveys were completed by patients between 2004–2005 and 2010–2011, and patients were classified as recurrent if they had a rising post-treatment PSA level (PSA ≥ 0.2 ng/mL), received secondary treatment (e.g., radiation therapy, androgen deprivation therapy, orchiectomy, or chemotherapy), had a positive lymph node or prostate bed biopsy, MRI, CT, or bone scan showing PCa, were told by a physician that their PCa had recurred, or died of PCa (confirmed by death certificate). A lethal phenotype category was defined as patients who developed metastasis or died of PCa.
Sample Preparation and RNA Extraction
Formalin-fixed paraffin-embedded (FFPE) tumor tissue blocks from RP samples were used to prepare H&E stained slides that were reviewed by a PCa pathologist to confirm the presence and location of prostate adenocarcinoma. Two 1-mm tumor tissue cores per patient were taken from the dominant lesion in areas containing ≥ 75% tumor cells. RNA was isolated using the RNeasy® FFPE Kit (Qiagen Inc., Valencia, CA), quantified using RiboGreen, and stored at -80°C. A 200ng RNA aliquot per patient was shipped to Illumina for genome-wide gene expression profiling [20]. Laboratory personnel were blinded to patient outcomes, which were evenly distributed across batches of tumor RNA.
Gene Expression Profiling
Expression profiling was conducted using the Whole-Genome DASL® (cDNA-mediated Annealing, Selection, Extension, and Ligation) HT Assay (Illumina, Inc., San Diego, CA). Briefly, tumor RNA was converted to cDNA using biotinylated oligo and random nonamer primers, and was subsequently immobilized to a streptavidin-coated solid support. Quantitative RT-PCR and analysis of housekeeping gene RPL13a were used for prequalification assessment of cDNA. Biotinylated cDNAs were annealed to assay-specific oligonucleotides to create PCR templates that were amplified using labeled and biotinylated universal primers. The labeled PCR products were captured on streptavidin paramagnetic beads, washed, and denatured to yield single-stranded fluorescent molecules. These were hybridized to the HumanHT-12 v4 Expression BeadChip and scanned using BeadArray® Reader to extract intensity images for >29,000 transcripts. We included a blind duplicate sample for six patients that were randomly distributed across the plates. The transcript correlations between duplicated samples ranged from 0.98 to 0.99. In addition, replicate tumor RNA samples from two patients were included on every plate, and the transcript correlations across plates were >0.99 for each subject.
Statistical Analysis
Batch effects were removed using ComBat [21] and gene expression data were quantile normalized and log2 transformed using R statistical computing software [22]. Gene expression levels were then calculated for: 1) men without evidence of PCa recurrence; 2) men with any evidence of PCa recurrence; and 3) a subgroup of patients who developed metastasis or died of PCa (Table 1). Time to recurrence was imputed for patients who died from PCa but whose date of recurrence was unknown [23], based on those with known time to recurrence who died of PCa. Follow-up time was calculated from the date of PCa diagnosis to the date of recurrence, PCa-specific death or death from another cause, or last follow-up.
TABLE I.
Characteristics of the Population-based Prostate Cancer (PCa) Patient Cohort According to Outcomes
| Characteristic | No Recurrence (n = 278; 72.6%) | Recurrencea (n = 105; 27.4%) | Lethal PCab (n = 27; 7.1%) | |||
|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |
| Mean age at diagnosis (±sd) | 58 (6.93) | 59 (6.96) | 58 (6.69) | |||
| Mean expression level of 30 cell cycle-regulated genes (±sd) | 8.82 (0.33) | 8.94 (0.38) | 9.13 (0.46) | |||
| Median duration of follow-up, yrs. | 12.4 | 12.2 | 12.5 | |||
| PSA at diagnosis (ng/mL): | ||||||
| 0 – 3.9 | 47 | (16.9) | 13 | (12.4) | 2 | (7.4) |
| 4 – 9.9 | 174 | (62.6) | 41 | (39.0) | 7 | (25.9) |
| 10 – 19.9 | 30 | (10.8) | 26 | (24.8) | 6 | (22.2) |
| 20+ | 11 | (4.0) | 16 | (15.2) | 8 | (29.6) |
| Missing | 16 | (5.8) | 9 | (8.6) | 4 | (14.8) |
| Pathological stage | ||||||
| Localized (T2) | 213 | (76.6) | 44 | (41.9) | 13 | (48.1) |
| Regional (T3) | 65 | (23.4) | 61 | (58.1) | 14 | (51.9) |
| Gleason score | ||||||
| <=6 | 157 | (56.5) | 24 | (22.9) | 5 | (18.5) |
| 7(3+4) | 93 | (33.5) | 49 | (46.7) | 11 | (40.7) |
| 7(4+3) | 15 | (5.4) | 14 | (13.3) | 5 | (18.5) |
| 8–10 | 13 | (4.7) | 18 | (17.1) | 6 | (22.2) |
| TMPRSS2-ERG fusion statusc | ||||||
| Negative | 106 | (40.9) | 42 | (44.2) | 15 | (60.0) |
| Positive | 153 | (59.1) | 53 | (55.8) | 10 | (40.0) |
| 1st Degree family history of PCa | ||||||
| No | 214 | (77.0) | 85 | (81.0) | 23 | (85.2) |
| Yes | 64 | (23.0) | 20 | (19.0) | 4 | (14.8) |
Includes patients with biochemical recurrence, secondary treatment, and metastatic or lethal PCa.
Includes patients with metastatic or lethal PCa.
Number of patients missing TMPRSS2-ERG fusion status: 19, 10, and 2, respectively in the three groups.
Low quality probes were filtered out with Illumina Human WGDASLv4.db in R Bioconductor. Of 31 cell cycle-regulated genes evaluated by Cuzick et al. [14,15], 30 were robustly expressed in our dataset (c18orf24 was filtered out due to low quality). For eight genes with multiple transcripts, we selected the transcript with the highest variability in the non-recurrence group for analysis. Expression levels for the 30 transcripts were used to calculate the mean gene expression level for each patient.
Cox proportional hazards models were used to calculate hazard ratios (HR) for time to PCa recurrence and time to PCa death according to the mean gene expression level, with and without adjusting for clinicopathological variables. Clinicopathological variables included age at diagnosis, Gleason score, diagnostic PSA level (using the natural logarithm of 1+PSA for consistency with earlier reports (categorized with a missing indicator variable)) [14,15], and pathologic tumor stage. Additional models were adjusted for the established cell proliferation marker Ki67 [24]. Secondary analyses evaluated mean gene expression level by time to recurrence in subsets of patients stratified by tumor TMPRSS2-ERG fusion status, first-degree PCa family history, and Gleason score (stratified into three groups: ≤6; 7=3+4; and ≥7=4+3). Kaplan-Meier plots were generated for both overall recurrence and lethal PCa outcomes using mean expression level divided into three categories: 1) < 25th percentile; 2) 25th to 75th percentile; and 3) ≥ 75th percentile. Log-rank tests were used to determine whether the time to event was significantly different between the highest (≥ 75%) and lowest (< 25%) categories. Receiver operating characteristic (ROC) curve analysis was performed to examine the sensitivity and specificity of models with mean gene expression level alone, clinicopathological variables alone, and mean gene expression level combined with clinicopathological variables, using a 5-fold cross validation, for both recurrence and lethal PCa outcomes. For these analyses, recurrence or lethal PCa outcome compared to non-recurrence was modelled as a binary outcome.
We next examined the expression levels of individual genes included in the 30 gene panel in relation to patient outcomes. The mean expression level of each transcript was compared between men without evidence of PCa recurrence and: 1) men with any evidence of PCa recurrence; and 2) a subgroup of patients with metastatic or lethal PCa, using a t test.
RESULTS
Mean gene expression level of the 30 transcripts was calculated for 383 PCa cases (mean age at diagnosis = 58 years) who were classified as having no evidence of PCa recurrence (73%) or having a recurrence event (27%) over an average follow-up of 12.3 years (Table 1). A subset of cases who progressed to metastatic or lethal PCa (7% of the total cohort) was also examined (Table 1). The average gene expression level was lower for the non-recurrence group (mean expression level = 8.82) compared to the recurrence group (mean expression level = 8.94, P = 0.005), and it was highest in men who progressed to metastatic or lethal PCa (mean expression level = 9.13, P = 0.002). The median time of follow-up or time to event was: 12.4 years for the non-recurrence group; 12.2 years for the recurrence group; and 12.5 years for the metastatic/lethal PCa group (Table 1).
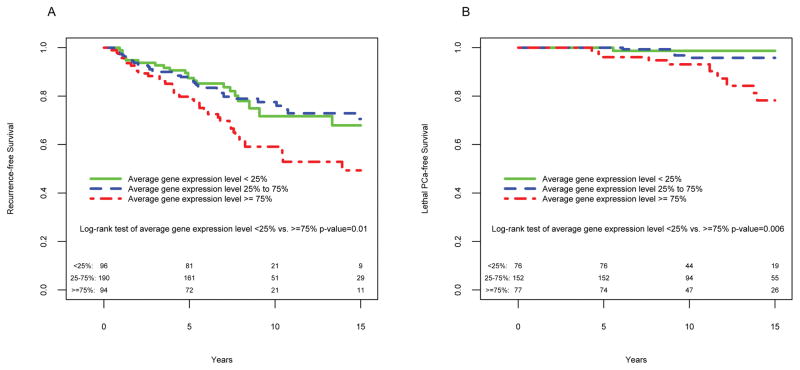
The hazard ratio (HR) associated with a change from the 25th to 75th percentile of mean expression level distribution (range 8.02–10.05, Suppl. Figure 1) in the univariate analysis was 1.43 (95% CI 1.12–1.84; P = 004) for time to recurrence and 2.41 (95% CI 1.57–3.71, P < 0.0001) for time to PCa death (Table 2). After adjusting for covariates (i.e., age at diagnosis, diagnostic PSA level, Gleason score, and pathological stage), the HRs decreased to 1.25 (95% CI 0.96–1.63; P = 0.10) for overall recurrence and 2.04 (95% CI 1.26–3.31; P = 0.004) for lethal outcomes. Addition of the cell proliferation marker Ki67 expression to the models did not significantly change the results (not shown). Mean gene expression level was weakly correlated with diagnostic PSA level (0.15) and Gleason score (0.14). Figure 1 provides Kaplan-Meier curves for time to recurrence and time to PCa metastasis or death for patients stratified by percentile of mean expression level distribution. Patients in the high (≥75%) versus low (<25%) expression group had a lower probability of recurrence-free survival (log-rank test P = 0.01) and lethal PCa-free survival (log-rank test P = 0.006) over a 15-year follow-up period.
TABLE II.
Cox Proportional Hazards Ratios (HR) for Time to Prostate Cancer (PCa) Recurrence and Time to Lethal PCa According to Mean Expression Level of 30 Cell Cycle-regulated Genes
| Time to recurrencea | Time to lethal PCab | |||
|---|---|---|---|---|
| Unadjusted HR (95% CI) | P-value | Unadjusted HR (95% CI) | P-value | |
| Mean gene expression levelc | 1.43 (1.12, 1.84) | 0.004 | 2.41 (1.57, 3.71) | <0.0001 |
| Adjusted HRd (95% CI) | P-value | Adjusted HRd (95% CI) | P-value | |
| Mean gene expression levelc | 1.25 (0.96, 1.63) | 0.10 | 2.04 (1.26, 3.31) | 0.004 |
| Age at diagnosis | 0.99 (0.96, 1.03) | 0.65 | 0.95 (0.88, 1.04) | 0.26 |
| Gleason score | ||||
| <=6 | reference | -- | reference | -- |
| 7(3+4) | 2.24 (1.33, 3.50) | 0.003 | 3.33 (1.09, 10.16) | 0.035 |
| 7(4+3) and 8–10 | 3.58 (1.95, 6.57) | <0.0001 | 7.91 (2.55, 24.51) | 0.0003 |
| Log(1+PSA) | 1.52 (1.11, 2.08) | 0.01 | 2.12 (1.13, 3.99) | 0.02 |
| Pathological stage | ||||
| Localized (T2) | reference | -- | reference | -- |
| Regional (T3) | 2.58 (1.67, 3.99) | <0.0001 | 1.27 (0.45, 3.62) | 0.65 |
Includes patients with biochemical recurrence, secondary treatment, and metastatic or lethal PCa.
Includes patients with metastatic or lethal PCa.
HRs are transformed to correspond to changes from the 25th to the 75th percentile of the distribution of gene expression profiles.
HR adjusted for age at diagnosis, Gleason score, diagnostic PSA, and pathological stage.
Fig. 1.
Kaplan-Meier analysis of A) time to prostate cancer (PCa) recurrence and B) time to lethal PCa according to percentile distribution of mean expression levels of 30 cell cycle-regulated genes
In secondary analyses, Cox proportional hazards models were used to evaluate mean gene expression level by time to PCa recurrence after stratifying by Gleason score, TMPRSS2-ERG fusion status, and PCa family history (Suppl. Table 3). The HR associated with mean expression level for patients with a Gleason score of ≤ 6 (HR=1.29; 95% CI 0.0.71–2.37) was not significantly different (P = 0.73) from those with higher Gleason scores of 7(3+4), HR=1.36 (95% CI 0.90–2.05) and Gleason 7(4+3) and 8–10 combined, HR=1.09 (95% CI 0.70–1.69). Patients with TMPRSS2-ERG fusion positive tumors had an HR for time to recurrence (1.61; 95% CI 1.13–2.31) similar to patients with fusion negative tumors (HR = 1.36; 95% CI 1.10–1.68; P = 0.67) in relation to mean expression level. HRs for expression level in association with time to recurrence did not differ substantially (P = 0.86) in patients without (HR = 1.42) versus with a family history of PCa (HR = 1.51). There were too few individuals in the lethal PCa subgroup to complete stratified analyses.
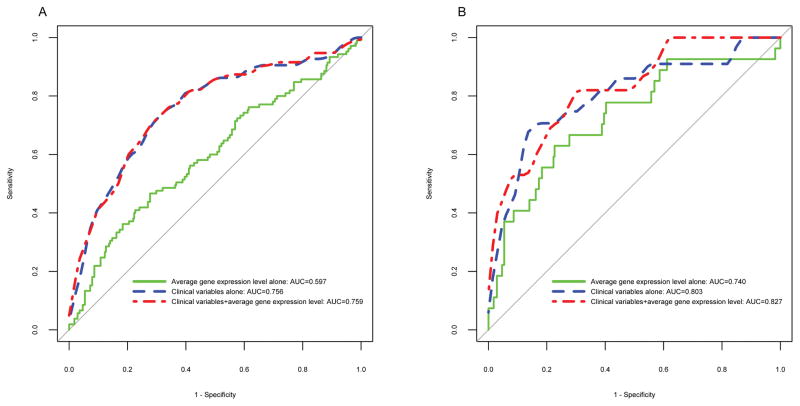
ROC curve analyses were used to examine specificity and sensitivity of mean gene expression level in comparison to clinicopathological variables for predicting PCa recurrence and lethal PCa (Figure 2). The expression level alone (AUC = 0.597) did not perform as well as the clinicopathological variables alone (AUC = 0.756) for predicting PCa recurrence, and when expression level was combined with clinical variables, there was a non-significant improvement (AUC = 0.759, P = 0.17). Similar results were obtained for predicting lethal PCa, with clinicopathological variables performing better (AUC = 0.803) than expression level alone (AUC = 0.740), and their combination showing only slight improvement (AUC = 0.827) over the clinicopathological variables (P = 0.18).
Fig. 2.
ROC curves for A) prostate cancer (PCa) recurrence and B) lethal PCa according to mean gene expression level alone, clinical variables alone, and in combination
For the individual gene analysis, the expression of TK1 exhibited the greatest statistically significant difference among the patient groups (Suppl. Table 4). The lowest level of expression was observed in patients with no evidence of PCa recurrence (9.54), with substantially higher levels seen in the recurrence group (9.96, P = 6.7×10−5) and in the lethal PCa group (10.5, P = 6.4×10−6). Nine other genes exhibited a similar pattern of statistically significant differences (P < 0.05) between the groups, with the lowest expression level for the non-recurrence group. Four transcripts (one in NUSAP1, ORC6L, PBK, and PTTG1) had a higher level of expression in the non-recurrence group compared to the other groups, however, these differences were not statistically significant (P > 0.05).
DISCUSSION
Our results demonstrate that the average gene expression level of 30 cell cycle-regulated genes is associated with time to PCa metastasis/death, independent of age, stage, PSA level, and Gleason score at diagnosis in a population-based cohort of men who underwent RP; however, this finding is based on a limited number of lethal events (n=27). The mean expression level was not associated with overall PCa recurrence after controlling for clinicpathological variables. These results are not totally consistent with findings from Cuzick et al. [14,15] who first described the mean expression level of cell cycle-regulated genes as a potential biomarker for predicting recurrence in a series of PCa patients in the U.S. who underwent RP at a single facility [14], and for predicting time to death in conservatively managed (i.e., watchful waiting) patients in the U.K. who were diagnosed by TURP [14] or needle biopsy [15] (Suppl. Table 1). Cuzick et al. [14,15] generated expression data for 31 cell cycle genes using an RT-PCR assay (Myriad Genetics, Inc.; Salt Lake City, UT), while we generated genome-wide transcriptome data and excluded one of the 31 genes (c18orf24) due to poor quality transcript data. The HRs reported by Cuzick et al. [15] for changes from the 25th to 75th percentile of the mean gene expression distribution for PCa-specific mortality (unadjusted HR = 2.56; HR adjusted for clinical variables = 1.96) are of similar magnitude and direction as ours (unadjusted HR = 2.41; adjusted HR = 2.41). Cuzick et al. [14] reported HRs for time to biochemical recurrence as a 1-unit change in mean gene expression level corresponding with an unadjusted HR = 1.89 and an adjusted HR = 1.77, while our results, which are not directly comparable due to scale differences in expression level, do not provide evidence of association with overall risk of PCa recurrence (unadjusted HR = 1.43; adjusted HR = 1.25, comparing the 25th to 75th percentile).
Other recent studies using the same gene expression panel (see Suppl. Table 1) include one by Cooperberg et al. [16] who studied a series of 413 RP patients treated at an academic medical center. They reported an unadjusted HR = 2.1 (95% CI 1.6–2.9) per unit increase in mean expression level, and an adjusted HR = 1.7 (95% CI 1.3–2.4), for biochemical recurrence or use of secondary treatment. Freedland et al. [25] calculated mean expression level for 141 African American and European American patients who underwent external beam radiation therapy (EBRT), and reported an adjusted HR of 2.11 (95% CI 1.05–4.25) for each 1-unit increase in score for time to biochemical recurrence, and an unadjusted HR of 3.77 (95% CI 1.34–10.4) for disease-specific mortality. Bishoff et al. [26] evaluated the mean gene expression level in biopsy samples from a series of RP patients (n=582), and reported adjusted HRs of 1.47 (95% CI 1.23–1.76) for recurrence and 4.19 (95% CI 2.08–8.45) for time to metastasis. These latter results, however, are based on small numbers of adverse events (i.e., 6 deaths in Freedland et al. [25] and 12 patients with metastatic disease in Bishoff et al. [26]). Our study examined the mean expression level of cell cycle-regulated genes in a population-based cohort, which may be more representative of the general PCa patient population in the U.S. than previous studies based on highly selected groups of hospital- or clinic-based patients from a single or a few facilities. To our knowledge, our study is the first to stratify patients based on tumor TMPRSS2-ERG fusion status or first-degree family history of PCa, and we found similar HRs for time to biochemical recurrence in these subgroups.
Despite differences across studies (Suppl. Table 1), most found similar associations (HRs of approximately 2.0) between mean expression level of selected cell cycle-regulated genes and risks of either PCa recurrence or lethal PCa, suggesting that expression level reflects tumor behavior. However, associations results may not directly translate into the gene expression profile having sufficient accuracy for classification of individual PCa patients. Pepe et al. [27] have shown that very strong associations on the order of ≥10-fold in magnitude (e.g., HRs ≥ 10.0) are usually required to accurately distinguish between individuals with versus without a particular disease-related outcome. Use of a biomarker test to guide treatment decisions for individual PCa patients also requires consideration of its classification performance (i.e., sensitivity, specificity, positive and negative predictive values) in terms of its intended clinical context and the extent to which its inclusion improves risk classification beyond existing prognostic factors (e.g., stage, Gleason score, PSA) [28]. To investigate the potential clinical application of the mean expression level of the 30-gene expression panel, we used ROC curve analyses for predicting both time to PCa recurrence and PCa-specific death. These results showed that expression level did not substantially improve the ability to predict patient outcomes beyond that of clinicopathological variables (age, Gleason score, stage, PSA) alone. Although the mean gene expression level was associated with a two-fold increase in the HR for lethal PCa, it had limited accuracy for stratifying individual patients into lower versus higher-risk groups for predicting subsequent recurrent or lethal PCa.
The majority of the cell cycle-regulated genes in the panel were overexpressed in tumor tissue from patients who experienced PCa recurrence compared to patients with no evidence of recurrence, with the highest expression level present in those with lethal PCa. The upregulation of cell cycle genes in proliferative tumors is well documented in the published literature for several different types of cancer [2–6]. These genes are necessary for normal function of the cell division cycle, as they play key roles in DNA replication and repair, and chromosomal segregation, and they are frequently deregulated during tumorigenesis [8]. In our study, there was a significant overexpression particularly of genes with peak expression levels during the G2 and G2/M phases of the cell cycle, such as CDC2, KIF11, KIF20A, PRC1, and TOP2A, with functions related to mitosis (see Suppl. Table 2). None of the genes examined in this study had significantly lower expression levels in the lethal PCa group, although previous studies have identified cell cycle-regulated genes that exhibit decreased expression levels in proliferative tumors and are associated with poor patient outcomes [7,9,10].
The top gene in the gene panel with significantly higher expression levels for the recurrence and lethal PCa patient groups compared to the non-recurrence group was thymidine kinase 1 (TK1). TK1 is a DNA repair enzyme that peaks during the S phase of the cell cycle [29], when DNA replication occurs. It is an established serum biomarker for several types of cancer (e.g., breast, colon, lung, and lymphoma [30–32]), and its overexpression in tumor compared to normal tissues was demonstrated across multiple cancer types, including PCa, along with an increase in expression level with tumor grade[33]. Our study also demonstrated that overexpression of FOXM1, KIF20A, and PRC1 transcripts was significantly (P = 0.001) associated with lethal PCa.
CONCLUSIONS
Our results confirm an association between the mean expression level of selected cell cycle-regulated genes and time to lethal PCa in a cohort of population-based patients treated with RP. However, mean expression level did not substantially improve the AUC and had less discriminatory accuracy than standard clinicopathological variables alone for predicting disease recurrence or lethal patient outcomes. Several of the cell cycle-regulated genes (e.g., TK1, FOXM1) were significantly upregulated in tumors with more aggressive behavior, these may contribute most to the cell cycle dysregulation involved in PCa progression.
Supplementary Material
Acknowledgments
We thank Drs. Beatrice Knudson and Antonio Hurado-Coll for their assistance with the pathology, and Illumina, Inc. for supplying and performing the gene expression arrays. Financial support: R01 CA056678, R01 CA092579, and P50 CA097186; and the Fred Hutchinson Cancer Research Center and the Prostate Cancer Foundation.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Mosley JD, Keri RA. Cell cycle correlated genes dictate the prognostic power of breast cancer gene lists. BMC Med Genomics. 2008;1:11. doi: 10.1186/1755-8794-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 4.LaTulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, Gerald WL. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62(15):4499–4506. [PubMed] [Google Scholar]
- 5.Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, Gascoyne RD, Grogan TM, Muller-Hermelink HK, Smeland EB, Chiorazzi M, Giltnane JM, Hurt EM, Zhao H, Averett L, Henrickson S, Yang L, Powell J, Wilson WH, Jaffe ES, Simon R, Klausner RD, Montserrat E, Bosch F, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Fisher RI, Miller TP, LeBlanc M, Ott G, Kvaloy S, Holte H, Delabie J, Staudt LM. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3(2):185–197. doi: 10.1016/s1535-6108(03)00028-x. [DOI] [PubMed] [Google Scholar]
- 6.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci U S A. 2004;101(25):9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, Botstein D. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13(6):1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitfield ML, George LK, Grant GD, Perou CM. Common markers of proliferation. Nat Rev Cancer. 2006;6(2):99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- 9.Engers R, Gabbert HE. Mechanisms of tumor metastasis: cell biological aspects and clinical implications. J Cancer Res Clin Oncol. 2000;126(12):682–692. doi: 10.1007/s004320000148. [DOI] [PubMed] [Google Scholar]
- 10.Rudiger M. Vinculin and alpha-catenin: shared and unique functions in adherens junctions. BioEssays. 1998;20(9):733–740. doi: 10.1002/(SICI)1521-1878(199809)20:9<733::AID-BIES6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 11.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lonning PE, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naderi A, Teschendorff AE, Barbosa-Morais NL, Pinder SE, Green AR, Powe DG, Robertson JF, Aparicio S, Ellis IO, Brenton JD, Caldas C. A gene-expression signature to predict survival in breast cancer across independent data sets. Oncogene. 2007;26(10):1507–1516. doi: 10.1038/sj.onc.1209920. [DOI] [PubMed] [Google Scholar]
- 13.Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel AB, van’t Veer LJ, Perou CM. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355(6):560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 14.Cuzick J, Swanson GP, Fisher G, Brothman AR, Berney DM, Reid JE, Mesher D, Speights VO, Stankiewicz E, Foster CS, Moller H, Scardino P, Warren JD, Park J, Younus A, Flake DD, 2nd, Wagner S, Gutin A, Lanchbury JS, Stone S. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12(3):245–255. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuzick J, Berney DM, Fisher G, Mesher D, Moller H, Reid JE, Perry M, Park J, Younus A, Gutin A, Foster CS, Scardino P, Lanchbury JS, Stone S Transatlantic Prostate G. Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br J Cancer. 2012;106(6):1095–1099. doi: 10.1038/bjc.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooperberg MR, Simko JP, Cowan JE, Reid JE, Djalilvand A, Bhatnagar S, Gutin A, Lanchbury JS, Swanson GP, Stone S, Carroll PR. Validation of a cell-cycle progression gene panel to improve risk stratification in a contemporary prostatectomy cohort. J Clin Oncol. 2013;31(11):1428–1434. doi: 10.1200/JCO.2012.46.4396. [DOI] [PubMed] [Google Scholar]
- 17.Stanford JL, Wicklund KG, McKnight B, Daling JR, Brawer MK. Vasectomy and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 1999;8(10):881–886. [PubMed] [Google Scholar]
- 18.Agalliu I, Salinas CA, Hansten PD, Ostrander EA, Stanford JL. Statin use and risk of prostate cancer: results from a population-based epidemiologic study. Am J Epidemiol. 2008;168(3):250–260. doi: 10.1093/aje/kwn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritz A, Ries L. SEER Program Code Manual. 3. National Cancer Institute, NIH; 1998. [Google Scholar]
- 20.April C, Klotzle B, Royce T, Wickham-Garcia E, Boyaniwsky T, Izzo J, Cox D, Jones W, Rubio R, Holton K, Matulonis U, Quackenbush J, Fan JB. Whole-genome gene expression profiling of formalin-fixed, paraffin-embedded tissue samples. PLoS One. 2009;4(12):e8162. doi: 10.1371/journal.pone.0008162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 22.R Core Development Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. [Google Scholar]
- 23.Harrell FE, editor. Regression modeling strategies: with applications to linear models, logistic regression, and survival analysis. xxii. New York, NY: Springer-Verlag; 2001. p. 568. [Google Scholar]
- 24.Fisher G, Yang ZH, Kudahetti S, Moller H, Scardino P, Cuzick J, Berney DM Transatlantic Prostate G. Prognostic value of Ki-67 for prostate cancer death in a conservatively managed cohort. Br J Cancer. 2013;108(2):271–277. doi: 10.1038/bjc.2012.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freedland SJ, Gerber L, Reid J, Welbourn W, Tikishvili E, Park J, Younus A, Gutin A, Sangale Z, Lanchbury JS, Salama JK, Stone S. Prognostic utility of cell cycle progression score in men with prostate cancer after primary external beam radiation therapy. Int J Radiat Oncol Biol Phys. 2013;86(5):848–853. doi: 10.1016/j.ijrobp.2013.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bishoff JT, Freedland SJ, Gerber L, Tennstedt P, Reid J, Welbourn W, Graefen M, Sangale Z, Tikishvili E, Park J, Younus A, Gutin A, Lanchbury JS, Sauter G, Brawer M, Stone S, Schlomm T. Prognostic utility of the cell cycle progression score generated from biopsy in men treated with prostatectomy. J Urol. 2014 doi: 10.1016/j.juro.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 27.Pepe MS, Janes H, Longton G, Leisenring W, Newcomb P. Limitations of the odds ratio in gauging the performance of a diagnostic, prognostic, or screening marker. Am J Epidemiol. 2004;159(9):882–890. doi: 10.1093/aje/kwh101. [DOI] [PubMed] [Google Scholar]
- 28.Feng Z. Classification versus association models: should the same methods apply? Scand J Clin Lab Invest Suppl. 2010;242:53–58. doi: 10.3109/00365513.2010.493387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He Q, Skog S, Tribukait B. Cell cycle related studies on thymidine kinase and its isoenzymes in Ehrlich ascites tumours. Cell Prolif. 1991;24(1):3–14. doi: 10.1111/j.1365-2184.1991.tb01506.x. [DOI] [PubMed] [Google Scholar]
- 30.Alegre MM, Robison RA, O’Neill KL. Thymidine kinase 1 upregulation is an early event in breast tumor formation. J Oncol. 2012;2012:575647. doi: 10.1155/2012/575647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He E, Xu XH, Guan H, Chen Y, Chen ZH, Pan ZL, Tang LL, Hu GZ, Li Y, Zhang M, Zhou J, Eriksson S, Fornander T, Skog S. Thymidine kinase 1 is a potential marker for prognosis and monitoring the response to treatment of patients with breast, lung, and esophageal cancer and non-Hodgkin’s lymphoma. Nucleosides Nucleotides Nucleic Acids. 2010;29(4–6):352–358. doi: 10.1080/15257771003738535. [DOI] [PubMed] [Google Scholar]
- 32.O’Neill KL, Zhang F, Li H, Fuja DG, Murray BK. Thymidine kinase 1--a prognostic and diagnostic indicator in ALL and AML patients. Leukemia. 2007;21(3):560–563. doi: 10.1038/sj.leu.2404536. [DOI] [PubMed] [Google Scholar]
- 33.Alegre MM, Robison RA, O’Neill KL. Thymidine Kinase 1: a universal marker for cancer. Cancer Clin Oncol. 2013;2(1):159–167. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.