Abstract
Despite advances in the surgical management of brain tumors, achieving optimal surgical results and identification of tumor remains a challenge. Raman spectroscopy, a laser-based technique that can be used to nondestructively differentiate molecules based on the inelastic scattering of light, is being applied toward improving the accuracy of brain tumor surgery. Here, the authors systematically review the application of Raman spectroscopy for guidance during brain tumor surgery. Raman spectroscopy can differentiate normal brain from necrotic and vital glioma tissue in human specimens based on chemical differences, and has recently been shown to differentiate tumor-infiltrated tissues from noninfiltrated tissues during surgery. Raman spectroscopy also forms the basis for coherent Raman scattering (CRS) microscopy, a technique that amplifies spontaneous Raman signals by 10,000-fold, enabling real-time histological imaging without the need for tissue processing, sectioning, or staining. The authors review the relevant basic and translational studies on CRS microscopy as a means of providing real-time intraoperative guidance. Recent studies have demonstrated how CRS can be used to differentiate tumor-infiltrated tissues from noninfiltrated tissues and that it has excellent agreement with traditional histology. Under simulated operative conditions, CRS has been shown to identify tumor margins that would be undetectable using standard bright-field microscopy. In addition, CRS microscopy has been shown to detect tumor in human surgical specimens with near-perfect agreement to standard H & E microscopy. The authors suggest that as the intraoperative application and instrumentation for Raman spectroscopy and imaging matures, it will become an essential component in the neurosurgical armamentarium for identifying residual tumor and improving the surgical management of brain tumors.
Keywords: brain metastasis, brain/tumor margin, coherent Raman scattering microscopy, glioma, primary brain tumors, Raman spectroscopy
Improvements in the surgical management of primary and metastatic brain tumors have resulted in greater extent of resection, less postoperative morbidity, and longer overall survival. Consequently, maximal safe re-section is the basic tenet of brain tumor surgery. Advanced surgical techniques such as electrophysiological mapping, intraoperative MRI, and fluorescence-guided surgery have been developed to improve safety and extent of tumor re-section.
Despite these surgical advances, studies analyzing patterns of failure after brain tumor surgery indicate that residual tumor near the resection cavity is the most common cause of tumor recurrence.40 For both malignant and low-grade gliomas, residual tumor burden is a major cause of morbidity and mortality.44,46 In instances of tumor located near eloquent cortex, extent of resection is limited by the need to preserve functional elements. However, even in tumors preoperatively deemed suitable for gross-total resection, at least a fraction of tumor is left behind in more than 75% of cases.37 This reflects the challenge of identifying tumor-infiltrated and noninfiltrated brain during surgery using current techniques. The implications of residual tumor burden are not limited to glioma. Large clinical investigations indicate that local recurrence after brain metastasis resection is the most common cause of treatment failure, with rates as high as 38%-45%.3,39
Therefore, technologies that enable assessment of a surgical cavity for residual tumor are needed. Label-free imaging modalities relying on intrinsic properties, such as the optical density or chemical composition of tissue, are among the most promising emerging approaches for tumor margin control. Here, we review the development of modalities based on the Raman effect, a phenomenon that occurs when molecular bonds absorb a portion of the energy of an incident photon. We detail the relevant scientific background of Raman-based technologies that are under evaluation as tools to improve the accuracy of brain tumor surgery.
Methods
Data Sources
We conducted a systematic review of the English-language literature, starting with a MEDLINE search using the terms Raman and brain tumor or glioma. The search resulted in 91 matches. The complete bibliography of accepted and rejected studies is available by request to the corresponding author.
Literature Screening and Catalog Construction
Study selection was accomplished through a 2-level screening approach. The first level consisted of the exclusion of all case reports, letters, comments, guidelines, reviews, abstract-only publications, articles not written in the English language, and MRI-based Raman technologies. The second level required that each article included dealt with one of the following: spectral characterization of neoplastic human brain tissue, imaging of primary brain tumor from either mouse models or human patients, identification of morphological features in Raman images, discrimination of normal from neoplastic tissue using Raman technology, or the use of Raman imaging to interrogate a cellular process disrupted in primary brain tumor cells. After second-level screening, a total of 34 articles were included: 25 articles on Raman spectroscopy and 9 on coherent Raman scattering (CRS) microscopy.
Spontaneous Raman Scattering: A Brief Overview
Photons can be scattered by molecules in 2 ways. Most are scattered elastically, meaning that their frequency and wavelength are preserved after interaction with a molecule. However, a small fraction of photons transfers energy to molecular bonds and is shifted to a new frequency in a process called the Raman effect, a discovery that resulted in the 1930 Nobel Prize in Physics.42 The difference in frequency of the scattered photon is called the Raman shift. A Raman shift can result in photons of lower frequency/lower energy (i.e., Stokes scattering) and higher frequency/higher energy photons (i.e., anti-Stokes scattering). This effect is very weak and is not usually observed under normal visual conditions. However, using narrow-band laser excitation and a sensitive spectrometer, Raman scattering can be measured.
The Raman spectrum of a molecule can be determined by measuring the Raman shifts caused by interaction with each of its constituent chemical bonds. Raman spectral peaks correspond to specific vibrational modes (e.g., stretching, bending, or scissoring) produced by a chemical bond. For example, the H-C= stretching vibrational mode in the H-C=C- bond plays an essential role in Raman spectroscopy of biological tissues due to the high concentration of fatty acids. Raman spectra can therefore be used as a means of chemical identification. The “vibrational fingerprint” of biological tissues reflects its chemical composition of nucleic acids, protein, and lipids, and is the sum of the vibrational spectra of the tissue constituents. This characteristic led to the hypothesis that the chemical differences between tumor and normal tissue may produce sufficiently distinct Raman spectra to allow accurate brain tumor identification and improve surgical results.
Application of Raman Spectroscopy in Brain Tissue
Early reports using Raman spectroscopy for brain imaging were limited to analyzing spontaneous Raman spectra to detect relative differences in the concentration of common molecules. In 1990, Tashibu was able to detect relative levels of water concentration in a normal versus edematous rat brain within a spectral region of 2800-3800 cm−1.48 This was possible because of a strong intensity peak at 3390 cm−1, corresponding to the O-H stretching vibrational mode of water. Use of this early technique was limited beyond this spectral range because of background interference and low sensitivity. Mizuno et al. improved on this technique by using near-infrared Fourier transform Raman spectroscopy to evaluate low- and high-grade glioma, vestibular schwannoma, and central neurocytoma, but found that tumor specimens were spectrally indistinguishable from gray matter.36
Later studies focused on distinguishing vital tumor from necrotic tissue in glioblastoma biopsy samples for real-time intraoperative brain biopsy guidance.23,25,29 A summary of the major Raman spectroscopy studies can be found in Table 1. Necrotic specimens have high levels of cholesterol and are easily distinguished from vital tumor tissue, with an accuracy approaching 100%. Other reports were able to further differentiate vital tumor from the invasive tumor margin and edematous white matter.2,30,32
TABLE 1. Summary of Raman spectroscopy techniques since 2002.
| Authors & Year | Title | Summary |
|---|---|---|
| Raman spectroscopy | ||
| Desroches et al., 2015 | Characterization of a Raman spectroscopy probe system for intraoperative brain tissue classifica tion |
A handheld Raman probe is used to differentiate necrosis from vital tissue (including tumor and normal brain tissue) with an accuracy of 87%. |
| Jermyn et al., 2015 | Intraoperative brain cancer detection with Raman spectroscopy in humans |
An intraoperative Raman spectroscopy probe is used to differentiate normal brain from dense tumor with 93% sensitivity and 91% specific ity. |
| Kalkanis et al., 2014 | Raman spectroscopy to distinguish grey matter, necrosis, and glioblastoma multiforme in frozen tissue sections |
Raman spectroscopy was used to differentiate gray matter, viable GBM, and necrosis in frozen specimens with 97.8% accuracy in samples without freeze artifacts, and 77.5% of samples with freeze artifacts. |
| Tanahashi et al., 2014 | Assessment of tumor cells in a mouse model of dif- fuse infiltrative glioma by Raman spectroscopy |
Principal component analysis was used to elucidate differences in the spectra of infiltrative glioma and normal brain with 98.3% sensitivity and 75% specificity. |
| Aguiar et al., 2013 | Discriminating neoplastic and normal brain tissues in vitro through Raman spectroscopy: a principal components analysis classification model |
Principal component analysis was able to discriminate normal tissue from tumor, and glioblastoma from other CNS neoplasms, with a sensitivity and specificity of 97.4% and 100%, respectively, in vitro. |
| Auner et al., 2013 | Conclusions and data analysis: a 6-year study of Raman spectroscopy of solid tumors at a major pediatric institute |
A database of Raman spectra from normal brain, kidney, and adrenal gland, and their malignancies, was compiled. Leave-one-out analysis predicted the presence of tumor with 85.5% accuracy in a test set not assuming tissue origin. |
| Gajjar et al., 2012 | Diagnostic segregation of human brain tumors using Fourier-transform infrared and/or Raman spectroscopy coupled with discriminant analysis |
Raman spectroscopy was capable of identifying tumor-specific changes in biochemical composition in formalin-fixed tumor samples. |
| Leslie et al., 2012 | Identification of pediatric brain neoplasms using Raman spectroscopy |
A support vector machine analysis was used to identify Raman spectra collected from various tumor subtypes and normal brain with ex tremely high accuracy (91%-100%). |
| Zhou et al., 2012 | Human brain cancer studied by resonance Raman spectroscopy |
Several specific molecular signatures were identified that distinguished the spectra of normal meningeal tissues from several primary and secondary brain neoplasms, with a sensitivity of 90.9% and specific ity of 100% when principal component analysis was employed. |
| Beljebbar et al., 2010 | Ex vivo and in vivo diagnosis of C6 glioblastoma development by Raman spectroscopy coupled to a microprobe |
Employed Raman spectra collected from ex vivo mouse tissue to differentiate normal tissue from tumor with 100% accuracy, and to delineate early from mature tumor tissue. |
| Kirsch et al., 2010 | Raman spectroscopic imaging for in vivo detection of cerebral brain metastases |
Demonstrates the first use of in vivo Raman spectral mapping of the brain surface to aid tumor resection in a mouse model. |
| Köhler et al., 2009 | Characterization of lipid extracts from brain tissue and tumors using Raman spectroscopy and mass spectrometry |
Demonstrated increased water and decreased lipid content in glioma versus healthy brain tissues in porcine and human samples, con firmed with mass spectroscopy. |
| Krafft et al., 2009 | Disease recognition by infrared and Raman spectroscopy |
Reviewed Raman spectroscopy applications for assessment of numer ous tissues and body fluids, as well as classification and supervised learning algorithms commonly used in analysis of Raman spectra. |
| Koljenović et al., 2005 | Tissue characterization using high wave number Raman spectroscopy |
Established that comparatively diagnostic information can be gleaned from high wave number and low wave number portions of the Raman spectrum from brain and bladder cancer samples in vitro. |
| Krafft et al., 2005 | Near infrared Raman spectra of human brain lipids | Demonstrated Raman spectral characteristics of 12 major brain lipids. |
| Hyperspectral Raman microscopy | ||
| Kast et al., 2015 | Identification of regions of normal grey matter and white matter from pathologic glioblastoma and necrosis in frozen sections using Raman imaging |
Raman spectra acquired grid-wise across a frozen section of brain tumor differentiated gray matter, white matter, tumor, and necrosis through molecular features. |
| Kast et al., 2014 | Raman molecular imaging of brain frozen tissue sections |
Frozen sections of brain tissue were mapped using grid-wise acquisition of Raman spectra, identifying boundaries of gray and white matter, necrosis, GBM, and infiltrating tumor. |
| Bergner et al., 2013 | Hyperspectral unmixing of Raman micro-images for assessment of morphological and chemical parameters in non-dried brain tumor specimens |
Both nuclear morphological characteristics and chemical composition as defined by hyperspectral Raman imaging may offer new ways to classify brain tumors. |
| Bergner et al., 2012 | Unsupervised unmixing of Raman microspectro- scopic images for morphochemical analysis of non-dried brain tumor specimens |
The hyperspectral unmixing algorithms N-FINDR and VCA were used to map abundances of cholesterol, cholesterol ester, nucleic acids, caro tene, proteins, and lipids in normal brain and several tumor subtypes based on hyperspectral Raman micrographs. |
| Krafft et al., 2012 | Advances in optical biopsy—correlation of malig nancy and cell density of primary brain tumors using Raman microspectroscopic imaging |
Demonstrated increased nucleic acid bends in high-grade glioma spectra, among other molecular differences correlating with structural features on H & E microscopy. |
| Amharref et al., 2007 | Discriminating healthy from tumor and necrosis tissue in rat brain tissue samples by Raman spectral imaging |
Demonstrated that Raman microspectroscopy can discriminate between healthy and tumoral brain tissue and yield spectroscopic markers as sociated with the proliferative and invasive properties of glioblastoma ex vivo. |
| Krafft et al., 2005 | Near infrared Raman spectroscopic mapping of native brain tissue and intracranial tumors |
Initial exploration of Raman spectroscopic mapping of frozen samples of brain tissue, meninges, and brain tumor, demonstrating measurable spectroscopic and structural differences. |
| Koljenović et al., 2002 | Discriminating vital tumor from necrotic tissue in human glioblastoma tissue samples by Raman spectroscopy |
Utilized Raman spectral maps of frozen tumor sections to differentiate viable from necrotic tumor via cluster analysis. |
CNS = central nervous system; GBM = glioblastoma multiforme; VCA = vertex component analysis.
Kalkanis and colleagues developed an imaging technique based on pseudocolor 300-μm and 25-μm square measurement grids mapped onto frozen brain tissue specimens.23 Red (1004 cm−1 channel), green (1300:1344 cm−1 channel), and blue (1600 cm−1 channel) were used for the color scheme. Relative intensities of each Raman feature determined the color for each pixel. For example, a pixel with a high concentration of protein would be red on the color scale because of the 1004-cm−1 phenylalanine peak. The single- and multichannel color maps were then used to discriminate white matter, gray matter, and tumor tissue. The diagnostic accuracy was approximately 90% using this simple multichannel imaging technique.
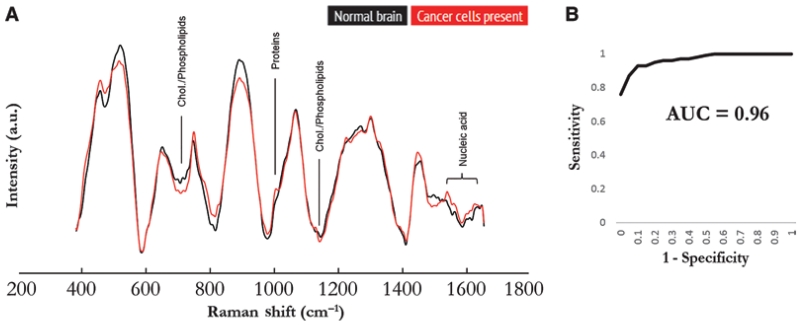
While the aforementioned studies used ex vivo specimens from animal models or human brain tumor specimens, fiberoptic Raman spectroscopy probes have been developed for intraoperative use.5,9,20,26 Fiberoptic probes have been previously used to measure Raman spectra in gastrointestinal, bladder, and cervical cancer surgery.10,18,35 Recently, a handheld probe was developed and used in a clinical trial to integrate Raman spectroscopy into the neurosurgical workflow.9,20 The handheld probe was placed in direct contact with the brain in the resection cavity. Acquisition time was 0.2 seconds for each area of interest measuring 0.5 mm in diameter. Image analysis indicated that 95% of the Raman spectra were generated by the first 1 mm of tissue below the surface. By analyzing differences in spectral peaks, the investigators were able to distinguish normal brain from tumor-invaded brain (> 15% tumor cell invasion) with an accuracy of 92%, sensitivity of 93%, and specificity of 91%. The Raman spectra of normal brain and tumor cell-infiltrated brain can be seen in Fig. 1. Accuracy did not significantly differ between low- and high-grade gliomas. These results provide a promising step toward translating Raman spectroscopy to real-time intraoperative use.
Fig. 1.
Raman spectra for discrimination of cancer tissue. A: Average Raman spectra of in vivo measurements for normal brain (all 66 spectra averaged) and tissue containing glioma cancer cells (all 95 spectra averaged). Corresponding molecular contributors are identified for the most significant differences between the spectra for normal and cancer tissues. Chol. = cholesterol. B: Receiver operating characteristic curve analysis of in vivo detection of glioma based on Raman spectroscopy, generated using the boosted trees classification method. AUC = area under the curve. From Jermyn M, Mok K, Mercier J, Desroches J, Pichette J, Saint-Arnaud K, et al: Intraoperative brain cancer detection with Raman spectroscopy in humans. Sci Transl Med 7:274ra219, 2015. Reprinted with permission from AAAS.
Brain Imaging With CRS Microscopy
The main limitation of spontaneous Raman spectroscopy is that the fraction of inelastically scattered photons is quite small. This results in long image acquisition times, increased image artifact, and poor resolution. To improve this technique, CRS was developed to increase signal intensity (Table 2). Two major forms of CRS microscopy have been developed and used in biomedical imaging—coherent anti-Stokes Raman scattering (CARS) and stimulated Raman scattering (SRS).11,13 CRS increases signal intensity by using a second excitation beam to coherently drive the vibrational frequency of Raman active chemical bonds. Because CRS produces a signal that is orders of magnitude greater (> 10,000-fold) than spontaneous Raman scattering, CRS microscopy is possible with submicron resolution and up to video-rate imaging speeds.12,43 Moreover, standard light microscopy requires thin sectioning to allow light within the visible spectrum to pass through the specimen. CRS microscopy avoids the necessity of thin-slicing because the nonlinear excitation of biological molecules provides intrinsic 3D sectioning.13
TABLE 2. Summary of coherent Raman scattering microscopy techniques.
| Authors & Year | Title | Summary |
|---|---|---|
| CARS microscopy | ||
| Camp et al., 2014 | High-speed coherent Raman fingerprint imaging of biological tissues |
Broadband CARS microscopy was used to image the entire biologi cally relevant Raman window (500-3500 cm−1), including the weak “fingerprint” region to increase sensitivity. |
| Galli et al., 2014 | Effects of tissue fixation on coherent anti-Stokes Raman scattering images of brain |
Formalin fixation does not significantly degrade chemical contrast in CARS imaging, though methanol-acetone fixation is incompat ible with subsequent CARS microscopy due to alterations in lipid content. |
| Uckermann et al., 2014 | Label-free delineation of brain tumors by coherent anti- Stokes Raman scattering microscopy in an orthotopic mouse model and human glioblastoma |
CARS imaging of the C-H vibrational mode enabled cellular-reso lution identification of tumor cells in frozen sections of orthotopic mouse models of GBM and brain metastases (melanoma, breast cancer). |
| Evans et al., 2007 | Chemically-selective imaging of brain structures with CARS microscopy |
Demonstrated the use of CARS microscopy to identify normal brain structures and primary glioma in fresh unfixed and unstained ex vivo brain tissue. |
| SRS microscopy | ||
| Hu et al., 2015 | Vibrational imaging of glucose uptake activity in live cells and tissues by stimulated Raman scattering |
Alkyne-labeled glucose was used to image energy utilization with subcellular resolution using SRS microscopy. |
| Ji et al., 2015 | Detection of human brain tumor infiltration with quantita tive stimulated Raman scattering microscopy |
Developed quantitative methods for measuring differences in cellular- ity, axonal density, and protein/lipid ratio using SRS microscopy. |
| Ji et al., 2013 | Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy |
An SRS microscope generating image contrast via the relative abun dance of lipid and protein facilitated tumor identification in a mouse model of GBM both in frozen sections and in vivo during resection. |
| Freudiger et al., 2012 | Multicolored stain-free histopathology with coherent Ra man imaging |
Multicolor images composed of lipid and protein vibrational modes detected by SRS microscopy were used to generate virtual histo- pathological images without sectioning or fixation. |
| Freudiger et al., 2008 | Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy |
Established SRS microscopy as a biomedical imaging modality capable of capturing the distributions of fatty acids in tissues and monitoring drug delivery in vivo. |
Evans and colleagues used CARS microscopy to image ex vivo samples of an orthotopic human astrocytoma mouse model.11,12 Images were obtained in 700 × 700-μm fields of view to produce a high-resolution mosaic image of the entire mouse brain in coronal sections. The histoarchitecture produced by CARS microscopy was confirmed by subsequently staining the same region with standard H & E stain. Imaging depth ranged from 25 to 80 μm, depending on tissue type and wavelength. CARS microscopy was capable of generating chemically selective images of lipid (2845 cm−1, CH2 symmetric stretching) and proteins (CH3 stretch, 2920 cm−1; amide I vibration, 2960 cm−1) within samples. Focusing on different peaks of the Raman spectra easily differentiated lipid-rich myelin or protein-rich neuronal cell bodies. A related broadband CARS technique has been developed that uses greater spectral breadth within the fingerprint region of the Raman spectra without compromising imaging speed or sensitivity.8 The biologically relevant Raman window (500-3500 cm−1) was used to image a xenograft glioblastoma mouse model. Pseudocolor broadband CARS microscopy was able to distinguish white matter, gray matter, and tumor, and provide nuclear resolution.
SRS has several advantages over CARS, including superior nuclear contrast, a linear relationship between signal intensity and chemical concentration, and a nondistorted spectrum nearly identical to spontaneous Raman allowing for quantitative chemical imaging. In 2008, Freudiger and colleagues published a landmark paper on stimulated Raman scattering microscopy for label-free biomedical imaging.13 Previous methods achieved a large Raman signal with a photodiode array in combination with a femto-second amplified laser system.41 The clinical applicability was halted due to laser-induced tissue damage.15 Freudiger et al. overcame this difficulty by implementation of a high-sensitivity detection scheme based on lock-in detection such that lower peak power, high-repetition rate lasers could be used. The high sensitivity for lipid detection by SRS is well suited for imaging the central nervous system due to the high concentration of lipid-rich myelin. For example, focusing on the 2845 cm−1 shift is ideal for visualizing axon bundles in the corpus callosum due to the high lipid content of myelin sheaths.
To make in vivo imaging possible, a novel microscope configuration was designed to allow for approximately 30% of the backscattered light to reach the objective, 3 times more than standard microscopy.43 This advance increased the imaging speed by 3 orders of magnitude to video-rate, eliminating motion artifact.
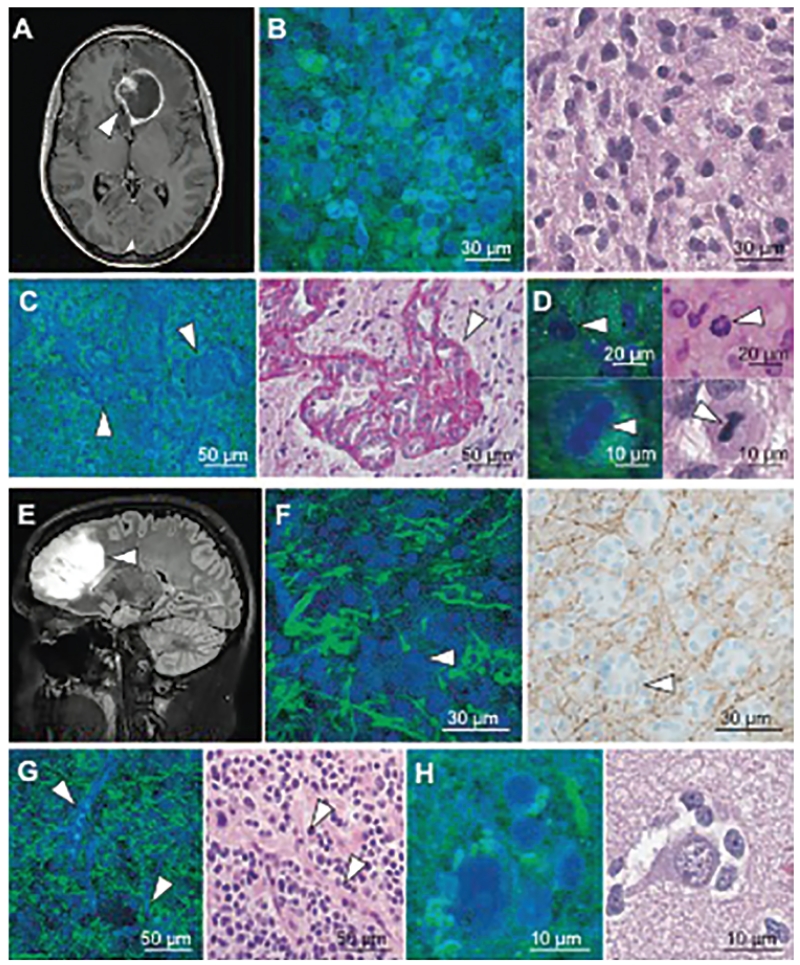
Ji and colleagues described the use of rapid, label-free SRS microscopy for in vivo imaging of brain tumors.22 A comparison of SRS microscopy versus traditional bright-field microscopy of high- and low-grade glioma can be found in Fig. 2. Video-rate SRS microscopy in combination with a human infiltrative glioblastoma xenograft mouse model was used. A “cranial window” model with clear coverslip allowed for direct visualization of the cortical surface that included normal brain and tumor invasion.38 Normal and tumor-infiltrated brain at the cortical surface were easily identified on SRS microscopy that were not visible on standard bright-field microscopy. The brain/tumor interface was visible using a novel blue-green color scheme to highlight contrasting histological features.14 Similar imaging results were noted after corticectomy and dissection, simulating intraoperative conditions during brain tumor surgery. Moreover, this study showed a near-perfect correlation (κ = 0.98) between SRS and H & E microscopy for detection of glioma infiltration based on neuropathologist assessment. These results indicate that in vivo SRS microscopy can approach the gold standard in histopathology.
Fig. 2.
SRS and traditional microscopy of intrinsic brain tumors. A: SRS imaging of a glioblastoma multiforme (arrowhead) demonstrating ring enhancement on MRI. B: Hypercellularity and nuclear atypia of viable tumor is apparent on both SRS (left) and H & E (right) microscopy. C: Microvascular proliferation creates tortuous vascular complexes evident on SRS microscopy (left, arrowheads) and is highlighted with periodic acid-Schiff staining (right, arrowhead). D: Mitotic figures are also visible (arrowheads) with SRS microscopy (left) and H & E staining (right). E and F: A nonenhancing, low-grade oligodendroglioma (arrowhead, E) consists of hypercellular tissue with nests of “fried-egg” morphology (arrowheads, F) causing minimal axonal disruption on SRS imaging (left), as confirmed through neurofilament immunostaining (right). G and H: “Chicken wire” blood vessels (arrowheads, G) imaged with SRS (left) and H & E (right) microscopy, and perineuronal satellitosis is visible in both SRS (left) and H & E (right) microscopy (H). From Ji M, Lewis S, Camelo-Piragua S, Ramkissoon SH, Snuderl M, Venneti S, et al: Detection of human brain tumor infiltration with quantitative stimulated Raman scattering microscopy. Sci Transl Med 7:309ra163, 2015. Reprinted with permission from AAAS.
Quantitative SRS microscopy was recently developed based on the alterations in tissue cellularity, axonal density, and protein/lipid ratio in tumor-infiltrated tissues.21 A classifier system based on these parameters was able to detect tumor infiltration with 97.5% sensitivity and 98.5% specificity. Quantitative SRS microscopy detected tumor infiltration in grossly normal brain, providing evidence that this technique could improve tumor detection during brain tumor surgery. A comparison of the accuracy of tumor detection using Raman spectroscopy and CRS microscopy can be found in Table 3.
TABLE 3. Summary of tumor detection accuracy for Raman-based technologies.
| Authors & Year | Accuracy (%) | Sensitivity (%) |
Specificity (%) |
|---|---|---|---|
| Raman spectroscopy | |||
| Desroches et al., 2015 | 87 | 84 | 89 |
| Jermyn et al., 2015 | 93 | 91 | |
| Kalkanis et al., 2014 | 97.8 (w/o freeze artifact); 77.5 (w/ freeze artifact) |
||
| Tanahashi et al., 2014 | 75 | 98.3 | |
| Aguiar et al., 2013 | 100 | 97.4 | |
| Auner et al., 2013 | 85.5 | ||
| Leslie et al., 2012 | 91-100 | ||
| Zhou et al., 2012 | 90.9 | 100 | |
| SRS microscopy | |||
| Ji et al., 2015 | 97.5 | 98.5 | |
| Ji et al., 2013 | 99.5 | ||
One barrier to translation of SRS microscopy into the clinical setting is determining how microscopic scale data, collected with small fields of view (400 × 400 μm) could be applied within the context of a large resection cavity. A previous clinical trial using intraoperative confocal microscopy for detection of low-grade glioma used a similarly sized field of view (475 × 475 μm).45 Using an iterative image-resect-image technique throughout tumor removal, additional operative time was 10 minutes for image acquisition and was not obstructive to surgical workflow. Based on previous advances in Raman-based technologies and our own preliminary work, our group has developed a clinical CRS microscopy system that is currently under evaluation for intraoperative use. We believe that Raman-based technologies are nearing a critical point of clinical translation where large-scale clinical trials can be planned to confirm promising preclinical results.
Conclusions
Raman spectroscopy and CRS microscopy are promising novel methods in brain tumor surgery that have been developed to improve the accuracy of tumor detection and better characterize tumor invasion and molecular features. Real-time in vivo Raman spectroscopy is a developing tool in brain tumor surgery with potential for integration into the neurosurgical workflow. CRS microscopy is a rapid, label-free imaging method capable of identifying tumor and delineating the brain/tumor interface. CRS microscopy has near-perfect agreement with standard H & E microscopy, and tumor infiltration can be quantified with precision. It is our hope that leaving residual tumor will become an operative strategy used only to reduce post-operative neurological morbidity, but never as a result of inadequate tumor identification. Translational research in Raman-based technology suggests that these methods will play an important role in improving the accuracy of brain tumor surgery.
Acknowledgments
We thank Holly Wagner for manuscript editing.
This research was supported by the National Institute of Biomedical Imaging and Bioengineering (R01EB017254 to X.S.X. and D.A.O.) and the NIH Director’s Transformative Research Award Program T-R01 (R01EB010244-01 to X.S.X.) of the National Institutes of Health.
ABBREVIATIONS
- CARS
coherent anti-Stokes Raman scattering
- CRS
coherent Raman scattering
- SRS
stimulated Raman scattering
Footnotes
Disclosures
The authors report the following. Dr. Freudiger: direct stock ownership in and employee of Invenio Imaging, Inc. Dr. Xie: direct stock ownership in Invenio, Inc. Dr. Orringer: consultant for and direct stock ownership in Invenio Imaging Inc. Invenio Imaging Inc. is a company focused on the commercialization of coherent Raman microscopes.
Author Contributions
Conception and design: Orringer. Acquisition of data: all authors. Analysis and interpretation of data: all authors. Drafting the article: Hollon, Lewis. Critically revising the article: Orringer, Hollon, Freudiger, Xie. Reviewed submitted version of manuscript: all authors. Approved the final version of the manuscript on behalf of all authors: Orringer. Study supervision: Orringer, Xie.
References
- 1.Aguiar RP, Silveira L, Jr, Falcão ET, Pacheco MT, Zângaro RA, Pasqualucci CA. Discriminating neoplastic and normal brain tissues in vitro through Raman spectroscopy: a principal components analysis classification model. Photomed Laser Surg. 2013;31:595–604. doi: 10.1089/pho.2012.3460. [DOI] [PubMed] [Google Scholar]
- 2.Amharref N, Beljebbar A, Dukic S, Venteo L, Schneider L, Pluot M, et al. Discriminating healthy from tumor and necrosis tissue in rat brain tissue samples by Raman spectral imaging. Biochim Biophys Acta. 2007;1768:2605–2615. doi: 10.1016/j.bbamem.2007.06.032. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong JG, Wronski M, Galicich J, Arbit E, Leibel SA, Burt M. Postoperative radiation for lung cancer metastatic to the brain. J Clin Oncol. 1994;12:2340–2344. doi: 10.1200/JCO.1994.12.11.2340. [DOI] [PubMed] [Google Scholar]
- 4.Auner AW, Kast RE, Rabah R, Poulik JM, Klein MD. Conclusions and data analysis: a 6-year study of Raman spectroscopy of solid tumors at a major pediatric institute. Pediatr Surg Int. 2013;29:129–140. doi: 10.1007/s00383-012-3211-6. [DOI] [PubMed] [Google Scholar]
- 5.Beljebbar A, Dukic S, Amharref N, Manfait M. Ex vivo and in vivo diagnosis of C6 glioblastoma development by Raman spectroscopy coupled to a microprobe. Anal Bioanal Chem. 2010;398:477–487. doi: 10.1007/s00216-010-3910-6. [DOI] [PubMed] [Google Scholar]
- 6.Bergner N, Krafft C, Geiger KD, Kirsch M, Schackert G, Popp J. Unsupervised unmixing of Raman microspectroscopic images for morphochemical analysis of non-dried brain tumor specimens. Anal Bioanal Chem. 2012;403:719–725. doi: 10.1007/s00216-012-5858-1. [DOI] [PubMed] [Google Scholar]
- 7.Bergner N, Medyukhina A, Geiger KD, Kirsch M, Schackert G, Krafft C, et al. Hyperspectral unmixing of Raman micro-images for assessment of morphological and chemical parameters in non-dried brain tumor specimens. Anal Bioanal Chem. 2013;405:8719–8728. doi: 10.1007/s00216-013-7257-7. [DOI] [PubMed] [Google Scholar]
- 8.Camp CH, Jr, Lee YJ, Heddleston JM, Hartshorn CM, Hight Walker AR, Rich JN, et al. High-speed coherent Raman fingerprint imaging of biological tissues. Nat Photonics. 2014;8:627–634. doi: 10.1038/nphoton.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desroches J, Jermyn M, Mok K, Lemieux-Leduc C, Mercier J, St-Arnaud K, et al. Characterization of a Raman spectroscopy probe system for intraoperative brain tissue classification. Biomed Opt Express. 2015;6:2380–2397. doi: 10.1364/BOE.6.002380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Draga RO, Grimbergen MC, Vijverberg PL, van Swol CF, Jonges TG, Kummer JA, et al. In vivo bladder cancer diagnosis by high-volume Raman spectroscopy. Anal Chem. 2010;82:5993–5999. doi: 10.1021/ac100448p. [DOI] [PubMed] [Google Scholar]
- 11.Evans CL, Xie XS. Coherent anti-stokes Raman scattering microscopy: chemical imaging for biology and medicine. Annu Rev Anal Chem (Palo Alto, Calif) 2008;1:883–909. doi: 10.1146/annurev.anchem.1.031207.112754. [DOI] [PubMed] [Google Scholar]
- 12.Evans CL, Xu X, Kesari S, Xie XS, Wong ST, Young GS. Chemically-selective imaging of brain structures with CARS microscopy. Opt Express. 2007;15:12076–12087. doi: 10.1364/oe.15.012076. [DOI] [PubMed] [Google Scholar]
- 13.Freudiger CW, Min W, Saar BG, Lu S, Holtom GR, He C, et al. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science. 2008;322:1857–1861. doi: 10.1126/science.1165758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freudiger CW, Pfannl R, Orringer DA, Saar BG, Ji M, Zeng Q, et al. Multicolored stain-free histopathology with coherent Raman imaging. Lab Invest. 2012;92:1492–1502. doi: 10.1038/labinvest.2012.109. Erratum in Lab Invest 92:1661, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Y, Wang H, Shi R, Cheng JX. Characterization of photo-damage in coherent anti-Stokes Raman scattering microscopy. Opt Express. 2006;14:3942–3951. doi: 10.1364/oe.14.003942. [DOI] [PubMed] [Google Scholar]
- 16.Gajjar K, Heppenstall LD, Pang W, Ashton KM, Trevisan J, Patel II, et al. Diagnostic segregation of human brain tumours using Fourier-transform infrared and/or Raman spectroscopy coupled with discriminant analysis. Anal Methods. 2012;5:89–102. doi: 10.1039/C2AY25544H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galli R, Uckermann O, Koch E, Schackert G, Kirsch M, Steiner G. Effects of tissue fixation on coherent anti-Stokes Raman scattering images of brain. J Biomed Opt. 2014;19:071402. doi: 10.1117/1.JBO.19.7.071402. [DOI] [PubMed] [Google Scholar]
- 18.Haka AS, Volynskaya Z, Gardecki JA, Nazemi J, Lyons J, Hicks D, et al. In vivo margin assessment during partial mastectomy breast surgery using Raman spectroscopy. Cancer Res. 2006;66:3317–3322. doi: 10.1158/0008-5472.CAN-05-2815. [DOI] [PubMed] [Google Scholar]
- 19.Hu F, Chen Z, Zhang L, Shen Y, Wei L, Min W. Vibrational imaging of glucose uptake activity in live cells and tissues by stimulated Raman scattering. Angew Chem Int Ed Engl. 2015;54:9821–9825. doi: 10.1002/anie.201502543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jermyn M, Mok K, Mercier J, Desroches J, Pichette J, Saint-Arnaud K, et al. Intraoperative brain cancer detection with Raman spectroscopy in humans. Sci Transl Med. 2015;7:274ra19. doi: 10.1126/scitranslmed.aaa2384. [DOI] [PubMed] [Google Scholar]
- 21.Ji M, Lewis S, Camelo-Piragua S, Ramkissoon SH, Snuderl M, Venneti S, et al. Detection of human brain tumor infiltration with quantitative stimulated Raman scattering microscopy. Sci Transl Med. 2015;7:309ra163. doi: 10.1126/scitranslmed.aab0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji M, Orringer DA, Freudiger CW, Ramkissoon S, Liu X, Lau D, et al. Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy. Sci Transl Med. 2013;5:201ra119. doi: 10.1126/scitranslmed.3005954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalkanis SN, Kast RE, Rosenblum ML, Mikkelsen T, Yurgelevic SM, Nelson KM, et al. Raman spectroscopy to distinguish grey matter, necrosis, and glioblastoma multiforme in frozen tissue sections. J Neurooncol. 2014;116:477–485. doi: 10.1007/s11060-013-1326-9. [DOI] [PubMed] [Google Scholar]
- 24.Kast R, Auner G, Yurgelevic S, Broadbent B, Raghunathan A, Poisson LM, et al. Identification of regions of normal grey matter and white matter from pathologic glioblastoma and necrosis in frozen sections using Raman imaging. J Neurooncol. 2015;125:287–295. doi: 10.1007/s11060-015-1929-4. [DOI] [PubMed] [Google Scholar]
- 25.Kast RE, Auner GW, Rosenblum ML, Mikkelsen T, Yurgelevic SM, Raghunathan A, et al. Raman molecular imaging of brain frozen tissue sections. J Neurooncol. 2014;120:55–62. doi: 10.1007/s11060-014-1536-9. [DOI] [PubMed] [Google Scholar]
- 26.Kirsch M, Schackert G, Salzer R, Krafft C. Raman spectroscopic imaging for in vivo detection of cerebral brain metastases. Anal Bioanal Chem. 2010;398:1707–1713. doi: 10.1007/s00216-010-4116-7. [DOI] [PubMed] [Google Scholar]
- 27.Köhler M, Machill S, Salzer R, Krafft C. Characterization of lipid extracts from brain tissue and tumors using Raman spectroscopy and mass spectrometry. Anal Bioanal Chem. 2009;393:1513–1520. doi: 10.1007/s00216-008-2592-9. [DOI] [PubMed] [Google Scholar]
- 28.Koljenović S, Bakker Schut TC, Wolthuis R, de Jong B, Santos L, Caspers PJ, et al. Tissue characterization using high wave number Raman spectroscopy. J Biomed Opt. 2005;10:031116. doi: 10.1117/1.1922307. [DOI] [PubMed] [Google Scholar]
- 29.Koljenović S, Choo-Smith LP, Bakker Schut TC, Kros JM, van den Berge HJ, Puppels GJ. Discriminating vital tumor from necrotic tissue in human glioblastoma tissue samples by Raman spectroscopy. Lab Invest. 2002;82:1265–1277. doi: 10.1097/01.lab.0000032545.96931.b8. [DOI] [PubMed] [Google Scholar]
- 30.Krafft C, Belay B, Bergner N, Romeike BF, Reichart R, Kalff R, et al. Advances in optical biopsy—correlation of malignancy and cell density of primary brain tumors using Raman microspectroscopic imaging. Analyst (Lond) 2012;137:5533–5537. doi: 10.1039/c2an36083g. [DOI] [PubMed] [Google Scholar]
- 31.Krafft C, Neudert L, Simat T, Salzer R. Near infrared Raman spectra of human brain lipids. Spectrochim Acta A Mol Biomol Spectrosc. 2005;6:1529–1535. doi: 10.1016/j.saa.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 32.Krafft C, Sobottka SB, Schackert G, Salzer R. Near infrared Raman spectroscopic mapping of native brain tissue and intracranial tumors. Analyst (Lond) 2005;130:1070–1077. doi: 10.1039/b419232j. [DOI] [PubMed] [Google Scholar]
- 33.Krafft C, Steiner G, Beleites C, Salzer R. Disease recognition by infrared and Raman spectroscopy. J Biophotonics. 2009;2:13–28. doi: 10.1002/jbio.200810024. [DOI] [PubMed] [Google Scholar]
- 34.Leslie DG, Kast RE, Poulik JM, Rabah R, Sood S, Auner GW, et al. Identification of pediatric brain neoplasms using Raman spectroscopy. Pediatr Neurosurg. 2012;48:109–117. doi: 10.1159/000343285. [DOI] [PubMed] [Google Scholar]
- 35.Mahadevan-Jansen A, Mitchell MF, Ramanujam N, Utzinger U, Richards-Kortum R. Development of a fiber optic probe to measure NIR Raman spectra of cervical tissue in vivo. Photochem Photobiol. 1998;68:427–431. [PubMed] [Google Scholar]
- 36.Mizuno AKH, Kawauchi K, Muraishi S, Ozaki Y. Near-infrared Fourier transform Raman spectroscopic study of human brain tissues and tumors. J Raman Spectrosc. 1994;25:25–29. [Google Scholar]
- 37.Orringer D, Lau D, Khatri S, Zamora-Berridi GJ, Zhang K, Wu C, et al. Extent of resection in patients with glioblastoma: limiting factors, perception of resectability, and effect on survival. J Neurosurg. 2012;117:851–859. doi: 10.3171/2012.8.JNS12234. [DOI] [PubMed] [Google Scholar]
- 38.Orringer DA, Chen T, Huang DL, Philbert M, Kopelman R, Sagher O. A technical description of the brain tumor window model: an in vivo model for the evaluation of intraoperative contrast agents. Acta Neurochir Suppl 109. 2011:259–263. doi: 10.1007/978-3-211-99651-5_41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patchell RA, Tibbs PA, Walsh JW, Dempsey RJ, Maruyama Y, Kryscio RJ, et al. A randomized trial of surgery in the treatment of single metastases to the brain. N Engl J Med. 1990;322:494–500. doi: 10.1056/NEJM199002223220802. [DOI] [PubMed] [Google Scholar]
- 40.Petrecca K, Guiot MC, Panet-Raymond V, Souhami L. Failure pattern following complete resection plus radiotherapy and temozolomide is at the resection margin in patients with glioblastoma. J Neurooncol. 2013;111:19–23. doi: 10.1007/s11060-012-0983-4. [DOI] [PubMed] [Google Scholar]
- 41.Ploetz E, Marx B, Klein T, Huber R, Gilch P. A 75 MHz light source for femtosecond stimulated Raman microscopy. Opt Express. 2009;17:18612–18620. doi: 10.1364/OE.17.018612. [DOI] [PubMed] [Google Scholar]
- 42.Raman C, Krishnan K. A new type of secondary radiation. Nature. 1928;121:501–502. [Google Scholar]
- 43.Saar BG, Freudiger CW, Reichman J, Stanley CM, Holtom GR, Xie XS. Video-rate molecular imaging in vivo with stimulated Raman scattering. Science. 2010;330:1368–1370. doi: 10.1126/science.1197236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanai N, Polley MY, McDermott MW, Parsa AT, Berger MS. An extent of resection threshold for newly diagnosed glioblastomas. J Neurosurg. 2011;115:3–8. doi: 10.3171/2011.2.jns10998. [DOI] [PubMed] [Google Scholar]
- 45.Sanai N, Snyder LA, Honea NJ, Coons SW, Eschbacher JM, Smith KA, et al. Intraoperative confocal microscopy in the visualization of 5-aminolevulinic acid fluorescence in low-grade gliomas. J Neurosurg. 2011;115:740–748. doi: 10.3171/2011.6.JNS11252. [DOI] [PubMed] [Google Scholar]
- 46.Smith JS, Chang EF, Lamborn KR, Chang SM, Prados MD, Cha S, et al. Role of extent of resection in the long-term outcome of low-grade hemispheric gliomas. J Clin Oncol. 2008;26:1338–1345. doi: 10.1200/JCO.2007.13.9337. [DOI] [PubMed] [Google Scholar]
- 47.Tanahashi K, Natsume A, Ohka F, Momota H, Kato A, Motomura K, et al. Assessment of tumor cells in a mouse model of diffuse infiltrative glioma by Raman spectroscopy. BioMed Res Int. 2014;2014:860241. doi: 10.1155/2014/860241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tashibu K. Analysis of water content in rat brain using Raman spectroscopy. No To Shinkei. 1990;42:999–1004. (Jpn) [PubMed] [Google Scholar]
- 49.Uckermann O, Galli R, Tamosaityte S, Leipnitz E, Geiger KD, Schackert G, et al. Label-free delineation of brain tumors by coherent anti-Stokes Raman scattering microscopy in an orthotopic mouse model and human glioblastoma. PLoS One. 2014;9:e107115. doi: 10.1371/journal.pone.0107115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Y, Liu CH, Sun Y, Pu Y, Boydston-White S, Liu Y, et al. Human brain cancer studied by resonance Raman spectroscopy. J Biomed Opt. 2012;17:116021. doi: 10.1117/1.JBO.17.11.116021. [DOI] [PMC free article] [PubMed] [Google Scholar]