Abstract
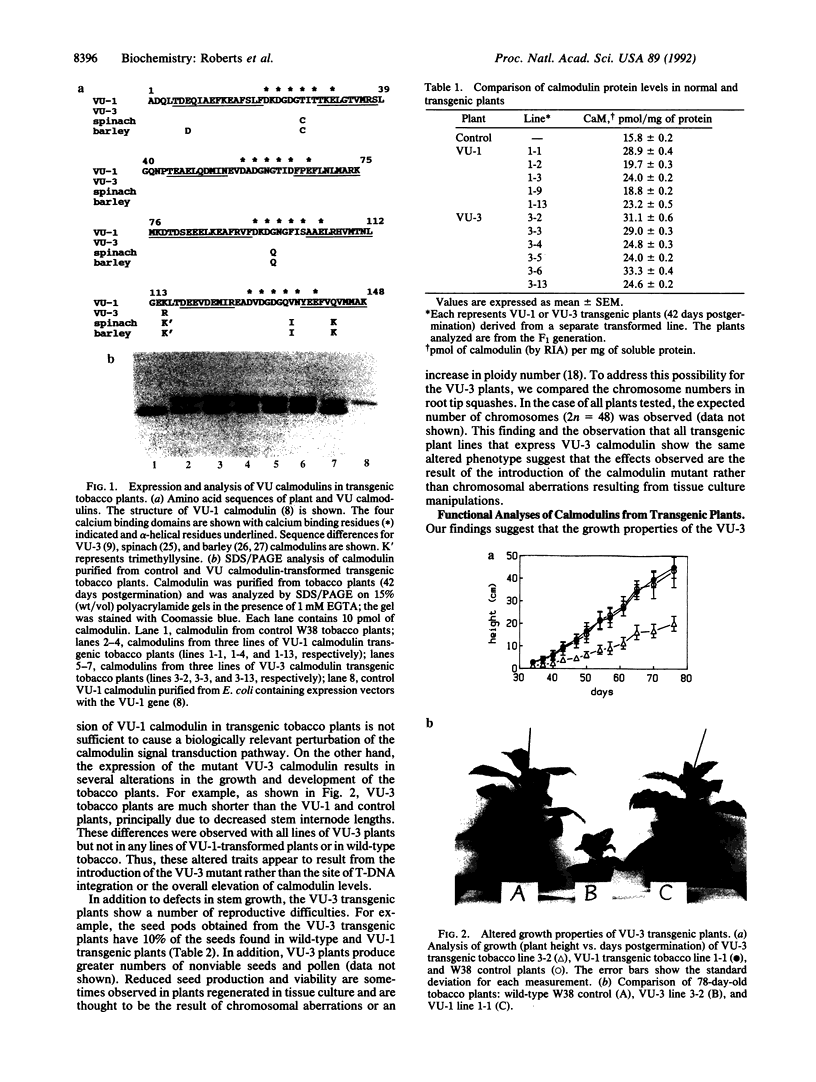
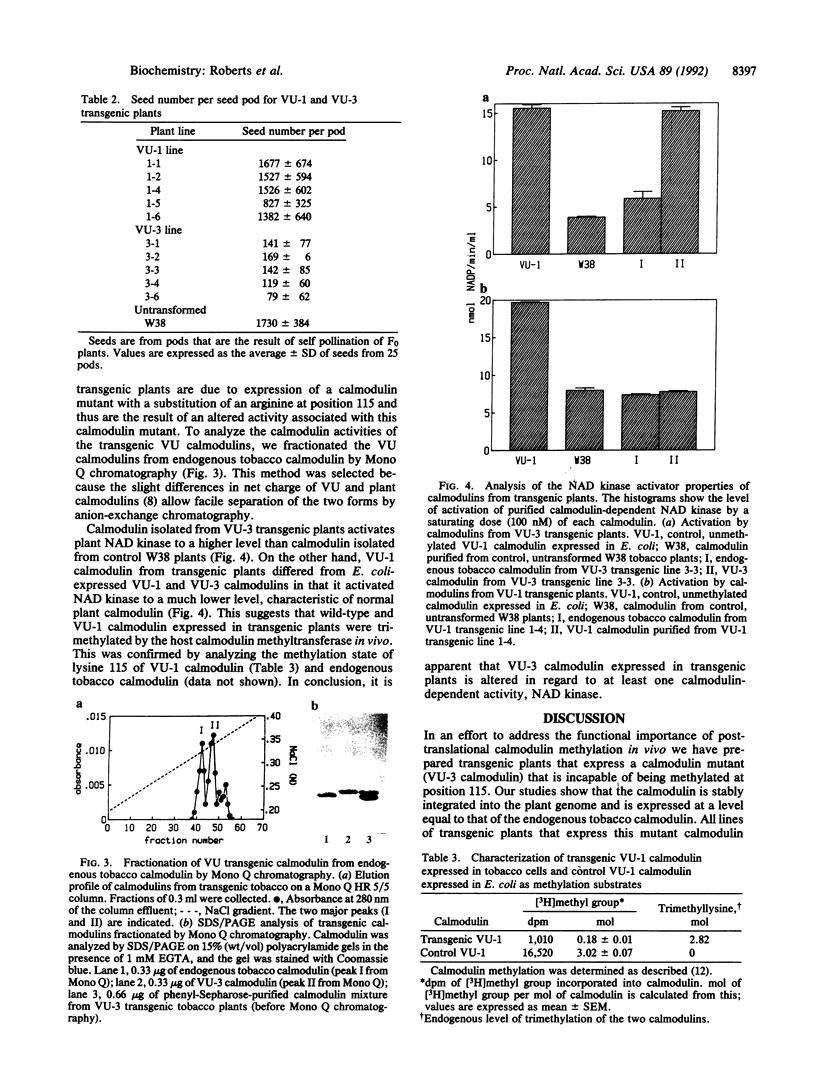
Transgenic plants were constructed that express two foreign calmodulins (VU-1 and VU-3 calmodulins) derived from a cloned synthetic calmodulin gene. VU-1 calmodulin, similar to endogenous plant calmodulin, possesses a lysine residue at position 115 and undergoes posttranslational methylation. VU-3 calmodulin is a site-directed mutant of VU-1 calmodulin that is identical in sequence except for the substitution of an arginine at position 115 and thus is incapable of methylation. Both calmodulin genes, under the control of the cauliflower mosaic virus 35S promoter, were expressed in transgenic tobacco. Foreign calmodulin protein accumulated in plant tissues to levels equivalent to that of the endogenous calmodulin. All transformed lines of VU-1 plants were indistinguishable from untransformed controls with respect to growth and development. However, all transformed lines of VU-3 plants were characterized by decreased stem internode growth, reduced seed production, and reduced seed and pollen viability. The data suggest that these phenotypes are the result of the expression of the calmodulin mutant rather than the position of transferred DNA insertion or the overall alteration of calmodulin levels. Analyses of the activity of the purified transgenic calmodulins suggest that calmodulin-dependent NAD kinase is among the potential targets that may have altered regulation in VU-3 transgenic plants.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- A simple and general method for transferring genes into plants. Science. 1985 Mar 8;227(4691):1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Anderson J. M., Cormier M. J. Calcium-dependent regulation of NAD kinase. Biochem Biophys Res Commun. 1978 Oct 16;84(3):595–602. doi: 10.1016/0006-291x(78)90747-7. [DOI] [PubMed] [Google Scholar]
- Braam J., Davis R. W. Rain-, wind-, and touch-induced expression of calmodulin and calmodulin-related genes in Arabidopsis. Cell. 1990 Feb 9;60(3):357–364. doi: 10.1016/0092-8674(90)90587-5. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Gopalakrishna R., Anderson W. B. Ca2+-induced hydrophobic site on calmodulin: application for purification of calmodulin by phenyl-Sepharose affinity chromatography. Biochem Biophys Res Commun. 1982 Jan 29;104(2):830–836. doi: 10.1016/0006-291x(82)90712-4. [DOI] [PubMed] [Google Scholar]
- Gregori L., Marriott D., Putkey J. A., Means A. R., Chau V. Bacterially synthesized vertebrate calmodulin is a specific substrate for ubiquitination. J Biol Chem. 1987 Feb 25;262(6):2562–2567. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Ling V., Zielinski R. E. Cloning of cDNA Sequences Encoding the Calcium-Binding Protein, Calmodulin, from Barley (Hordeum vulgare L.). Plant Physiol. 1989 Jun;90(2):714–719. doi: 10.1104/pp.90.2.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas T. J., Iverson D. B., Schleicher M., Watterson D. M. Structural characterization of a higher plant calmodulin : spinacia oleracea. Plant Physiol. 1984 Jul;75(3):788–795. doi: 10.1104/pp.75.3.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshak D. R., Clarke M., Roberts D. M., Watterson D. M. Structural and functional properties of calmodulin from the eukaryotic microorganism Dictyostelium discoideum. Biochemistry. 1984 Jun 19;23(13):2891–2899. doi: 10.1021/bi00308a007. [DOI] [PubMed] [Google Scholar]
- Morino H., Kawamoto T., Miyake M., Kakimoto Y. Purification and properties of calmodulin-lysine N-methyltransferase from rat brain cytosol. J Neurochem. 1987 Apr;48(4):1201–1208. doi: 10.1111/j.1471-4159.1987.tb05647.x. [DOI] [PubMed] [Google Scholar]
- Muto S., Miyachi S. Properties of a Protein Activator of NAD Kinase from Plants. Plant Physiol. 1977 Jan;59(1):55–60. doi: 10.1104/pp.59.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. H., Roberts D. M. Analysis of the state of posttranslational calmodulin methylation in developing pea plants. Plant Physiol. 1990 Jul;93(3):880–887. doi: 10.1104/pp.93.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D. M., Burgess W. H., Watterson D. M. Comparison of the NAD Kinase and Myosin Light Chain Kinase Activator Properties of Vertebrate, Higher Plant, and Algal Calmodulins. Plant Physiol. 1984 Jul;75(3):796–798. doi: 10.1104/pp.75.3.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D. M., Crea R., Malecha M., Alvarado-Urbina G., Chiarello R. H., Watterson D. M. Chemical synthesis and expression of a calmodulin gene designed for site-specific mutagenesis. Biochemistry. 1985 Sep 10;24(19):5090–5098. doi: 10.1021/bi00340a020. [DOI] [PubMed] [Google Scholar]
- Roberts D. M., Rowe P. M., Siegel F. L., Lukas T. J., Watterson D. M. Trimethyllysine and protein function. Effect of methylation and mutagenesis of lysine 115 of calmodulin on NAD kinase activation. J Biol Chem. 1986 Feb 5;261(4):1491–1494. [PubMed] [Google Scholar]
- Rowe P. M., Wright L. S., Siegel F. L. Calmodulin N-methyltransferase. Partial purification and characterization. J Biol Chem. 1986 May 25;261(15):7060–7069. [PubMed] [Google Scholar]
- Schleicher M., Lukas T. J., Watterson D. M. Further Characterization of Calmodulin from the Monocotyledon Barley (Hordeum vulgare). Plant Physiol. 1983 Nov;73(3):666–670. doi: 10.1104/pp.73.3.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T., Imai Y., Yamamoto M. Substitution at position 116 of Schizosaccharomyces pombe calmodulin decreases its stability under nitrogen starvation and results in a sporulation-deficient phenotype. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9737–9741. doi: 10.1073/pnas.86.24.9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T., Yamamoto M. Analysis and in vivo disruption of the gene coding for calmodulin in Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3580–3584. doi: 10.1073/pnas.84.11.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Töpfer R., Matzeit V., Gronenborn B., Schell J., Steinbiss H. H. A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Res. 1987 Jul 24;15(14):5890–5890. doi: 10.1093/nar/15.14.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson D. M., Iverson D. B., Van Eldik L. J. Spinach calmodulin: isolation, characterization, and comparison with vertebrate calmodulins. Biochemistry. 1980 Dec 9;19(25):5762–5768. doi: 10.1021/bi00566a015. [DOI] [PubMed] [Google Scholar]






