Abstract
Background
Starch and protein are two major components of polished rice, and the amylose and protein contents affect eating and cooking qualities (ECQs). In the present study, genome-wide association study with high-quality re-sequencing data was performed for 10 ECQs in a panel of 227 non-glutinous rice accessions and four derived panels.
Results
Population structure accounted for high phenotypic variation in three routine panels and had minor effects on subspecies-based panels. Using the mixed linear model method based on the P + K model, we detected 29, 24, 16, 17, and 29 loci that were significant for ECQ parameters in each of the five panels. Some of these loci were close to starch synthesis-related genes. Two quantitative trait loci (QTLs) (chr.9: 15417525 ~ 15474876; 17538294 ~ 18443016) for several starch paste viscosity properties detected in four panels were close to the isoamylase 3 gene, one QTL (chr.1: 30627943 ~ 31668474) for consistency detected in three panels was close to the starch synthase IV-1 gene. The QTL (chr.7: 1118122 ~ 1967247) for breakdown (BD), detected in the whole panel and japonica panel, and one QTL (chr.7: 25312126 ~ 26540950) for BD and setback (SB), detected in the whole panel and indica panel, may be specific gene alleles in japonica or indica panels. One previously detected QTL (chr.11: 22240707 ~ 22563596) for protein content and one new QTL (chr.5: 7756614 ~ 8042699) for many ECQ traits detected in more than two panels, may represent valuable targets for future cloning of the underlying genes.
Conclusions
This study detected minor-effect QTLs affecting ECQs, and may increase our understanding of the genetic differences regulating the formation of ECQ between indica and japonica varieties.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-3000-z) contains supplementary material, which is available to authorized users.
Keywords: Rice, Eating and cooking quality, Subpopulations, GWAS
Background
Improving grain quality and grain yield are among the most important objectives in rice breeding programs. Apparent amylose content (AAC) is known to affect the eating and cooking quality (ECQ) of rice [1]. However, the rapid viscosity analyser (RVA) profile can distinguish eating quality rice varieties with similar AACs by evaluating the breakdown (BD), setback (SB), and consistency (CS) viscosities [2]. In general, rice varieties with good eating quality have a BD of more than 100 rapid visco units (RVU) and an SB of less than 25 RVU [3]. Therefore, starch paste viscosities play an essential role in estimating the eating, cooking, and processing qualities of rice, and have received more and more attention in rice breeding programs [4, 5].
The Wx gene encodes granule bound starch synthase I (GBSSI), which is responsible for amylose synthesis. Starch paste viscosity properties are predominantly controlled by the Wx gene, as well as minor-effect genes with various additive effects [6–8]. The starch paste viscosity of glutinous rice differs from that of non-glutinous rice due to its lack of amylose [1]. Since GBSSI is inactive in glutinous varieties, the starch paste properties are thought to be controlled by other genes. The pullulanase (PUL) gene plays an important role in the control of peak viscosity (PV), hot paste viscosity (HPV), cold paste viscosity (CPV), BD, peak time (PeT), and pasting temperature (PTemp) in glutinous rice [4]. Soluble starch synthase IIa (SSIIa) is responsible for HPV, CPV, BD, SB, PeT, and PTemp, as well as the thermal properties of glutinous rice [9]. Polymorphisms in both SBE1 and SBE3 loci account for 70 % of the observed variations in HPV and CPV, and account for 40 % in both PV and CS in waxy rice [10]. Many populations derived from bi-parental crosses with similar AACs were developed to identify QTLs for starch paste viscosities and eliminate the Wx effects in non-glutinous accessions [7, 11]. A QTL cluster close to SSIIa that had major effects on the alkali spreading value (ASV), pasting time, and minor effects on gel consistency, AAC, PV, CPV, BD, and SB was identified [7]. The branching enzyme IIb (BEIIb or SBE3) gene cluster is responsible for PV, HPV, CPV, BD, CS, and gel consistency [7] . Another method to avoid the effects of the Wx gene on starch paste viscosity was performed to detect more QTLs within sub-populations of the same Wx background [8, 12].
Association mapping is a powerful method to connect structural genomics and phenomics in plants, provided that information on population structure and linkage disequilibrium (LD) are available [13]. Association mapping, especially candidate-gene association mapping, is commonly used to detect QTLs for starch qualities [4, 9, 14, 15]. Tian et al. [14] found that the starch synthesis-related genes (SSRGs) cooperated with each other to form a regulatory network that controls the ECQs and defines correlations among AAC, ASV, and gel consistency. Yan et al. [4] genotyped waxy rice with 17 SSRGs using 43 gene-tagged molecular markers, and found that 10 SSRGs are involved in controlling the RVA profile parameters. However, candidate-gene association mapping will miss other unknown loci, and genome-wide association studies (GWAS) are comprehensive approaches to systemically search the genome for causal genetic variations [16]. The distribution of functional alleles is strongly correlated with population structure since LD can be caused by an admixture of subpopulations, which leads to false-positive results, particularly in populations with a small number of samples [17]. An ideal sample with a subtle population structure and familial relatedness is the best panel to control for false-negatives [17]. Moreover, subdividing the diversity panel to analyze subpopulations independently using the mixed model provides a reasonable solution to lower false-positive or -negative rates [18]. For example, Huang et al. [19] chose only the indica landrace population to conduct GWAS for 14 agronomic traits. Huang et al. [20] collected phenotypic data of flowering time and grain-related traits for a GWAS on the indica or japonica subpopulations and the whole population, respectively.
Two subspecies of rice (Oryza sativa L.), indica and japonica, differ in several morphological and physiological characteristics including grain shape, shattering, apiculus hair length, leaf color, seed dormancy, cold tolerance, phenol, and sensitivity to potassium chlorate [21–23]. Moreover, japonica and indica rice varieties vary in starch physicochemical properties, which strongly influences rice cooking and processing methods for food and industrial applications [24]. The Wxa allele is predominant in non-waxy indica cultivars with high AACs (22–29 %), whereas the Wxb allele, with low AACs (12–19 %), is common to the non-waxy japonica variety [25–27]. The splice donor mutation is prevalent in temperate japonica rice but rare or absent in indica and tropical japonica rice [28]. The indica type SSIIa can convert S-type amylopectin to L-type amylopectin by elongating short chains of DP (degree of polymerization) ≤10 to form chains of DP 13–25 [24]. The japonica S-type amylopectin has enriched short branch chains of DP ≤10 and depleted intermediate chains of 12 ≤DP ≤24 compared with the indica L-type amylopectin, whereas the proportion of cluster interconnecting long chains of DP ≥25 are comparable in the two varieties [29]. Bao et al. [30] classified 56 glutinous rice varieties into two groups according to the gelatinization temperature (GT), corresponding to high GT and low GT, and found that all 15 high GT accessions belonged to indica rice. Despite the two genes (Wx and SSIIa) being well-studied, the exact genetic effects of other SSRGs in shaping grain quality and starch paste viscosities in different rice subpopulations remain unclear [4].
In the present study, we divided the 227 non-glutinous rice accessions into two panels and two ecotype based panels, and explored the genetics of 10 ECQ parameters in different panels and ecotypes based on genome-wide association mapping with high-quality re-sequencing data. These results will increase our understanding of the mechanism of ECQ properties in different ecotypes and how minor-effect QTLs function on ECQs.
Results
Phenotypic variation and contribution of population structure to phenotypic variation
Population structure was evaluated using the ADMIXTURE program, which estimates individual ancestry and admixture proportions assuming that K populations exist based on a maximum-likelihood method [31]. Because rice (Oryza sativa L.) has two subspecies, it was not surprising that indica and japonica subpopulations could be identified. However, the indica subpopulation could be further divided into indica, aus, as well as some admixtures (admix), while the japonica subpopulation could be divided into temperate japonica, tropical japonica, and some admixtures (Additional file 1: Figure S1). Principal component analysis (PCA) plots of the first two components (Fig. 1) clearly differentiated rice accessions into different subpopulations, which corresponded with the population structure results (Fig. 1; Additional file 1: Figure S1). All 10 phenotypic variations in the admixture group showed moderate levels. The aus group had the highest AAC, PeT, SB, and CS, but the lowest PV, HPV, CPV, BD, and PTemp. The indica group had a wide phenotypic variation range with regards to AAC, PV, HPV, CPV, BD, SB, and CS compared to the other groups, and its AAC, PV, CPV, SB, and CS mid-values were higher than those of the japonica group. Temperate japonica had the lowest AAC, SB, CS, and PeT, and the highest PTemp. The phenotypic variation of tropical japonica was between that of indica and temperate japonica. However, there was no significant difference in protein content (PRO) among groups (Fig. 2).
Fig. 1.
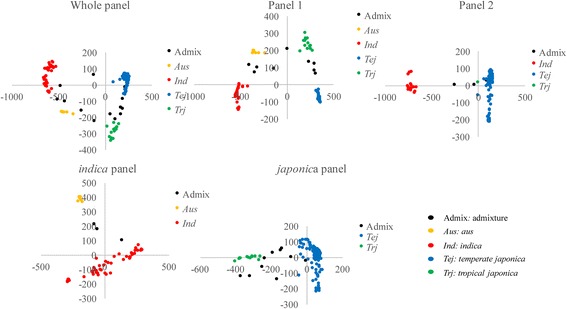
Principal component analysis (first two components) in 5 panels
Fig. 2.
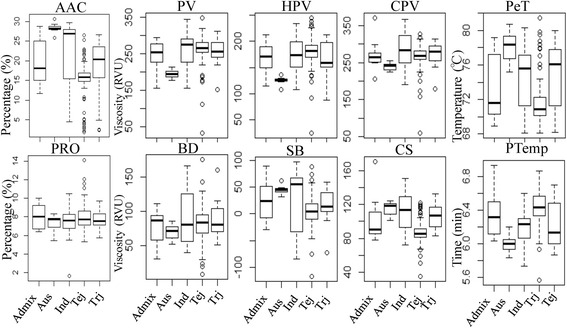
Phenotypic variation among the different ecotype groups
With respect to the whole panel, a wide range in all traits was observed for AAC (10.2 ~ 30.7 %), PRO (1.6 ~ 14.1 %), PV (32.7 ~ 347.8RVU), HPV (24.2 ~ 245.3 RVU), CPV (59.6 ~ 371.5 RVU), BD (8.4 ~ 179.5 RVU), SB (−117.1 ~ 97.2 RVU), CS (28.6 ~ 170.6 RVU), PeT (3.7 ~ 6.9 min), and PTemp (68.1 ~ 81.5 °C) (Table 1). When comparing the indica panel with the japonica panel, the AAC, CPV, SB, CS, and PTemp of the indica panel were significantly higher than those of the japonica panel. The PRO, HPV, and PeT of the indica panel were significantly lower than those of the japonica panel, whereas PV and BD were similar between the two panels (Table 1). Furthermore, the AAC, PRO, SB, CS, and PTemp of Panel 1 were significantly higher than those of Panel 2. Panel 1 had lower PV, HPV, BD, and PeT than Panel 2 (Additional file 1: Table S1).
Table 1.
Phenotypic variation in different panels and ecotype (indica and japonica panels) contribution to phenotypic variation
| AAC | PRO | PV | HPV | CPV | BD | SB | CS | PeT | PTemp | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Means ± SD1 | 18.5 ± 3.8 | 7.8 ± 1.3 | 260.2 ± 37.6 | 176.0 ± 27.0 | 271.6 ± 32.9 | 84.1 ± 27.3 | 11.4 ± 35.5 | 95.1 ± 15.4 | 6.4 ± 0.2 | 72.7 ± 3.0 | |
| whole | Range | 10.2 ~ 30.7 | 1.6 ~ 14.1 | 32.7 ~ 347.8 | 24.2 ~ 245.3 | 59.6 ~ 371.5 | 8.4 ~ 179.5 | −117.1 ~ 97.2 | 28.6 ~ 170.6 | 3.7 ~ 6.9 | 68.1 ~ 81.5 |
| panel | CV2 | 20.7 | 16.0 | 14.5 | 15.4 | 12.1 | 32.4 | 311.0 | 16.2 | 3.1 | 4.1 |
| R square | 0.429 | 0.030 | 0.100 | 0.122 | 0.084 | 0.029 | 0.078 | 0.365 | 0.277 | 0.224 | |
| P value | <.0001 | 0.1547 | 0.0001 | <.0001 | 0.0007 | 0.1689 | 0.0014 | <.0001 | <.0001 | <.0001 | |
| Means ± SD1 | 24.0 ± 5.8a | 7.5 ± 1.4b | 254.1 ± 42.0a | 169.4 ± 30.0b | 281.8 ± 42.9a | 84.8 ± 37.0a | 27.7 ± 51.8a | 112.4 ± 20.1a | 6.2 ± 0.2b | 75.0 ± 3.2a | |
| indica | Range | 12.9 ~ 30.7 | 1.6 ~ 10.5 | 155.6 ~ 344.0 | 107.2 ~ 234.5 | 191.2 ~ 371.5 | 40.2 ~ 166.9 | −84.5 ~ 97.2 | 72.5 ~ 170.6 | 5.7 ~ 6.6 | 68.1 ~ 80.7 |
| panel | CV2 | 24.1 | 18.7 | 16.5 | 17.7 | 15.2 | 43.6 | 187.0 | 17.9 | 2.9 | 4.3 |
| R square | 0.107 | 0.007 | 0.372 | 0.325 | 0.214 | 0.092 | 0.036 | 0.014 | 0.131 | 0.139 | |
| P value | 0.044 | 0.824 | 0.002 | 0.006 | 0.044 | 0.286 | 0.625 | 0.828 | 0.021 | 0.016 | |
| Means ± SD1 | 16.7 ± 2.7b | 7.9 ± 1.2a | 262.6 ± 35.1a | 179.2 ± 26.7a | 267.78 ± 29.2b | 83.4 ± 23.4a | 5.2 ± 26.6b | 88.4 ± 11.9b | 6.4 ± 0.2a | 71.9 ± 2.8b | |
| japonica | Range | 10.2 ~ 26.7 | 5.3 ~ 14.1 | 32.7 ~ 347.8 | 24.2 ~ 245.3 | 59.6 ~ 326.5 | 8.4 ~ 179.5 | −117.1 ~ 87.5 | 28.6 ~ 132.5 | 3.7 ~ 6.9 | 68.1 ~ 81.5 |
| panel | CV2 | 16.0 | 15.2 | 13.4 | 14.9 | 10.9 | 28.1 | 509.6 | 13.5 | 3.3 | 3.9 |
| R square | 0.119 | 0.008 | 0.001 | 0.005 | 0.016 | 0.003 | 0.020 | 0.146 | 0.026 | 0.050 | |
| P value | <.0001 | 0.537 | 0.937 | 0.698 | 0.287 | 0.774 | 0.195 | <.0001 | 0.127 | 0.017 |
1. SD: standard deviation; 2. CV: coefficient of variation; 3: different letters mean significant difference between indica and japonica panels (P < 0.05)
Given that population structure is a main factor affecting GWAS, we evaluated phenotypic variation among different subspecies groups (indica/japonica) in the whole panel (Fig. 2), as well as the population structure contribution for phenotypic variation in each panel (Table 1 and Additional file 1: Table S1).
In the whole panel, population structure explained 42.9 (AAC), 10.0 (PV), 12.2 (HPV), 8.4 (CPV), 7.8 (SB), 36.5 (CS), 27.7 (PeT), and 22.4 % (PTemp) of phenotypic variation (P <0.01) (Table 1). However, population structure affected only two traits in the indica panel (PV and HPV) and two traits in the japonica panel (AAC and CS) at the significance level of P <0.01, and affected 6 traits (AAC, PV, HPV, CPV, PeT, and PTemp) in indica panel and 3 traits (AAC, CS, and PTemp) in japonica panel at significance level of P <0.05 (Table 1). In Panel 1, except for BD, population structure accounted for large phenotypic variation explanation (PVE), ranging from 13.8 % (PTemp) to 57.4 % (AAC) at a significance level of P <0.001. In Panel 2, population structure explained 37.5 (PV), 46.0 (BD), 25.7 (SB), 22.6 (CS), and 25.3 % (PeT) phenotypic variation (P <0.01) (Additional file 1: Table S1).
GWAS of 10 ECQ parameters
There were 29, 24, 16, 17, and 29 loci identified in the whole panel, Panel 1, Panel 2, indica panel, and japonica panel, respectively (Tables 2, 3, and 4, Fig. 3).
Table 2.
Loci detected for eating and cooking qualities in the whole panel
| Chr. | Position | P value | MAFa | R squareb | |
|---|---|---|---|---|---|
| AAC | 6 | 1765761 | 3.87 × 10−18 | 0.288 | 0.087 |
| PRO | 5 | 13312843 | 6.43 × 10−10 | 0.058 | 0.153 |
| 11 | 22562237 | 2.14 × 10−8 | 0.071 | 0.124 | |
| PV | 5 | 7756614 | 3.40 × 10−6 | 0.235 | 0.070 |
| 6 | 1765761 | 3.50 × 10−7 | 0.285 | 0.085 | |
| HPV | 5 | 7944999 | 2.25 × 10−5 | 0.071 | 0.067 |
| 6 | 1750498 | 1.37 × 10−6 | 0.077 | 0.088 | |
| 7 | 3359832 | 9.88 × 10−5 | 0.199 | 0.056 | |
| 9 | 17538294 | 7.44 × 10−5 | 0.071 | 0.058 | |
| CPV | 1 | 31640680 | 1.05 × 10−6 | 0.277 | 0.087 |
| 6 | 1750498 | 6.88 × 10−8 | 0.077 | 0.108 | |
| 9 | 17539188 | 1.53 × 10−6 | 0.071 | 0.084 | |
| BD | 6 | 1765761 | 3.24 × 10−8 | 0.285 | 0.105 |
| 7 | 1967427 | 4.05 × 10−6 | 0.173 | 0.071 | |
| 7 | 26540950 | 3.34 × 10−6 | 0.093 | 0.073 | |
| SB | 4 | 24293919 | 3.44 × 10−7 | 0.166 | 0.069 |
| 6 | 1765761 | 1.69 × 10−2 | 0.285 | 0.140 | |
| 7 | 1967427 | 2.11 × 10−7 | 0.173 | 0.072 | |
| 7 | 25326697 | 5.68 × 10−7 | 0.080 | 0.066 | |
| CS | 1 | 30634461 | 2.42 × 10−6 | 0.261 | 0.041 |
| 6 | 1765761 | 8.64 × 10−8 | 0.285 | 0.054 | |
| 9 | 15474876 | 4.59 × 10−6 | 0.215 | 0.039 | |
| PeT | 1 | 31330790 | 1.39 × 10−7 | 0.265 | 0.087 |
| 2 | 18003109 | 6.84 × 10−8 | 0.268 | 0.091 | |
| 5 | 8042699 | 1.06 × 10−6 | 0.173 | 0.074 | |
| 6 | 21450052 | 1.67 × 10−7 | 0.058 | 0.085 | |
| 8 | 21593355 | 2.64 × 10−7 | 0.270 | 0.082 | |
| PTemp | 6 | 7002095 | 1.99 × 10−9 | 0.219 | 0.065 |
| 12 | 17937512 | 1.59 × 10−6 | 0.157 | 0.041 |
a: MAF: minor allele frequency; b: R square was calculated by R square of model with SNP minus R square of model without SNP
Table 3.
Loci detected for eating and cooking qualities in Panel 1 and Panel 2
| Trait | Panel1 | Panel 2 | ||||||
|---|---|---|---|---|---|---|---|---|
| Chr. | Position | P value | MAFa | R squareb | P value | MAFa | R squareb | |
| AAC | 6 | 1765761 | 5.29 × 10−11 | 0.436 | 0.152 | |||
| 6 | 1585864 | 9.21 × 10−8 | 0.052 | 0.231 | ||||
| 9 | 19310549 | 2.32 × 10−7 | 0.143 | 0.214 | ||||
| PRO | 5 | 12559930 | 1.57 × 10−7 | 0.110 | 0.242 | |||
| 11 | 22563596 | 2.47 × 10−6 | 0.092 | 0.190 | ||||
| PV | 1 | 5102566 | 6.31 × 10−5 | 0.155 | 0.118 | |||
| 2 | 29640874 | 7.77 × 10−5 | 0.136 | 0.118 | ||||
| 5 | 7944999 | 4.15 × 10−5 | 0.086 | 0.125 | ||||
| 6 | 1666386 | 2.86 × 10−5 | 0.177 | 0.133 | ||||
| 7 | 16503569 | 1.62 × 10−5 | 0.194 | 0.139 | ||||
| HPV | 1 | 3541593 | 1.72 × 10−5 | 0.164 | 0.169 | |||
| 1 | 27287504 | 3.51 × 10−5 | 0.082 | 0.127 | ||||
| 2 | 29640874 | 5.22 × 10−5 | 0.136 | 0.121 | ||||
| 5 | 7944999 | 2.88 × 10−5 | 0.086 | 0.159 | ||||
| 6 | 1750498 | 5.19 × 10−6 | 0.141 | 0.157 | ||||
| 7 | 16506178 | 2.63 × 10−5 | 0.181 | 0.161 | ||||
| 11 | 27792076 | 6.92 × 10−6 | 0.147 | 0.186 | ||||
| CPV | 1 | 3541593 | 4.78 × 10−6 | 0.164 | 0.178 | |||
| 4 | 27772775 | 1.59 × 10−5 | 0.336 | 0.130 | ||||
| 6 | 1750498 | 3.78 × 10−5 | 0.141 | 0.118 | ||||
| 7 | 16506178 | 4.68 × 10−6 | 0.181 | 0.178 | ||||
| 9 | 15417525 | 2.31 × 10−5 | 0.282 | 0.125 | ||||
| 11 | 192690 | 4.16 × 10−5 | 0.136 | 0.116 | ||||
| BD | 2 | 8596549 | 5.57 × 10−5 | 0.138 | 0.102 | |||
| 6 | 1765761 | 1.16 × 10−5 | 0.441 | 0.183 | ||||
| 6 | 1985864 | 9.85 × 10−5 | 0.052 | 0.095 | ||||
| SB | 4 | 24056499 | 1.09 × 10−6 | 0.125 | 0.165 | |||
| 6 | 1765761 | 2.39 × 10−8 | 0.441 | 0.261 | ||||
| CS | 1 | 30627943 | 1.98 × 10−5 | 0.214 | 0.111 | |||
| 6 | 1765761 | 2.41 × 10−6 | 0.441 | 0.138 | ||||
| 9 | 15474876 | 2.19 × 10−5 | 0.277 | 0.110 | ||||
| 11 | 24255295 | 1.96 × 10−5 | 0.118 | 0.111 | ||||
| PeT | 2 | 24259382 | 7.68 × 10−5 | 0.086 | 0.115 | |||
| 2 | 27305077 | 1.11 × 10-5 | 0.055 | 0.131 | ||||
| 3 | 33893179 | 1.12 × 10−5 | 0.068 | 0.131 | ||||
| 6 | 21443081 | 8.35 × 10−6 | 0.059 | 0.135 | ||||
| 10 | 23087051 | 9.69 × 10−6 | 0.132 | 0.133 | ||||
| PTemp | 6 | 6757255 | 1.54 × 10−8 | 0.491 | 0.217 | |||
| 6 | 21712093 | 1.11 × 10−6 | 0.118 | 0.155 | ||||
| 8 | 9459681 | 4.28 × 10−5 | 0.233 | 0.141 | ||||
a: MAF: minor allele frequency; b: R square was calculated by R square of model with SNP minus R square of model without SNP
Table 4.
Loci detected for eating and cooking qualities in indica and japonica panels
| Trait | indica panel | japonica panel | ||||||
|---|---|---|---|---|---|---|---|---|
| Chr. | Position | P value | MAFa | R squareb | P value | MAFa | R squareb | |
| AAC | 6 | 1529682 | 2.15 × 10−7 | 0.331 | 0.079 | |||
| 6 | 1765761 | 1.69 × 10−13 | 0.146 | 0.279 | ||||
| PRO | 2 | 3118861 | 1.87 × 10−5 | 0.098 | 0.076 | |||
| 4 | 6536977 | 1.37 × 10−5 | 0.055 | 0.079 | ||||
| 5 | 6814906 | 3.10 × 10−5 | 0.122 | 0.072 | ||||
| 6 | 2576841 | 2.23 × 10−5 | 0.458 | - | ||||
| 7 | 1631971 | 9.32 × 10−7 | 0.073 | 0.102 | ||||
| 8 | 2755270 | 4.39 × 10−5 | 0.280 | - | ||||
| 10 | 9032827 | 4.38 × 10−6 | 0.207 | 0.089 | ||||
| 11 | 22240707 | 3.60 × 10−5 | 0.093 | - | ||||
| 11 | 22563596 | 2.89 × 10−5 | 0.064 | 0.073 | ||||
| PV | 6 | 1665483 | 2.96 × 10−5 | 0.322 | 0.127 | |||
| 6 | 1765761 | 6.42 × 10−6 | 0.145 | 0.110 | ||||
| 8 | 2885700 | 6.78 × 10−5 | 0.087 | 0.085 | ||||
| 11 | 5061851 | 1.46 × 10−5 | 0.084 | 0.101 | ||||
| HPV | 3 | 26688241 | 7.17 × 10−5 | 0.449 | 0.625 | |||
| 5 | 7944999 | 6.99 × 10−5 | 0.096 | 0.089 | ||||
| 6 | 1750498 | 1.50 × 10−6 | 0.280 | 0.387 | ||||
| 9 | 18431810 | 6.09 × 10−5 | 0.142 | 0.090 | ||||
| 9 | 18729438 | 6.47 × 10−5 | 0.203 | 0.250 | ||||
| 11 | 3161548 | 2.46 × 10−5 | 0.078 | 0.101 | ||||
| 11 | 27792076 | 3.84 × 10−5 | 0.229 | 0.096 | ||||
| CPV | 6 | 1750498 | 1.24 × 10−6 | 0.280 | 0.402 | |||
| 9 | 18443016 | 1.28 × 10−6 | 0.054 | 0.133 | ||||
| BD | 6 | 1721792 | 9.99 × 10−5 | 0.339 | 0.097 | |||
| 7 | 1118122 | 1.51 × 10−5 | 0.054 | 0.111 | ||||
| 7 | 26540950 | 6.68 × 10−5 | 0.339 | 0.103 | ||||
| 11 | 3735389 | 4.38 × 10−5 | 0.398 | 0.109 | ||||
| 11 | 20805844 | 1.84 × 10-5 | 0.160 | 0.109 | ||||
| SB | 1 | 31774927 | 4.37 × 10−5 | 0.205 | 0.092 | |||
| 3 | 8987316 | 2.53 × 10−5 | 0.075 | 0.098 | ||||
| 6 | 1617418 | 6.01 × 10−6 | 0.339 | 0.133 | ||||
| 6 | 1765761 | 1.82 × 10−8 | 0.145 | 0.183 | ||||
| 7 | 25312126 | 9.18 × 10−5 | 0.297 | 0.095 | ||||
| 11 | 3781133 | 9.23 × 10−5 | 0.356 | 0.095 | ||||
| CS | 1 | 31668474 | 5.17 × 10−6 | 0.202 | 0.085 | |||
| 6 | 1759451 | 6.46 × 10−7 | 0.120 | 0.102 | ||||
| PeT | 1 | 23829740 | 5.22 × 10−5 | 0.322 | 0.323 | |||
| 2 | 4054094 | 1.84 × 10−6 | 0.111 | 0.093 | ||||
| 3 | 33737410 | 3.27 × 10−7 | 0.060 | 0.107 | ||||
| 5 | 7996572 | 1.96 × 10−5 | 0.057 | 0.073 | ||||
| PTemp | 1 | 2609442 | 2.80 × 10−6 | 0.084 | 0.055 | |||
| 4 | 33873772 | 5.72 × 10−6 | 0.051 | 0.052 | ||||
| 6 | 6722646 | 1.29 × 10−7 | 0.356 | 0.295 | ||||
| 6 | 6747298 | 3.71 × 10−6 | 0.482 | 0.054 | ||||
| 9 | 8085354 | 1.09 × 10−5 | 0.090 | 0.049 | ||||
a: MAF: minor allele frequency; b: R square was calculated by R square of model with SNP minus R square of model without SNP
Fig. 3.
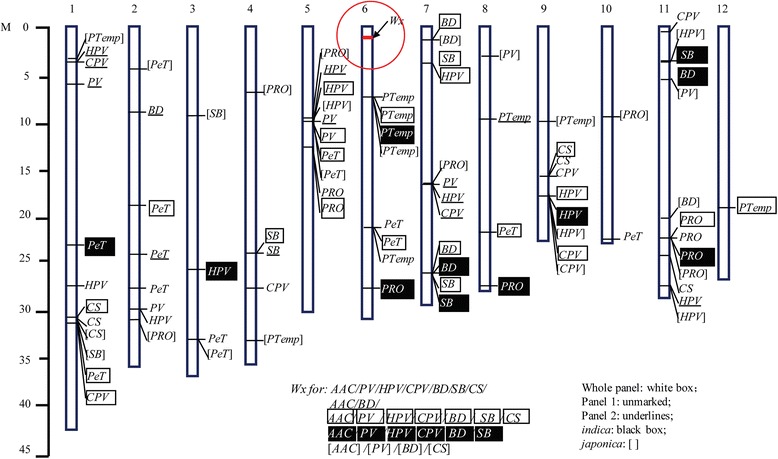
Distribution of the significant loci for ECQs in five panels (The locus in the red circle indicates the Wx gene for AAC, PV, HPV, CPV, BD, SB, and CS in whole panel and Panel 1, for AAC and BD in panel 2; for AAC, PV, HPV, CPV, BD, and SB in indica panel, for AAC, PV, BD, and CS in japonica panel. Loci with white box, unmarked, underlines, black box, and brackets were detected in the whole panel, Panel 1, Panel 2, indica and japonica panels, respectively.)
Whole panel
AAC and PRO
Only one locus (chr.6: 1765761) for AAC was detected on chromosome 6. Two loci (chr.5: 13312843; chr.11: 22562237) for PRO were detected on chromosomes 5 and 11, accounting for 15.3 and 12.4 % PVE, respectively (Table 2).
PV, HPV, CPV, BD, SB, and CS
Two, four, three, three, four, and three loci were detected for PV, HPV, CPV, BD, SB, and CS, respectively. The major locus (chr.6: 1765761) was detected for PV, BD, SB, and CS, and the nearby locus (chr.6: 1750498) was the most significant site that linked with HPV and CPV. One locus (chr.5: 7756614) for PV was close to the locus (chr.5: 7944999) for HPV. One locus (chr.1: 31640680) for CPV was close to the locus (chr.1: 30634461) for CS. One locus (chr.9: 17538294) for HPV was close to one locus (chr.9: 17539188) for CPV. One locus (chr.7: 1967427) was detected for both BD and SB. One (chr.7: 26540950) for BD and one (chr.7: 25326697) for SB were close to each other (Table 2).
PeT and PTemp
A total of five loci located on chromosomes 1, 2, 5, 6, and 8, were detected for PeT. Two loci (chr.6: 7002095; chr.12: 17937512) for PTemp on chromosomes 6 and 12, accounted for 6.5 and 4.1 % of PVE, respectively (Table 2).
Panel 1 and Panel 2
AAC and PRO
One and two loci for AAC were detected in Panel 1 and Panel 2, respectively. The locus (chr.6: 1765761) and locus (chr.6: 1585864) were major QTLs for AAC in Panel1 and Panel 2, respectively. Another locus (chr.9:19310549) for AAC was detected only in Panel 2. Two loci for PRO, located on chromosomes 5 and 11, were identified in Panel 1. No locus was detected for PRO in Panel 2 (Table 3).
PV, HPV, CPV, BD, SB, and CS
In Panel 1, GWAS detected 2, 3, 4, 1, 1, and 4 loci for PV, HPV, CPV, BD, SB, and CS, respectively. Similar to the whole panel GWAS results, the locus (chr.6: 1765761) had major effects on BD, and SB, and CS, the locus (chr.6: 1750498) was detected for HPV and CPV, whereas the locus (chr.6: 1666386) was the most significant site for PV. The locus (chr.2: 29640874) for both PV and HPV accounted for 11.8 and 12.1 % PVE, respectively. One locus (chr.9: 15417525) for CPV and one (chr.9: 15474876) for CS was close to each other (Table 3).
In Panel 2, we detected 3, 4, 2, 2, and 1 locus for PV, HPV, CPV, BD, and SB, respectively. No locus was detected for CS. One locus (chr.5: 7944999) was significant for both PV and HPV. One locus (chr.1: 3541593) was detected for both HPV and CPV, while the locus (chr.7: 16503569) for PV was close to one locus (chr.7: 16506178) for both HPV and CPV on chromosome 7 (Table 3).
PeT and PTemp
In Panel 1, we detected four loci for PeT on chromosomes 2, 3, 6, and 10, while two loci (chr.6: 6757255 and chr.6: 21712093) for PTemp were detected on chromosome 6, accounting for 21.7 and 15.5 % PVE, respectively (Table 3).
In Panel 2, only one locus (chr.2: 24259382) for PeT and one locus (chr.8: 9459681) for PTemp were detected on chromosomes 2 and 8, respectively (Table 3).
indica and japonica panels
AAC and PRO
One locus (chr.6: 1529682) and one (chr.6: 1765761) for AAC were detected in indica and japonica panels, respectively. Three and six loci for PRO were detected in indica and japonica panels, respectively. One locus (chr.11: 22240707) for PRO in indica panel was close to the locus (chr.11: 22563596) detected in japonica panel (Table 4).
PV, HPV, CPV, BD, SB, and CS
In indica panel, one locus for PV, three for HPV, one for CPV, three for BD, and three for SB were detected and no locus was detected for CS. The locus (chr.6: 1750498) was a major QTL for both HPV and CPV, whereas loci for PV (chr.6: 1665483), BD (chr.6: 1721792), and SB (chr.6: 1617418) were close to each other in the indica panel. One locus (chr.9: 18729438) for HPV was close to the locus (chr.9: 18443016) for CPV in indica panel. One locus (chr.7: 26540950) for BD was close to the locus (chr.7: 25312126) for SB. The QTL (chr.11: 3735389) for BD was close to one QTL (chr.11: 3781133) for SB in the indica panel (Table 4).
Three loci for PV, four for HPV, one for CPV, two for BD, three for SB, and two for CS were identified in the japonica panel. The QTL (chr.6: 1765761) for PV and SB was close to the locus (chr.6: 1759451) for CS. One locus (chr.9: 18431810) for HPV was close to the locus (chr.9: 18443016) for CPV, one QTL (chr.1: 31774927) for SB was close to one QTL (chr.1: 31668474) for CS in japonica panel (Table 4).
PeT and PTemp
One locus (chr.1: 23829740) for PeT and one locus (chr.6: 6722646) for PTemp were detected in the indica panel, accounting for 32.3 and 29.5 % PVE, respectively.
Three loci for PeT and four for PTemp were detected in the japonica panel. The locus (chr.5: 7996572) for PeT were close to the locus detected in the whole panel. The locus (chr.6: 6747298) for PTemp was close to the loci in the whole panel, Panel 1, and the indica panel (Table 4).
Discussion
Effect of population structure on GWAS
Given that linkage disequilibrium can be caused by the admixture of subpopulations and may result in false-positives if not correctly controlled in the statistical analysis, estimation of population structure must be included in the association analysis [17]. In this study, to determine whether different networks of alleles were associated with ECQ trait variation in the different panels, we performed GWAS on each of the four panels independently and in the panel as a whole. The present study allowed us to compare GWAS results derived from different panels.
The effect of population structure on phenotypic variation was estimated based on analysis of variance (ANOVA) (Table 1 and Additional file 1: Table S1). For the whole panel, eight of 10 traits were strongly affected by population structure. For Panel 1 and 2, nine and five traits were strongly affected by population structure (P <0.01), respectively, indicating that Panel 2 (breeding lines) experienced artificial selection, which was less structured than Panel 1. However, only two traits were significantly affected by population structure in the indica and japonica panels at significance level of P <0.01, suggesting that association mapping in the subpopulations, e.g. indica or japonica subpopulations, with simple population structures can lower false-positives. indica and japonica varietal groups should be properly treated for association analysis, which may explain why transgressive offspring occur in divergent subpopulation crosses [18] .
QTL comparison among different panels
For convenience, we regarded those SNPs that were detected in different panels with the physical distance less than 2.0 Mb as the same QTL, and these SNPs were regarded as alleles.
Similar to previous studies, the Wx gene is responsible for AAC in these five panels. The single nucleotide polymorphism (SNP) (chr.6: 1765761) for AAC is the Wxa/Wxb allele (a SNP at the splice site of intron 1), which was present in the whole panel, Panel 1, and japonica panel. The Wx gene was also the major QTL for AAC in the other two panels, but the Wxa/Wxb allele was not the most significant association site, suggested that different traits maybe preferred linked with different SNPs. Besides the Wx gene, one minor effect locus (chr.9: 19310549) for AAC was detected in Panel 2 (consisted of breeding lines); this locus was close to the QTL flanked by BPH and RM160 [32] (Fig. 3).
One QTL (chr.5: 12559930 ~ 13312843) for PRO detected in the whole panel and Panel 1 was close to the protein content QTL qPRO5, which was detected by Xu et al. [8] in the M201-Wx subpopulation. The other QTL (chr.11: 22562237 ~ 22563596) for PRO detected in the whole, Panel 2, indica, and japonica panels was close to pro11, which is flanked by RM209 and RM229 [33].
For starch paste viscosity parameters, a specific SNP (chr.6: 1765761), recognized as the Wxa/Wxb allele, was responsible for 4 (PV, BD, SB, and CS), 3 (BD, SB, and CS), and 2 (PV and SB) in the whole panel, Panel 1, and japonica panel, respectively. There were also some SNPs close to Wx gene that significantly associated with RVA parameters. However, only one Wx gene linked SNP was detected for BD in Panel 2, that may due to the reason that most Korean breeding lines were low-AAC varieties with less diversity. The GWAS results in our study may increase our understanding of the genetic control of starch properties in South Korea japonica rice varieties with low AACs and the same Wx allele. Besides the Wx gene, 12 QTLs were identified in two panels, 3 QTLs were detected in 3 panels, and 2 QTLs were detected in 4 panels (Fig. 3). The QTL (chr.5: 7756614 ~ 8042699) was detected for PV in the whole panel and Panel 2, for HPV in the whole panel, Panel 2 and japonica panel, and for PeT in the whole panel and japonica panel. This locus has not been detected in previous studies, and is a potentially novel locus specifically present in japonica rice varieties. The QTL (chr.9: 17538294 ~ 18443016) for HPV in the whole panel, indica, and japonica panels, and for CPV in the whole panel and japonica panel, was close to qPV9 for PV, HPV, and CPV [8], which is close to the isoamylase 3 gene (Os09g0469400, chr.9: 17851431 ~ 17868668). The QTL (chr.7: 1118122 ~ 1967427) for BD in the whole panel and japonica panel and SB in the whole panel, was close to qSB7 [7]. The QTL (chr.7: 2532697 ~ 26540950) for BD and SB in the whole panel and indica panel was close to the QTL (linked with RM6420) for PV, HPV, CPV, and SB [34], and close to the QTL for SB and CS [8]. The QTL (chr.4: 24056499 ~ 24293919) for SB, detected in the whole panel and Panel 2, was close to the QTL for FV (final viscosity, CPV), PKT, SBV, and TV (HPV), and is linked with RM3785 [35]. The QTL (chr.1: 30627943 ~ 31774927) for CS in the whole panel, Panel 1, and japonica panel, and three loci for SB (japonica panel), PeT (whole panel), and CPV (whole panel) was 0.21 Mb from the SSIV-1 gene (Os01g0720600, chr.1:30032428 ~ 30041425), indicating that SSIV-1 may influence starch qualities through affecting CPV, SB, CS, and PeT. The QTL (chr.9: 15417525 ~ 15474876) for CS in the whole panel and Panel 1, and for CPV in Panel 1, was close to qCS9 described by Yan et al. [7]. The QTL (chr.3: 33737410 ~ 33893179) for PeT was detected in Panel 1 and the japonica panel, and was close to Btemp (peak temperature) flanked by RM203 and RM422 [15]. The QTL (chr.6: 6722646 ~ 7002095) for PTemp, detected in the whole panel, Panel 1, indica, and japonica panels, was close to the ALK gene (Os06g0229800, chr.6: 6748398 ~ 6753302) [36], which encodes starch synthase IIa (SSIIa).
QTLs detected in specific panels
Panel 1
The locus (chr.1: 27287504) for HPV, detected only in the Panel 1, was about 1.9 Mb away from the ADP-glucose pyrophosphorylase large subunit 2 (AGPL2) gene (Os01g0633100, chr.1:25353805 ~ 25361883). The locus (chr.2: 29640874) for PV and CPV was about 1.6 Mb from the starch synthase II-2 (SSII-2 or SSIIb) gene (Os02g0744700, chr.2:31233292 ~ 31238210). The locus (chr.11: 24255295) for CS was close to qSB11b (Yan et al. 2014). The locus (chr.4: 27772775) for CPV was close to the QTL for PV and FV, which is linked with RM255 [35].
Panel 2
The QTL (chr.7: 16503569 ~ 16506178) for PV, HPV, and CPV was close to qHPV7 and qCPV7, which are linked with RM418 [37]. The locus (chr.1:3541593) for HPV and CPV, and locus (chr.1: 5102566) for PV were close to the QTL for fa (DP <12) and HPV, which are linked with RM7278 [35]. The locus (chr.2: 8596549) for BD was close to qBD2 [7]. The locus (chr.2: 24259382) for PeT was close to the qBTemp2 for peak temperature [7].
indica panel
The locus (chr.3: 26688241) for HPV was close to the qHPV3.2 [8].
japonica panel
The locus (chr.4: 6536977) for PRO was close to the qPC-4 [38]. The locus (chr.8: 2885700) for PV was close to the qPV8 [39] and qPKV8 [7]. The QTL (chr.2: 4054094) for PeT was close to the qPeT2 [34].
Conclusions
Overall, we investigated the genetics of 10 rice ECQ parameters by GWAS mapping in the whole panel and four derived panels. GWAS within subspecies panels (indica/japonica) cannot only lower false-positives influenced by population structure, but can also increase our understanding of ECQ differences between indica and japonica subspecies. Comparison of GWAS results indicated that some loci were common in different panels, while some were in specific panels. Loci detected in more than one panel were potentially reliable QTLs for ECQs, and these QTLs should be properly used in molecular breeding to improve rice ECQs, particularly in the Korean japonica rice with similar AACs and the same Wx allele.
Methods
Materials
A total of 227 non-glutinous rice accessions from 295 rice accessions (Additional file 2: Table S2) and their whole genome re-sequencing data [40] were used in this study. Here, we defined the 227 accessions as a whole panel for GWAS, among which the core collection of 110 diverse rice accessions was defined as Panel 1, which comprises 57 Korean accessions and 53 rice accessions collected form 27 countries worldwide. The remaining 117 accessions were considered as Panel 2, which consists of breeding lines, one USDA rice accession, three Japan breeding lines, and 113 Korean breeding lines released between 1967 and 2011. Furthermore, all 227 accessions were divided into the indica panel (59) and japonica panel (168), since indica/japonica species contribute the most to the population structure. The whole panel was planted in 2014 during the growing season from early May to September at Kongju National University farm, Yeasan Gun, South Korea. A randomized block design with two replications for each line was used in field experiments.
After being air-dried and stored at room temperature for 3 months, all rice samples were de-hulled to brown rice, then milled into white rice, afterwards ground and passed through a 100-mesh sieve to obtain milled-grain flour. All samples were stored in cooling rooms at 4 °C until use.
Phenotypic data evaluation
AAC and protein content
Determination of AAC was performed using the iodine staining method [41]. The absorbance of the solution was measured at 620 nm against the blank solution with a spectrophotometer. AAC was calculated using a standard curve made from rice samples with known AACs. PRO was measured using the Kjeldahl method with a Kjeltec-Foss 70 2400 Auto analyzer using 5.95 as the nitrogen-to-protein conversion factor.
RVA profile parameters
The RVA profile of rice flour was determined using a Rapid Visco Analyser (RVA-3, Newport Scientific, Warrie wood, Australia). According to the method of AACC61-02, 3.0 g of rice flour (12 % m. b.) was placed in an aluminum canister, after which 25.0 g of distilled water was added. The heating profile was set as follows: (1) the temperature was held at 50 °C for 1 min; (2) the temperature was linearly ramped up to 95 °C until 4.8 min; (3) the temperature was held at 95 °C until 7.3 min; (4) the temperature was linearly ramped down to 50 °C at 11.1 min; and (5) held at 50 °C until 12.5 min.
Four primary parameters were obtained from the pasting curve: PV, HPV, CPV, and PeT. Three secondary parameters were calculated from the primary parameters: BD (PV minus HPV), SB (CPV minus PV), and CS (CPV minus HPV). The viscosity parameters were measured in RVU. PTemp was determined according to the method proposed by Bao [42].
Statistical analyses of phenotypic data
Analysis of variance (ANOVA) was performed on the different panels to determine the phenotypic variation and population structure influence on phenotypic variation using the general linear model procedure (PROC GLM) by the SAS program (version 9.3, SAS Institute Inc., Cary, NC). In the model, population structure was set as the fixed independent variable, and 10 phenotypic data points were set as the dependent variables.
Population structure evaluation and principal component analysis
The genome re-sequencing data of the 227 accessions were collected from the 295 rice whole genome re-sequencing data, which were previously obtained using HiSeq 2000 and HiSeq 2500 [40, 43]. After removing SNPs with a minimum frequency <0.05 by filter alignment using TASSEL 5.0, there were 2,071,925, 1,229,263, 1,563,570, 1,178,226, and 574,169 high-quality SNPs remaining for Panel 1, Panel 2, the whole panel, the indica panel, and the japonica panel, respectively. Population structure was evaluated by the ADMIXTURE program, which estimates individual ancestry and admixture proportions assuming that K populations exist based on a maximum-likelihood method [31]. We ran the program with the script:
admixture --cv genotype.bed k
k ranged from 1 to 5. Principal component analysis (PCA) of each panel was calculated by TASSEL 5.0 with default parameters by using high-quality SNPs.
Genome-wide association mapping of five panels
To control for false-positives, the P (PCA) + K model, based on a mixed linear model (MLM), was used for genome-wide association mapping for the five panels by GAPIT software [44]. Here the principal components (PCs) were used to control for population structure in GWAS. We used the default parameters for running the GAPIT software: GAPIT < − myGAPIT(Y = myY, G = myG, PCA.total = 3, Model.selection = TRUE). myY is the phenotype matrix and myG is genotype matrix. SNP sites with the lowest P value in the peak region (P <10−4) were considered significant SNPs (or loci) for phenotypic variation. Otherwise, sites with regions having only one SNP significant were considered false-positives. Loci with a physical distance less than 2.0 Mb were considered the same locus.
Abbreviations
AAC, apparent amylose content; BD, breakdown; CPV, cold paste viscosity; CS, consistency; ECQ, eating and cooking quality; GWAS, genome-wide association study; HPV, hot paste viscosity; PeT, peak time; PRO, protein content; PTemp, pasting temperature; PV, peak viscosity; PVE, phenotypic variation explanation; QTL, quantitative trait locus; RVA, rapid viscosity analyser; RVU, rapid visco units; SB, setback; SNP, single nucleotide polymorphism; SSRGs, starch synthesis related genes.
Acknowledgements
We thank Dr. Kyu-Won Kim for his technical supports on data analysis.
Funding
This work was carried out with the support of “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01116101)” Rural Development Administration, Republic of Korea. This research was supported by Bio-industry Technology Development Program, Ministry of Agriculture, Food and Rural Affairs (115078–2).
Availability of data and material
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable requests. Additional file 2: Table S2 was added to show the source of the rice accessions used in this study, and the phylogenetic data was uploaded to the Tree base (http://purl.org/phylo/treebase/phylows/study/TB2:S19653?x-access-code=fe69ee8b3f634b20bc4224523aa87639&format=html).
Authors’ contributions
JB and YP conceived and designed the study; FX measured phenotype data; QH performed data analysis; FX, JB and YP drafted the manuscript. All authors critically reviewed and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable
Ethics approval and consent to participate
Not applicable
Additional files
Table S1. Phenotypic variation in different panels and contribution of population structure to phenotypic variation. Figure S1 Population structure of the whole panel. (DOCX 6297 kb)
Table S2. The sources of the rice varieties used in this study. (DOC 32 kb)
References
- 1.Bao JS. Toward understanding the genetic and molecular bases of the eating and cooking qualities of rice. Cereal Food World. 2012;57(4):148–156. doi: 10.1094/CFW-57-4-0148. [DOI] [Google Scholar]
- 2.Bao JS. Progress in studies on inheritance and improvement of rice starch quality. Mol Plant Breed. 2007;5(6):1–20. [Google Scholar]
- 3.Shu Q, Wu D, Xia Y, Gao M. Relationship between RVA profile character and eating quality in Oryza sativa L. Scientia Agricultura Sinica. 1998;31(3):25–29. [Google Scholar]
- 4.Yan CJ, Tian ZX, Fang YW, Yang YC, Li JA, Zeng SY, Gu SL, Xu CW, Tang SZ, Gu MH. Genetic analysis of starch paste viscosity parameters in glutinous rice (Oryza sativa L.) Theor Appl Genet. 2011;122(1):63–76. doi: 10.1007/s00122-010-1423-5. [DOI] [PubMed] [Google Scholar]
- 5.He Y, Han Y, Jiang L, Xu C, Lu J, Xu M. Functional analysis of starch-synthesis genes in determining rice eating and cooking qualities. Mol Breeding. 2006;18(4):277–290. doi: 10.1007/s11032-006-5505-7. [DOI] [Google Scholar]
- 6.Bao JS, Zheng XW, Xia YW, He P, Shu QY, Lu X, Chen Y, Zhu LH. QTL mapping for the paste viscosity characteristics in rice (Oryza sativa L.) Theor Appl Genet. 2000;100(2):280–284. doi: 10.1007/s001220050037. [DOI] [Google Scholar]
- 7.Yan B, Tondi Yacouba N, Chen J, Wang Y, Gao G, Zhang Q, Liu X, He Y. Analysis of minor quantitative trait loci for eating and cooking quality traits in rice using a recombinant inbred line population derived from two indica cultivars with similar amylose content. Mol Breeding. 2014;34(4):2151–2163. doi: 10.1007/s11032-014-0170-8. [DOI] [Google Scholar]
- 8.Xu F, Sun C, Huang Y, Chen Y, Tong C, Bao JS. QTL mapping for rice grain quality: a strategy to detect more QTLs within sub-populations. Mol Breeding. 2015;35(4):1–11. doi: 10.1007/s11032-015-0296-3. [DOI] [Google Scholar]
- 9.Xu F, Zhang G, Tong C, Sun X, Corke H, Sun M, Bao JS. Association mapping of starch physicochemical properties with starch biosynthesizing genes in waxy rice (Oryza sativa L.) J Agr Food Chem. 2013;61(42):10110–10117. doi: 10.1021/jf4029688. [DOI] [PubMed] [Google Scholar]
- 10.Han YP, Xu ML, Liu XY, Yan CJ, Korban SS, Chen XL, Gu MH. Genes coding for starch branching enzymes are major contributors to starch viscosity characteristics in waxy rice (Oryza sativa L.) Plant Sci. 2004;166(2):357–364. doi: 10.1016/j.plantsci.2003.09.023. [DOI] [Google Scholar]
- 11.Bao JS, Wu YR, Hu B, Wu P, Cui HR, Shu QY. QTL for rice grain quality based on a DH population derived from parents with similar apparent amylose content. Euphytica. 2002;128(3):317–324. doi: 10.1023/A:1021262926145. [DOI] [Google Scholar]
- 12.Yang F, Chen Y, Tong C, Huang Y, Xu F, Li K, Corke H, Sun M, Bao JS. Association mapping of starch physicochemical properties with starch synthesis-related gene markers in nonwaxy rice (Oryza sativa L.) Mol Breeding. 2014;34(4):1747–1763. doi: 10.1007/s11032-014-0135-y. [DOI] [Google Scholar]
- 13.Jin L, Lu Y, Xiao P, Sun M, Corke H, Bao JS. Genetic diversity and population structure of a diverse set of rice germplasm for association mapping. Theor Appl Genet. 2010;121(3):475–487. doi: 10.1007/s00122-010-1324-7. [DOI] [PubMed] [Google Scholar]
- 14.Tian Z, Qian Q, Liu Q, Yan M, Liu X, Yan C, Liu G, Gao Z, Tang S, Zeng D, et al. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc Natl Acad Sci U S A. 2009;106(51):21760–21765. doi: 10.1073/pnas.0912396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Liu W, Xu Y, He Y, Luo L, Xing Y, Xu C, Zhang Q. Genetic basis of 17 traits and viscosity parameters characterizing the eating and cooking quality of rice grain. Theor Appl Genet. 2007;115(4):463–476. doi: 10.1007/s00122-007-0580-7. [DOI] [PubMed] [Google Scholar]
- 16.Zhu C, Gore M, Buckler ES, Yu J. Status and prospects of association mapping in plants. Plant Genome. 2008;1(1):5–20. doi: 10.3835/plantgenome2008.02.0089. [DOI] [Google Scholar]
- 17.Yu J, Buckler ES. Genetic association mapping and genome organization of maize. Curr Opin Chem Biol. 2006;17(2):155–160. doi: 10.1016/j.copbio.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Zhao K, Tung C-W, Eizenga GC, Wright MH, Ali ML, Price AH, Norton GJ, Islam MR, Reynolds A, Mezey J, et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun. 2011;2:467. doi: 10.1038/ncomms1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 2010;42(11):961–U976. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- 20.Huang X, Zhao Y, Wei X, Li C, Wang A, Zhao Q, Li W, Guo Y, Deng L, Zhu C, et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet. 2012;44(1):32–39. doi: 10.1038/ng.1018. [DOI] [PubMed] [Google Scholar]
- 21.Lin Z, Griffith ME, Li X, Zhu Z, Tan L, Fu Y, Zhang W, Wang X, Xie D, Sun C. Origin of seed shattering in rice (Oryza sativa L.) Planta. 2007;226(1):11–20. doi: 10.1007/s00425-006-0460-4. [DOI] [PubMed] [Google Scholar]
- 22.Kovach MJ, Sweeney MT, McCouch SR. New insights into the history of rice domestication. Trends Genet. 2007;23(11):578–587. doi: 10.1016/j.tig.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 23.Oka HI, Morishima H. Phylogenetic differentiation of cultivated rice.23. potentiality of wild progenitors to evolve the indica and japonica types of rice cultivars. Euphytica. 1982;31(1):41–50. doi: 10.1007/BF00028305. [DOI] [Google Scholar]
- 24.Nakamura Y, Francisco PB, Hosaka Y, Sato A, Sawada T, Kubo A, Fujita N. Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol Biol. 2005;58(2):213–227. doi: 10.1007/s11103-005-6507-2. [DOI] [PubMed] [Google Scholar]
- 25.Yamanaka S, Nakamura I, Watanabe KN, Sato YI. Identification of SNPs in the waxy gene among glutinous rice cultivars and their evolutionary significance during the domestication process of rice. Theor Appl Genet. 2004;108(7):1200–1204. doi: 10.1007/s00122-003-1564-x. [DOI] [PubMed] [Google Scholar]
- 26.Hirano HY, Eiguchi M, Sano Y. A single base change altered the regulation of the Waxy gene at the posttranscriptional level during the domestication of rice. Genome Biol Evol. 1998;15(8):978–987. doi: 10.1093/oxfordjournals.molbev.a026013. [DOI] [PubMed] [Google Scholar]
- 27.Wang ZY, Zheng FQ, Shen GZ, Gao JP, Snustad DP, Li MG, Zhang JL, Hong MM. The amylose content in rice endosperm is related to the posttranscriptional regulation of the Waxy gene. Plant J. 1995;7(4):613–622. doi: 10.1046/j.1365-313X.1995.7040613.x. [DOI] [PubMed] [Google Scholar]
- 28.Olsen KM, Caicedo AL, Polato N, McClung A, McCouch S, Purugganan MD. Selection under domestication: Evidence for a sweep in the rice Waxy genomic region. Genetics. 2006;173(2):975–983. doi: 10.1534/genetics.106.056473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura Y, Sakurai A, Inaba Y, Kimura K, Iwasawa N, Nagamine T. The fine structure of amylopectin in endosperm from Asian cultivated rice can be largely classified into two classes. Starch-Starke. 2002;54(3–4):117–131. doi: 10.1002/1521-379X(200204)54:3/4<117::AID-STAR117>3.0.CO;2-2. [DOI] [Google Scholar]
- 30.Bao JS, Corke H, Sun M. Genetic diversity in the physicochemical properties of waxy rice (Oryza sativa L) starch. J Sci Food Agr. 2004;84(11):1299–1306. doi: 10.1002/jsfa.1750. [DOI] [Google Scholar]
- 31.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Ma W, Wang Z, Guo Y, Liu F, Wu H. Study on integration of QTLs related to amylose content in rice. Journal of Northeast Agricultural University. 2014;45(3):8–14. [Google Scholar]
- 33.Aluko G, Martinez C, Tohme J, Castano C, Bergman C, Oard J. QTL mapping of grain quality traits from the interspecific cross Oryza sativa × O. glaberrima. Theor Appl Genet. 2004;109(3):630–639. doi: 10.1007/s00122-004-1668-y. [DOI] [PubMed] [Google Scholar]
- 34.Hsu YC, Tseng MC, Wu YP, Lin MY, Wei FJ, Hwu KK, Hsing YI, Lin YR. Genetic factors responsible for eating and cooking qualities of rice grains in a recombinant inbred population of an inter-subspecific cross. Mol Breeding. 2014;34(2):655–673. doi: 10.1007/s11032-014-0065-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao WG, Chung JW, Kwon SW, Lee JH, Ma KH, Park YJ. Association analysis of physicochemical traits on eating quality in rice (Oryza sativa L.) Euphytica. 2013;191(1):9–21. doi: 10.1007/s10681-012-0820-z. [DOI] [Google Scholar]
- 36.Gao Z, Zeng D, Cui X, Zhou Y, Yan M, Huang D, Li J, Qian Q. Map-based cloning of the ALK gene, which controls the gelatinization temperature of rice. Sci China Ser C. 2003;46(6):661–668. doi: 10.1360/03yc0099. [DOI] [PubMed] [Google Scholar]
- 37.Cho YC, Suh JP, Yoon MR, Baek MK, Won YJ, Lee JH, Park HS, Baek SH, Lee JH. QTL mapping for paste viscosity characteristics related to eating quality and QTL-NIL development in japonica rice (Oryza sativa L.) Plant Breed Biot. 2013;1(4):333–346. doi: 10.9787/PBB.2013.1.4.333. [DOI] [Google Scholar]
- 38.Zheng L, Zhang W, Chen X, Ma J, Chen W, Zhao Z, Zhai H, Wan J. Dynamic QTL analysis of rice protein content and protein index using recombinant inbred lines. J Plant Biol. 2011;54(5):321–328. doi: 10.1007/s12374-011-9170-y. [DOI] [Google Scholar]
- 39.Kobayashi A, Tomita K. QTL detection for stickiness of cooked rice using recombinant inbred lines derived from crosses between japonica rice cultivars. Breeding Sci. 2008;58(4):419–426. doi: 10.1270/jsbbs.58.419. [DOI] [Google Scholar]
- 40.He Q, Yu J, Kim TS, Cho YH, Lee YS, Park YJ. Resequencing reveals different domestication rate for BADH1 and BADH2 in rice (Oryza sativa) PLoS One. 2015;10(8):e0134801. doi: 10.1371/journal.pone.0134801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bao JS, Shen S, Sun M, Corke H. Analysis of genotypic diversity in the starch physicochemical properties of nonwaxy rice: apparent amylose content, pasting viscosity and gel texture. Starch‐Stärke. 2006;58(6):259–267. doi: 10.1002/star.200500469. [DOI] [Google Scholar]
- 42.Bao JS. Accurate measurement of pasting temperature by the rapid visco-analyser: a case study using rice flour. Rice Sci. 2008;15(1):69–72. doi: 10.1016/S1672-6308(08)60022-0. [DOI] [Google Scholar]
- 43.Kim TS, He Q, Kim KW, Yoon MY, Ra WH, Li FP, Tong W, Yu J, Win HO. Choi. et al. Genome-wide resequencing of KRICE_CORE reveals their potential for future breeding, as well as functional and evolutionary studies in the post-genomic era. BMC Genomics. 2016;17(1):1. doi: 10.1186/s12864-016-2734-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z. GAPIT: genome association and prediction integrated tool. Bioinformatics. 2012;28(18):2397–2399. doi: 10.1093/bioinformatics/bts444. [DOI] [PubMed] [Google Scholar]


