Abstract
A missense mutation in CIZ1 (c.790A > G, p.S264G) was linked to autosomal dominant cervical dystonia in a large multiplex Caucasian pedigree (OMIM614860, DYT23). CIZ1 is a p21(Cip1/Waf1) -interacting zinc finger protein, widely expressed in neural and extra-neural tissues, and plays a role in DNA synthesis at the G1/S cell-cycle checkpoint. The role of CIZ1 in the nervous system and relative contributions of gain- or loss- of function to the pathogenesis of CIZ1-associated dystonia remain indefinite. Using relative quantitative reverse transcriptase-PCR, cerebellum showed the highest expression levels of Ciz1 in adult mouse brain, over two fold higher than liver, and higher than striatum, midbrain and cerebral cortex. Overall, neural expression of Ciz1 increased with postnatal age. A Ciz1 gene-trap knock-out (KO) mouse model (Ciz1−/−) was generated to examine the functional role(s) of CIZ1 in the sensorimotor nervous system and contributions of CIZ1 to cell-cycle control in the mammalian brain. Ciz1 transcripts were absent in Ciz1−/− mice and reduced by approximately 50% in Ciz1+/− mice. Ciz1−/− mice were fertile but smaller than wild-type (WT) littermates. Ciz1−/− mice did not manifest dystonia, but exhibited mild motoric abnormalities on balance, open-field activity, and gait. To determine the effects of germline KO of Ciz1 on whole-genome gene expression in adult brain, total RNA from mouse cerebellum was harvested from 6 10-month old Ciz1−/− mice and 6 age- and gender- matched WT littermates for whole-genome gene expression analysis. Based on whole-genome gene-expression analyses, genes involved in cellular movement, cell development, cellular growth, cellular morphology and cell-to-cell signaling and interaction were up-regulated in Ciz1−/− mice. The top up-regulated pathways were metabolic and cytokine-cytokine receptor interactions. Down-regulated genes were involved in cell cycle, cellular development, cell death and survival, gene expression and cell morphology. Down-regulated networks included those related to metabolism, focal adhesion, neuroactive ligand-receptor interaction, and MAPK signaling. Based on pathway analyses, transcription factor 7-like 2 (TCF7L2), a member of the Wnt/β-catenin signaling pathway, was a major hub for down-regulated genes, whereas NF-κB was a major hub for up-regulated genes. In aggregate, these data suggest that CIZ1 may be involved in the post-mitotic differentiation of neurons in response to external signals and changes in gene expression may compensate, in part, for CIZ1 deficiency in our Ciz1−/− mouse model. Although CIZ1 deficiency was associated with mild motor abnormalities, germline loss of Ciz1 was not associated with dystonia on the C57BL/6J background.
Keywords: CIZ1, Knock-out mouse, Dystonia, Cell cycle, Cerebellum, Gene expression
1. Introduction
Genetic factors play an important role in the development of isolated dystonia, a common movement disorder characterized by involuntary and sustained muscle contractions causing twisting and repetitive movements (Albanese et al., 2013, LeDoux, 2012b; LeDoux, 2012a). To date, >20 monogenic inherited dystonias and dystonia-related disorders (DYT1-25) have been reported in Online Mendelian Inheritance in Man (OMIM) (Xiao et al., 2014). Moreover, dystonia can also be a presenting or prominent clinical manifestation of numerous hereditary neurological diseases (LeDoux, 2012b). In 2012, a missense variant (c.790A > G, p.S264G) in Cip1-interacting zinc finger protein 1 (CIZ1) was identified in a large multiplex pedigree with adult-onset isolated cervical dystonia (Xiao et al., 2012). CIZ1 is a p21 (Cip1/Waf1)-interacting zinc finger protein expressed in brain and involved in DNA synthesis and cell-cycle control (Mitsui et al., 1999, Coverley et al., 2005, Ainscough et al., 2007, Copeland et al., 2010, Copeland et al., 2015). CIZ1 is also widely expressed in extra-neural tissues and may contribute to post-mitotic differentiation of spermatocytes (Greaves et al., 2012). Although CIZ1 promotes DNA replication at the G1/S checkpoint of the cell cycle (Coverley et al., 2005), its role in post-mitotic neurons with the central nervous system (CNS) remains largely unknown (Herrup, 2013; Greaves et al., 2012). Recently, another missense variant (c.2357C > T, p.T786I) in CIZ1 was identified in a German patient of unknown family history (Dufke et al., 2015). Moreover, Exon 2 indels in CIZ1 may be enriched in subjects with dystonia in comparison to controls (Xiao et al., 2014, Dufke et al., 2015). However, neither nonsense nor large deletion mutations leading to haploinsufficiency have been identified in subjects with dystonia (Xiao et al., 2014). Therefore, the relative contributions of gain- or loss-of-function to the pathogenesis of CIZ1-associated dystonia and cell-cycle control within the adult nervous system remain indeterminate (LeDoux et al., 2013).
Besides its potential roles in dystonia and the maintenance and/or differentiation of post-mitotic neurons within the CNS, the biology and pathobiology of CIZ1 appear to play a role in several other neural (Alzheimer disease, medulloblastoma, and neuroblastoma) and extraneural diseases (breast, lung, prostate, and colon cancer; and rheumatoid arthritis) (Yin et al., 2013, Warder and Keherly, 2003, Nishibe et al., 2013, Liu et al., 2015; Wu et al., 2016; Judex et al., 2003). For examples, altered splicing in Exon 8 of CIZ1 was found to be associated with Alzheimer disease (Dahmcke et al., 2008), and Exon 4 is skipped in Ewing tumor and neuroblastoma cell lines (Rahman et al., 2007, Rahman et al., 2010). More importantly, CIZ1 is related to the progression of several human cancers and a CIZ1 variant is a circulating biomarker of early-stage lung cancer (Wang et al., 2014, Zhang et al., 2015, Liu et al., 2015; Higgins et al., 2012, Yin et al., 2013). To provide a platform for mechanistic studies of CIZ1 biology and pathobiology, we have generated and phenotypically characterized a Ciz1−/− gene-trap mouse model.
2. Materials and methods
2.1. Developmental expression of Ciz1 transcripts
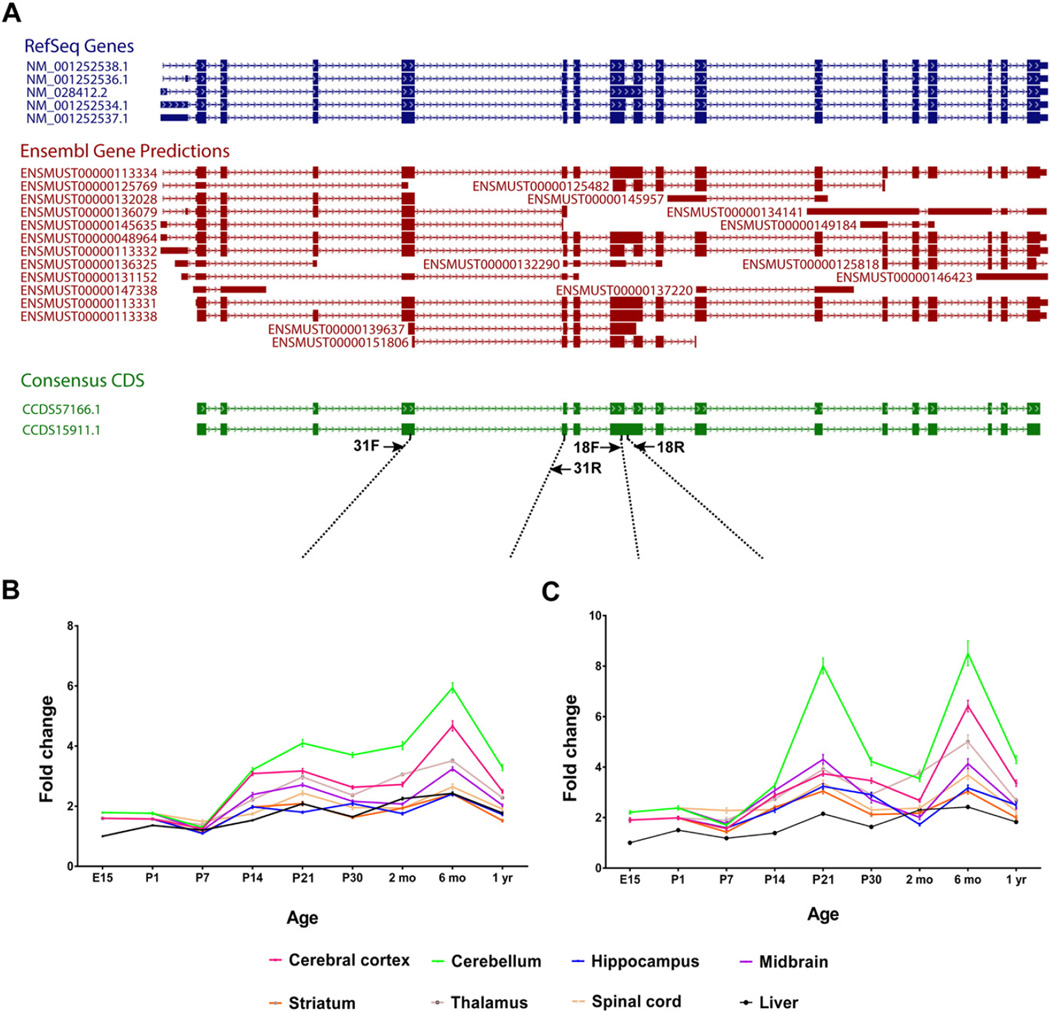
Expression patterns of Ciz1 transcripts in C57BL/6J mice were established with relative quantitative reverse transcriptase-PCR (QRT-PCR) using tissues from cerebral cortex, cerebellum, hippocampus, ventral midbrain, striatum, thalamus, spinal cord, and liver. We examined 6 mice (3 male, 3 female) at each of 9 developmental time points (E15, P1, P7, P14, P21, P30, 2 mo, 6 mo, and 1 yr). TaqMan-based QRT-PCR was performed with Ambion's RETROscript® Reverse Transcription Kit and a LightCycler® 480 System (Roche, Indianapolis, IN, USA). Two primer pairs were designed to examine expression of both Consensus Coding Sequence (CCDS) isoforms (CCDS57166.1 [2376 nt] and CCDS15911.1 [2538 nt]), in aggregate, or the longer isoform (CCDS15911.1, 2538 nt), in isolation, with β-actin as the endogenous control (Fig. 1A, Supplemental Table 1). As seen in Fig. 1A, primers 31F and 31R should amplify all RefSeq and most predicted Ensembl isoforms. In contrast, only RefSeq isoform NM_028412.2 and a subset of predicted Ensembl isoforms should be amplified with primers 18F and 18R. Data were normalized to E15 liver. Detailed methods are provided in previous work from our laboratory (Xiao et al., 2012).
Fig. 1.
(A) Schematic representation of Ciz1 isoforms. Two distinct primer pairs were designed to examine expression of both CCDS isoforms, in aggregate, or the longer isoform (CCDS15911.1), in isolation, with β-actin as the endogenous control. Primers pairs: 31F/31R and 18F/18R. Relative to E15 liver, neural expression of Ciz1 isoforms amplified by primer pair 31F/31R (B) and primer pair 18F/18R (C) increase with increasing postnatal age. Among those neural structures subjected to analysis, Ciz1 expression was highest in cerebellum.
2.2. Generation of Ciz1−/− gene-trap mice
All mouse experiments were performed in accordance with the National Institutes of Health's Guidelines for the Care and Use of Laboratory Animals and approved by our Institutional Animal Care and Use Committee. Ciz1+/− mice were generated by the Texas A&M Institute for Genomic Medicine with the gene trapped clone IST13830B6 (Ciz1Gt(IST13830B6)Tigm) which has an insertion fragment localized to Intron 1 of Ciz1 isoform NM_028412.2. Upon establishment of germline transmission, Ciz1 gene-trap mice were bred to C57BL/6J mice for at least 6 generations. All experiments used Ciz1+/+ littermates on the same genetic background. Three primers (Supplemental Table 1) were used for PCR-based genotyping of Ciz1 mutant mice with one primer located within the gene trap vector (V76F) and the two other primers located within Intron 1 of Ciz1 and flanking the gene trap insertion. The Mouse Direct PCR kit from Biotool (Houston, TX, USA) was used for genotyping with the following cycling conditions: 95 °C for 5 min, 35 cycles at 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 30 s. The WT allele yields a 406 bp amplicon whereas the gene-trap allele generates a 292 bp amplicon.
2.3. Hematology and serum chemistry
Retro-orbital venous blood was acquired from WT and Ciz1−/− mice as a terminal procedure after pentobarbital overdose (200 mg/kg, intraperitoneal). Blood was collected into Becton Dickinson Microtainer™ K2EDTA and clot activator tubes for hematological and serum chemistry studies, respectively. Complete blood counts with differentials and platelet counts were acquired with a VetScan HM5 Hematology System (Abaxsis, Union City, CA, USA). Serum chemistry studies were performed by IDEXX BioResearch (West Sacramento, CA, USA).
2.4. Ciz1 expression in gene-trap mice
Relative levels of mouse Ciz1 mRNA were determined in 6 brain regions (cerebral cortex, cerebellum, hippocampus, striatum, thalamus, and midbrain), cervical spinal cord, and liver harvested from 3 male mice (3 mo) of each genotype (Ciz1+/−, Ciz1−/−, and WT). TaqMan-based QRT-PCR was performed using two primer pairs and probes (Supplemental Table 1) with a LightCycler® 480 System (Roche, Indianapolis, IN, USA). Mouse β-actin and hypoxanthine-guanine phosphoribosyltransferase (HGPRT) were used as endogenous controls.
2.5. Qualitative Western blot analysis
For qualitative Western blots, whole brains from adult (3 mo) WT, Ciz1+/− and Ciz1−/− mice (n = 3 mice/genotype) were dissected to isolate the cerebellum and cerebral cortex. The tissues were lysed with ice-cold IP lysis buffer from Pierce™ Classic Magnetic IP/Co-IP Kit (ThermoFisher Scientific, Grand Island, NY, USA) containing Halt™protease and phosphatase inhibitor cocktail (ThermoFisher Scientific, Grand Island, NY, USA) using a Teflon glass homogenizer. Lysed samples were microcentrifuged for 15 min at 14,000 rpm and the supernatants were collected. Supernatants were pre-cleared with protein A/G magnetic beads for 3–4 h at 4 °C using a magnetic separation rack. Total protein (500 µg) was immunoprecipitated overnight at 4 °C with constant rocking using 4 µg of mouse anti-CIZ1 antibody (ab172442, Abcam, Cambridge, MA, USA) and pulled down the next morning with 25 µL of protein A/G magnetic beads for 4–6 h at 4 °C. Beads were washed 4 times in with ice-cold IP lysis buffer, and bound proteins eluted in 2× SDS sample buffer. For Western blots, proteins were separated by SDS-PAGE (4–12% Bis-Tris gels) and transferred to PVDF membranes using a BioRad (Berkeley, CA, USA) wet transfer system. Subsequently, membranes were blocked for 2 h in 5% non-fat dry milk and incubated overnight with mouse anti-CIZ1 primary antibody (1:10,000; ab172442, Abcam) in phosphate-buffered saline with Tween 20 (PBST) containing 5% non-fat dry milk. Membranes were washed three times with 1 × PBST for 15 min and then incubated with horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 h with constant rocking at room temperature. Signal was detected using enhanced chemiluminescence (Amersham, Pittsburgh, PA).
2.6. Immunohistochemistry
Adult (5 mo old) Ciz1−/− mice (n = 4) and WT littermates (n = 4) were overdosed with intraperitoneal pentobarbital (100 mg/kg) and perfused transcardially, first with ice cold saline and then 4% paraformaldehyde in 0.1M phosphate buffer (PB, pH 7.4). Brains were removed and post-fixed with 4% paraformaldehyde and cryoprotected with 30% sucrose in 0.1M phosphate-buffered saline (PBS). The spleen and other organs (kidney, lung, liver, and colon) were blocked and embedded in paraffin. WT and Ciz1−/− hindbrains were sectioned on a cryostat at 40 µm. The spleen was sectioned at 5 µm on a microtome. Paraffin sections were deparaffinized, rehydrated, treated with an antigen unmasking solution (Citric Acid Based H-3300, Vector Laboratories, Burlingame, CA, USA) and rinsed with distilled water and PBS. Endogenous peroxidases were quenched with 1% H2O2 in PBS and sections were rinsed with PBS followed by blocking with 2% non-fat dry milk and 0.3% Triton X-100. Sections were incubated with rabbit anti-CIZ1 antibody (1:500, A301-496A, Bethyl Laboratories, Montgomery, TX, USA) overnight, rinsed and incubated with biotinylated goat anti-rabbit antibody (Jackson ImmunoResearch Laboratories) for 4 h. After rinsing with PBS × 3, sections were developed using the Vectastain ABC Kit from Vector Laboratories and DAB (3.3′-diaminobenzidine). Sections were mounted onto Superfrost™ Plus glass slides (Thermo Fisher Scientific, Waltham, MA, USA) and counter stained with cresyl violet to identify the anatomical locations of CIZ1-immunoreactivity (IR). Contiguous sections from the spleen were stained with hematoxylin and eosin.
2.7. Behavioral assessments
Adult (3 mo) Ciz1−/− mice (n = 15 male and 17 female) and gender-matched Ciz1+/+ littermates (n = 16 male and 15 female) were subjected to a battery of motor and behavioral examinations including open-field activity, rotarod, vertical rope climbing, raised-beam task, grip strength, gait analysis (DigiGait™), dominance tube, and cross-maze. Mice were examined weekly for evidence of dystonia or other involuntary movements during open-field activity and routine handling until 10 mo of age and then euthanized for whole-genome gene expression analyses. Righting reflex assays were performed prior to weaning in independent groups of Ciz1+/+ (n = 3 males and 2 females) and Ciz1−/− (n = 3 males and 1 female) mice.
Open-field activity assay
Mice were placed in commercially-available activity monitors (MED Associates, Inc., Georgia, VT, USA) for 10-min sessions. The activity monitors measured 27 × 27 cm, with 16 infrared photocell beams equally spaced in the x and y axes of the horizontal plane, 1 cm from the floor of the chambers. An additional array of 16 photocells was situated 5 cm above the floor to track rearing. All tests were conducted in the dark. One hour prior to testing, cages were moved from the housing racks to a quiet anteroom adjacent to the testing room. Following this period of habituation, animals were removed from their home cage, immediately placed in the center of the open field and allowed to freely explore the apparatus for a test interval of 10 min. Animals were scored for a number of behaviors including total distance traveled (cm). The data recorded during testing was scored in post-session analyses using commercially-available software (Activity Monitor 5.1, Med Associates). The testing apparatus was cleaned with a 70% ethanol solution and allowed to air dry between mice. Mice were given 1 trial at the same time daily for 3 consecutive days. Median values of activity parameters were used for statistical comparisons.
Rotarod
Mice were acclimated to a Rotamex-5 rotarod (Columbus Instruments, Columbus, OH, USA) rotating at 5 rpm for 5 min on the day prior to data acquisition. On the following day, mice were exposed to a 30 s acclimation period at 4 rpm followed by an acceleration of 4 rpm every 30 s to a target of 40 rpm at 5 min. Mice were given 3 trials at the same time each day for 5 consecutive days. Median values were used for statistical comparisons.
Vertical rope climb
Mice were acclimated to a vertical, 40-cm long, 10-mm thick nylon rope prior to testing. The bottom of the rope was suspended 15 cm above a padded base and the top entered into a darkened escape box. Three trials with a 5-min inter-trial interval were completed for each mouse. Median times were used for statistical analysis.
Raised-beam task
Mice were acclimated to an 80-cm long, 20-mm wide beam elevated 50 cm above a padded base. A 60 W lamp at the start served as an aversive stimulus, whereas the opposite end of the beam entered a darkened escape box. Transversal time and number of slips were measured as mice traversed the beam. After initial testing with a 20-mm diameter square beam, mice were given follow-up tests using supplementary round (12-mm and 9-mm diameter) and square (12-mmand 9-mmdiameter) beams. All testing was performed in triplicate and median values were used for subsequent statistical analyses.
Grip strength analysis
To measure grip strength, mice were held by the scruff of the neck with one hand and the base of the tail with the other hand. Mice were then free to grasp a metal grid attached to a force meter (Columbus Instruments, Columbus, OH, USA) as they were moved along the axis of the grid. Maximal strength (g) with which mice pulled the grid was measured in triplicate trials with a minimal inter-trial interval of 5 min. Median values were used for subsequent statistical analyses.
Gait analysis
The DigiGait™imaging system consists of a motorized treadmill with a digital camera positioned below a transparent belt (Mouse Specifics Inc., Boston, MA). For each mouse, the location and timing of each paw contact on the belt was automatically recorded at a belt speed of 20 cm/s for 5 s. A minimum of 800 video frames collected at 160 frames/s was digitized and gait parameters were calculated by the accompanying software (Supplemental Fig. 1). Three trials with 30 min inter-trial intervals were completed for each mouse. Median values were used for statistical analysis.
Righting reflex
Mice (Postnatal Day 6 [P6] to P14) were placed in the supine position and then released. The time required for all four limbs to contact the tabletop is measured for three trials.
Cross-Maze analysis
The testing apparatus consisted of 4 radial arms, each 25 cm-long, and a camera along with ANY-maze software (Stoelting Co., Wood Dale, IL, USA). Mice were placed into the apparatus for 5 min and entries into each arm were recorded sequentially using an ABCD format with each letter representing an individual arm of the maze. Alternation was counted if mice went into radial arms sequentially (% alternations = 100 × total alternations / [total entries − 3]).
Dominance tube test
This test was used for identifying a general abnormality in social interaction through the measurement of aggression. Ciz1+/+ and Ciz1−/− mice were released into opposite ends of a dark, narrow tube (30-cm long). Ciz1+/+ and Ciz1−/− mice interacted in tube and the more dominant or aggressive mouse forced its opponent out of the tube. When one animal had all four paws out of the tube, it was declared the loser while the animal remaining inside the tube was the winner, ending the match. The number of wins was reported as a percentage of the total number of matches. The match was deemed a draw when both mice remained in the tube >120 s.
2.8. Whole-genome gene expression analysis
Total RNA from mouse cerebellum was isolated from6 adult (10mo) Ciz1−/− mice (3 males and 3 females) and 6 age- and gender- matched Ciz1+/+ controls (Ambion™ TRI Reagent®, ThermoFisher Scientific). This time point was chosen for analysis of gene expression given that the incidence of cervical dystonia in humans is highest in middle-aged adults (Xiao et al., 2012, Vemula et al., 2013). The quality of total RNA was assessed with a NanoDrop® ND-1000 spectrophotometer (NanoDrop Technologies) and RNA integrity was verified with an Agilent 2100 Bioanalyzer using the Agilent RNA 6000 Nano kit. Whole-genome gene expression data were generated with the Affymetrix GeneChip® Mouse Gene 2.0 ST Array (Santa Clara, CA, USA). This array was designed using data from RefSeq (release 51), Ensembl (release 65) and lncRNA db and provides comprehensive coverage of over 30,000 mRNA transcripts and 2000 lincRNA transcripts. The 2.0 ST Array was designed with a median of 22 25-mer probes per transcript. Each array includes background antigenomic probes, poly-A controls and hybridization controls. Target RNA was first reverse transcribed into cDNA, followed by in-vitro transcription to generate biotin-labelled cRNA for subsequent hybridization. Hybridized target cRNA were stained with streptavidin phycoerythrin and scanned using an Affymetrix GeneArray Scanner.
Data were processed using Affymetrix Expression Console software that incorporates the Robust Multi-array Average (RMA) normalization algorithm. Genes were annotated using Affymetrix MoGene 2.0 ST, V.1, release 35 annotation files from NetAffx™ server of Affymetrix. The RMA-normalized .chp files were summarized further with GeneSpring GX® 13.1.1 (Agilent® Technologies, Santa Clara, CA). Scatter plots were used to access the reproducibility of gene expression within genotypes (Supplemental Figs. 2 & 3). The t-test statistic was used in identifying significantly dysregulated genes (p ≤ 0.05) betweenWT and Ciz1−/− mice. Differential gene expression was also analyzed using a False Discovery Rate (FDR) corrected p ≤ 0.05 (Benjamini and Hochberg, 1995). A heat map created using unsupervised hierarchical clustering with average linkage and Euclidean distance (Supplemental Fig. 4) and a volcano plot (Supplemental Fig. 5) were used to visualize differential gene expression. Using WebGestalt (WEB-based GEne SeT AnaLysis Toolkit) and Ingenuity Pathway Analysis (IPA), we investigated the effects of differentially regulated genes on pathways (KEGG, Kyoto Encyclopedia of Genes and Genomes) and molecular/cellular networks (Zhang et al., 2005) using a stepwise collection of fold changes (1.10 [10%], 1.15 [15%], 1.20 [20%], and 1.25 [25%]). Data is provided for KEGG and IPA pathways enriched by genes with fold changes of 1.10 and 1.20, respectively.
To validate data obtained with the Affymetrix GeneChip® Mouse Gene 2.0 ST Array, we selected 6 up-regulated and 6 down-regulated genes, moderately expressed in cerebellum, for QRT-PCR. Using Taqman probes, QRT-PCRwas performed on the Roche LightCycler® 480 system with primers designed on the Roche Universal ProbeLibrary Assay Design Center (https://lifescience.roche.com/shop/en/us/overviews/brand/universal-probe-library). A total of 16 cerebellar RNA samples from WT and Ciz1−/− mice (8 males and 8 females in each group, including the 6 samples used for whole-genome gene expression analysis) were employed for validation with β-actin as the endogenous control. Technical triplicates were performed for all samples and median values were utilized for statistical analysis.
2.9. Statistics
ANOVA with post-hoc tests was used to determine the effects of genotype and gender on parametric behavioral measures. The Mann-Whitney test was used to determine the effects of genotype within gender for a non-parametric behavioral measure (slips on the raised beam task). Two-tailed t-tests were used to determine the effects of genotype on hematology and chemistry parameters. Fisher's exact test was used to determine the effects of genotype on the results of dominance tube testing. An alpha (α) of 0.05 was chosen for statistical significance.
3. Results
3.1. Developmental expression of Ciz1
Data were analyzed by normalizing Ciz1 expression to E15 liver (Figs. 1B–C). Dissection of all brain regions, readily identifiable in older mice (midbrain, cerebral cortex, striatum, cerebellum, thalamus, hippocampus, and spinal cord), was not possible at E15 and P1. There was a significant effect of region on expression of Ciz1 (p < 0.0001, for both B and C). However, there was no effect of gender on expression of either 31F/31R isoforms (B) or 18F/18R isoforms (C). At P21 and later postnatal time points, expression of Ciz1 isoforms amplified by 31F/31R was highest in cerebellum; with a peak at 6 mo that was approximately 2.5 fold higher than liver. Cerebellar expression of isoforms amplified by 18F/18R showed two peaks, one at P21 and the other at 6 mo. Brain expression of Ciz1 increased with increasing postnatal age (p < 0.0001, for both B [31F/31R] and C [18F/18R]).
3.2. Basic characterization of Ciz1 gene-trap mice
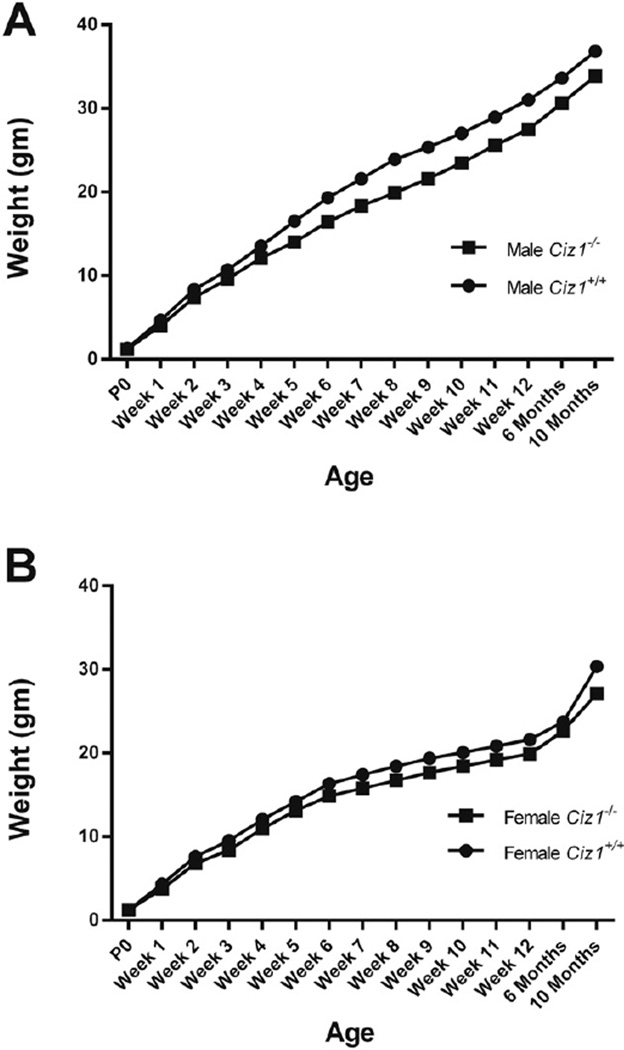
Male and female Ciz1+/− and Ciz1−/− mice were fertile and pups of all genotypes (Ciz1+/+, Ciz1+/−, Ciz1−/−) and both genders were born at normal Mendelian ratios. Ciz1−/− mice showed no dysmorphic features or gross pathological abnormalities of the brain. As shown in Table 2, there were significant effects of gender (F1,59 = 254, p < 0.0001) and genotype (F1,59 = 24, p < 0.0001) on weights in 3-mo old mice. Overall, Ciz1−/− mice were 10 to 15% smaller than their WT littermates at 3 mo. However, there was no evidence for progressive weight loss in Ciz1−/− mice up to 10 mo (Fig. 2). There were no statistically significant differences in hematological or serum chemistry parameters between WT and Ciz1−/− mice (Supplemental Table 2). However, there were notable trends towards higher monocyte and neutrophil counts in Ciz1−/− mice in comparison to WT littermates (raw p = 0.03, for both).
Table 2.
Effects of genotype and gender on weight and behavioral measures in 3-month old mice. Ambulatory count, the total number of X + Y photo beam breaks while in ambulatory movement status. Stereotypic count, any partial-body movements that occur within the ambulatory box such as grooming, head-weaving or scratching. Vertical count, number of periods of continuous Z photo beam breaks. Jump count, the number of times that the mouse leaves the photo beam array for a period of time. Ambulatory episodes, the number of times the mouse has started moving after the resting delay has expired. Values are means ± SEM except for dominance tube.
| Male | Female | |||
|---|---|---|---|---|
| Ciz1+/+(n = 16) | Ciz1−/−(n = 15) | Ciz1+/+(n = 15) | Ciz1−/−(n = 17) | |
| Weight (g) | 31.0 ± 0.8 | 27.5 ± 0.9* | 21.6 ± 0.6 | 19.9 ± 0.4* |
| Grip strength (g) | 329.2 ± 7.7 | 289.3 ± 9.8* | 288.9 ± 7.9 | 268.5 ± 8.8* |
| Grip strength/weight | 10.6 ± 0.6 | 10.5 ± 0.6 | 13.4 ± 0.7 | 13.5 ± 0.8 |
| Dominance tube | 27% | 71%* | 39% | 59% |
| Cross maze score (%) | 33.5 ± 5.1 | 31.2 ± 7.5 | 32.1 ± 5.9 | 30.5 ± 8.1 |
| Rope climbing (s) | 5.0 ± 0.7 | 5.3 ± 1.1 | 4.8 ± 1.4 | 4.5 ± 1.3 |
| Rope climbing/weight | 0.16 ± 0.02 | 0.19 ± 0.04 | 0.22 ± 0.06 | 0.23 ± 0.06 |
| Open field activity | ||||
| Distance traveled (cm) | 1858.2 ± 57.4 | 2180.7 ± 79.3* | 2452.3 ± 94.8 | 1895.8 ± 103.2* |
| Ambulatory count | 897.3 ± 36.0 | 1165.8 ± 55.2* | 1280.1 ± 100.1 | 908.6 ± 89.6* |
| Stereotypic count | 2597.3 ± 102.0 | 2684.7 ± 121.3 | 2582.0 ± 117.2 | 2108.0 ± 99.4* |
| Vertical count | 190.5 ± 11.7 | 125.0 ± 9.2* | 141.6 ± 10.6 | 96.8 ± 8.1* |
| Jump count | 57.4 ± 4.0 | 45.9 ± 3.1* | 39.6 ± 3.7 | 30.2 ± 2.9* |
| Average velocity (cm/s) | 36.2 ± 1.3 | 45.5 ± 1.9* | 43.2 ± 2.5 | 44.1 ± 3.1 |
| Ambulatory episodes | 85.4 ± 4.1 | 104.2 ± 4.2* | 104.2 ± 4.2* | 83.9 ± 4.5* |
| DigiGait™ | ||||
| Propel (s) forelimb | 0.125 ± 0.005 | 0.123 ± 0.003 | 0.114 ± 0.004 | 0.110 ± 0.005 |
| Propel (s) hindlimb | 0.180 ± 0.006 | 0.172 ± 0.004 | 0.166 ± 0.005 | 0.157 ± 0.006 |
| Stride length (cm) forelimb | 6.85 ± 0.21 | 6.57 ± 0.11 | 6.25 ± 0.10 | 5.93 ± 0.12 |
| Stride length (cm) hindlimb | 7.07 ± 0.21 | 6.95 ± 0.11 | 6.57 ± 0.14 | 6.25 ± 0.16* |
| Stride Frequency (steps/s) forelimb | 3.05 ± 0.10 | 3.15 ± 0.05 | 3.27 ± 0.18 | 3.48 ± 0.15 |
| Stride Frequency (steps/s) hindlimb | 2.94 ± 0.08 | 3.03 ± 0.07 | 3.15 ± 0.12 | 3.44 ± 0.14* |
| Stance width (cm) forelimb | 1.46 ± 0.04 | 1.44 ± 0.04 | 1.50 ± 0.06 | 1.41 ± 0.11 |
| Stance width (cm) hindlimb | 2.73 ± 0.07 | 2.49 ± 0.04* | 2.33 ± 0.09 | 2.23 ± 0.08 |
| Step angle (deg) forelimb | 68.08 ± 2.23 | 66.48 ± 1.70 | 67.53 ± 2.01 | 65.24 ± 2.30 |
| Step angle (deg) hindlimb | 55.60 ± 2.09 | 56.32 ± 1.00 | 64.11 ± 2.00 | 58.77 ± 1.89* |
| Paw Area (cm2) forelimb | 0.22 ± 0.01 | 0.19 ± 0.02* | 0.20 ± 0.01 | 0.19 ± 0.01 |
| Paw Area (cm2) hindlimb | 0.45 ± 0.01 | 0.40 ± 0.02* | 0.38 ± 0.02 | 0.34 ± 0.02* |
p < 0.05, for effect of genotype within gender.
Fig. 2.
Growth curves for (A) male and female (B) Ciz1−/− mice (n = 15 males and 17 females), and Ciz1+/+ littermates (n = 16 males and 15 females).
Ciz1 transcripts were reduced by approximately 50% in Ciz1+/− mice and absent in Ciz1−/− mice (Table 1). Expression of Ciz1 in 3 month-old brain was highest in cerebellum, about two-fold higher than liver. The relative expression of Ciz1 was similar with two independent pairs of PCR primers: cerebellum > cerebral cortex > midbrain > thalamus > striatum > spinal cord > hippocampus.
Table 1.
Expression of Ciz1 in gene-trap mice.
| Tissue | Ciz1+/+ | Ciz1+/− | Ciz1−/− | |||
|---|---|---|---|---|---|---|
| Primers 1* | Primers 2** | Primers 1 | Primers 2 | Primers 1 | Primers 2 | |
| Liver | 1.00 ± 0.07 | 1.00 ± 0.02 | 0.62 ± 0.03 | 0.54 ± 0.02 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Cerebral cortex | 1.67 ± 0.03 | 1.59 ± 0.02 | 0.97 ± 0.02 | 0.90 ± 0.02 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Cerebellum | 1.91 ± 0.08 | 2.12 ± 0.07 | 1.16 ± 0.04 | 1.16 ± 0.02 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Hippocampus | 1.03 ± 0.02 | 0.97 ± 0.06 | 0.66 ± 0.06 | 0.49 ± 0.03 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Midbrain | 1.52 ± 0.06 | 1.54 ± 0.02 | 0.86 ± 0.04 | 0.79 ± 0.01 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Striatum | 1.13 ± 0.07 | 1.03 ± 0.01 | 0.80 ± 0.02 | 0.67 ± 0.02 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Thalamus | 1.41 ± 0.06 | 1.38 ± 0.04 | 0.85 ± 0.02 | 0.78 ± 0.01 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Spinal cord | 1.19 ± 0.05 | 1.25 ± 0.03 | 0.72 ± 0.02 | 0.68 ± 0.03 | 0.00 ± 0.00 | 0.00 ± 0.00 |
Primers 1, Ciz1_Q31F/R, using β-actin as the endogenous control.
Primers 2, Ciz1_Q40F/R, using hypoxanthine-guanine phosphoribosyltransferase (HGPRT) as the endogenous control. QRT-PCR values are referenced to wild-type (Ciz1+/+) liver and presented as means ± SEM (n = 3 mice/genotype).
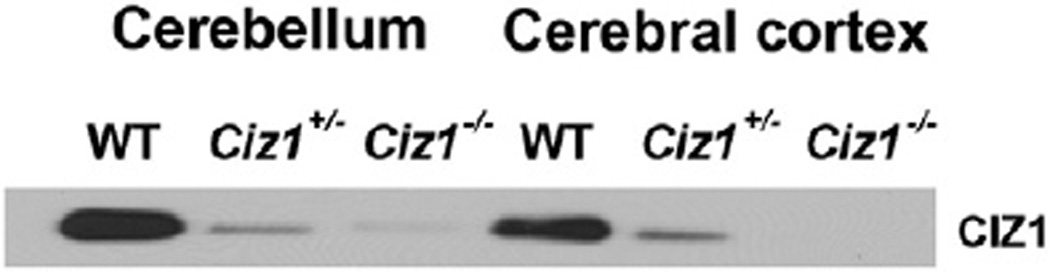
Qualitative Western blotting was compatible with the QRT-PCR results (Fig. 3). However, despite attempts with several commercially available antibodies (Bethyl Laboratories A301-496A and A301-497A, Aviva ARP34296-P050, Santa Cruz sc-107,184, Abcam ab172442, and Novus NBP2-33890) generated against CIZ1, we were unable to generate Western blot and immunohistochemical data without some degree of non-specific immunoreactivity (IR) in Ciz1−/− mice (Supplemental Figs. 6 & 7). Moreover, none of the antibodies produced convincing staining of CIZ1 immunoreactivity without background staining in WT mouse brain. In contrast, non-specific immunoreactivity was relatively minimal in extra-neural organs such as the spleen (Supplemental Fig. 7).
Fig. 3.
Qualitative Western blotting of immunoprecipitated lysates from either cerebellum or cerebral cortex from adult (3 mo) WT, Ciz1+/−, and Ciz1−/− mice for detection of mouse CIZ1.
3.3. Sensorimotor and behavioral analyses
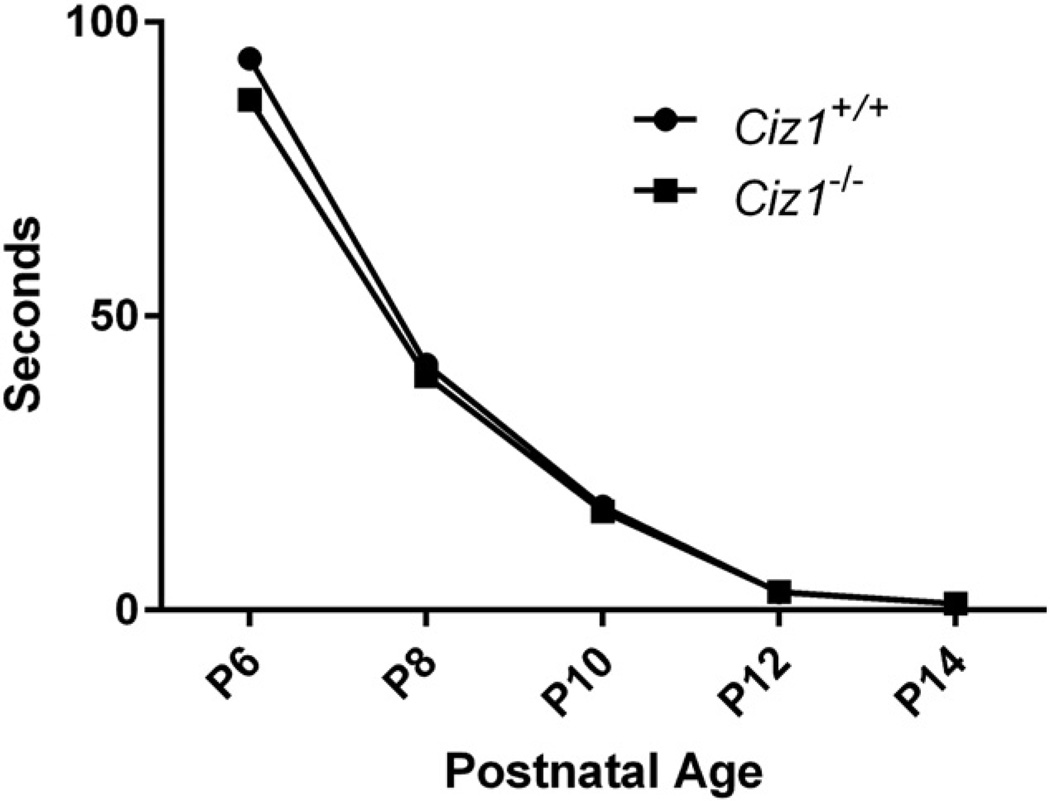
No evidence of dystonia was noted while mice were routinely observed in their home cages, during open field behavior, and while handling from the early postnatal period through 10 mo of age. WT and Ciz1−/− mice showed no overt differences on righting in the preweanling period (Fig. 4). There were significant effects of gender (F1,59 = 24.9, p < 0.0001) and genotype (F1,59 = 24.3, p < 0.0001) on grip strength (Table 2). Male WT mice exhibited greater grip strength than Ciz1−/− littermates (mean difference = 40 g, p < 0.0001). The effect of genotype was smaller in female mice (mean difference = 20 g, p = 0.021). However, after normalized by weight, the effect of genotype on grip strength was no longer apparent (p = 0.99). Male Ciz1−/− mice were more aggressive than their WT littermates in the tube dominance test (p = 2.7E-05). However, the effect of genotype was not significant in female mice (p = 0.090). There were no effects of genotype on rope climbing or cross maze scores.
Fig. 4.
Righting reflex for Ciz1−/− mice (n = 3 males and 1 female) and Ciz1+/+ littermates (n = 3 males and 2 females).
There were significant genotype*gender interactions on open-field assays of distance traveled, ambulatory count, and ambulatory episodes (p < 0.0001, for all). In aggregate, male Ciz1−/− mice were more active than WT littermates whereas the reverse was apparent for female Ciz1−/− mice (Table 2). In contrast, there were large effects of genotype on vertical counts (F1,59 = 14.8, p = 0.0003) and jump counts (F1,59 = 33.9, p = 0.0001) but no effect of the genotype*gender interaction with both male and female WT mice showing significantly more vertical and jump counts than their gender-matched Ciz1−/− littermates.
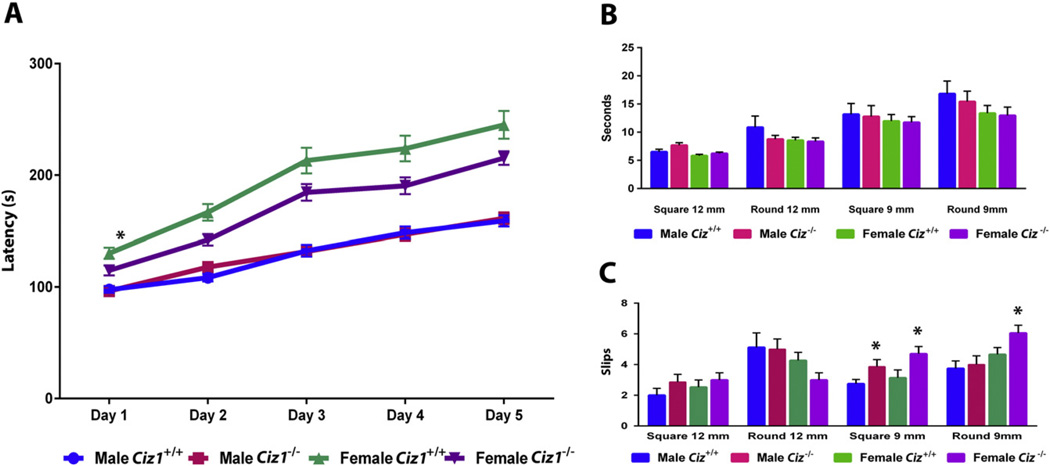
DigiGait™ analyses (Supplemental Fig. 1) showed moderate effects of genotype (Table 2). Female Ciz1−/− mice had shorter stride lengths (p = 0.016) and higher stride frequencies (p < 0.0001) than WT gender-matched littermates. Hindpaw areas were smaller in Ciz1−/− mice than WT littermates (F1,59 = 31.1, p < 0.0001). Overall, hindpaw stance width was narrower in Ciz1−/− mice than WT littermates (F1,59 = 71.2, p < 0.0001) although the difference between Ciz1−/− and WT females did not reach statistical significance (p = 0.075). On the rotarod, WT and Ciz1−/− mice showed increased latencies to fall from Day 1 through Day 5 (Fig. 5). There was no overall effect of genotype but female Ciz1−/− mice tended to fall off the rotarod at shorter latencies than WT females (p = 0.020). Female mice also had more slips on the 9-mm square (p = 0.012) and 9-mm round beams (p = 0.102). There were no significant effects of gender or genotype on beam traversal times.
Fig. 5.
Performance of Ciz1−/− mice (n = 15 males and 17 females) and Ciz1+/+ littermates (n = 16 males and 15 females) on the rotarod (A) and raised beam tasks (B and C). Beam traversal times (B) and slips (C) were recorded for 4 different beams. *Significant effect of genotype within gender (p < 0.05).
3.4. Whole-genome gene expression analysis
As shown in Supplemental Figs. 2 & 3, biological replicates showed high linear correlations throughout the ranges of gene expression for WT and Ciz1−/− cerebellar samples. The relative symmetric volcano plot (Supplemental Fig. 5) and stepwise decline in the number of dysregulated genes at specific fold changes (Table 3) also attests to the quality of our gene expression analyses. At an FDR of ≤0.05, only 3 genes showed significantly altered expression (Ciz1 [−3.11×], Dtx4 [−1.12×], and 1110008P14Rik [1.76×]). Dtx4 encodes an E3 ubiquitin ligase. It is not known if CIZ1 is a target of DTX4. In mouse, 1110008P14Rik is a 2-exon, protein-coding gene located on Chr. 2. Its human ortholog, C9orf16, is a 3-exon gene located on the forward strand of Chr. 9q34.1 adjacent to CIZ1. Without correction for family-wise error, we found 94 and 53 genes that were up- or down-regulated ≥1.25×, respectively (Table 3, Supplemental Table 3). At a fold-change of 1.5×, 9 genes showed down-regulation and 10 genes were up-regulated in Ciz1−/− cerebellum (Table 4). QRT-PCR corroborated the direction and relative magnitude of differential expression for 9 out of the 12 genes that were assayed (Table 5).
Table 3.
Numbers of dysregulated genes at specific fold changes in Ciz1−/− mice.
| Fold Change | Genes dysregulated (p < 0.05) | |
|---|---|---|
| Up | Down | |
| 1.10× (10%) | 679 | 673 |
| 1.15× (15%) | 329 | 276 |
| 1.20× (20%) | 175 | 124 |
| 1.25× (25%) | 94 | 53 |
Table 4.
Dysregulated genes in Ciz1−/− mice.
| Symbol | Gene name and description | Fold change (> 1.5 and p < 0.05) |
|---|---|---|
| Ciz1 | CDKN1A interacting zinc finger protein 1 | −3.11 |
| Mid1 | Midline 1 | −2.02 |
| Slc17a6 | Solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 |
−1.94 |
| Gm15726 | Predicted gene 15,726 | −1.90 |
| Kcnc2 | Potassium voltage gated channel, Shaw-related subfamily, member 2 |
−1.60 |
| Gm22956 | Predicted gene 22956 | −1.60 |
| Nr4a2 | Nuclear receptor subfamily 4, group A, member 2 | −1.60 |
| C1ql3 | C1q-like3 | −1.53 |
| Cd24a | CD24a antigen | −1.52 |
| mt-Ty | Mitochondrially encoded tRNA tyrosine | 1.56 |
| Gm10325 | Predicted gene 10,325 | 1.58 |
| Tomm5 | Translocase of outer mitochondrial membrane 5 homolog (Yeast) |
1.59 |
| mt-Tn | Mitochondrially encoded tRNA asparagine | 1.60 |
| Gm20236 | Predicted gene 20,236 | 1.64 |
| mt-Tt | Mitochondrially encoded tRNA threonine | 1.73 |
| 1110008P14Rik | RIKEN cDNA 1110008P14 gene | 1.76 |
| Prl2c3 | Prolactin family 2, sub family C, member 3 | 1.94 |
| Gm10002 | Predicted gene 10,002 | 2.54 |
| Gm21464 | Predicted gene 21,464 | 3.14 |
Table 5.
Validation of whole-genome gene expression with QRT-PCR.
| Gene name |
Protein | Function | Fold change (p value)* | |
|---|---|---|---|---|
| Microarray | QRT-PCR | |||
| Nup35 | Nucleoporin 35 | Component of the nuclear pore complex | 1.48 ± 0.10 (0.019) | 1.07 ± 0.07 (0.445) |
| Zbtb16 | Zinc finger protein 145 | Transcription factor | 1.41 ± 0.19 (0.041) | 2.46 ± 0.43 (0.003) |
| Entpd4 | Ectonucleoside triphosphate diphosphohydrolase 4 | Hydrolase activity | 1.35 ± 0.05 (0.0004) | 1.27 ± 0.11 (0.042) |
| Npy | Pro-neuropeptide Y | Involved in feeding and secretion of gonadotrophin-releasing hormone |
1.29 ± 0.12 (0.019) | 1.11 ± 0.08 (0.253) |
| Mpp4 | MAGUK p55 subfamily member 4 | Synaptic function | 1.25 ± 0.03 (0.00003) | −1.04 ± 0.05 (0.206) |
| Creld2 | Cysteine-rich with EGF-like domains 2 | Calcium ion binding | 1.23 ± 0.08 (0.008) | 1.27 ± 0.08 (0.045) |
| Psat1 | Phosphoserine aminotransferase | Phosphoserine aminotransferase activity | −1.27 ± 0.04 (0.002) | −1.11 ± 0.07 (0.066) |
| Psat1 | Phosphoserine aminotransferase | Phosphoserine aminotransferase activity | −1.27 ± 0.04 (0.002) | −1.11 ± 0.07 (0.066) |
| Nkain2 | Sodium/potassium-transporting ATPase subunit beta-1-interacting protein 2 |
Regulation of sodium ion transport | −1.27 ± 0.04 (0.004) | −1.52 ± 0.14 (0.018) |
| Zfhx4 | Zinc finger homeobox protein 4 | Transcriptional regulation | −1.34 ± 0.06 (0.013) | −1.82 ± 0.08 (0.011) |
| Nr4a1 | Nuclear receptor subfamily 4 group A member 1 | Orphan nuclear receptor | −1.36 ± 0.05 (0.031) | −1.52 ± 0.05 (0.008) |
| Opalin | Oligodendrocytic myelin paranodal and inner loop protein | Integral component of membrane | −1.37 ± 0.07 (0.010) | −1.45 ± 0.06 (0.015) |
| Nr4a2 | Nuclear receptor subfamily 4 group A member 2 | Transcriptional regulator | −1.60 ± 0.07 (0.005) | −1.75 ± 0.06 (0.006) |
Means ± SEM (p value).
Genes showing the greatest up-regulation (Table 4) included a nuclear encoded mitochondrial protein (Tomm5) and 3 mitochondrial transfer RNAs (tRNAs). The top down-regulated genes included those encoding an E3-ubiquitin ligase (Mid1), two channel proteins (vesicular glutamate cotransporter [Slc17a6] and voltage gated potassium channel [Kcnc2]), a complement protein (C1ql3) and cell adhesion molecule (Cd24a). Overall, a relatively large percentage of up- or down-regulated genes are involved in metabolic pathways (Supplemental Tables 3–5). Other enriched KEGG pathways for up-regulated genes included cytokine-cytokine receptor interactions and signal transduction (Supplemental Table 4). Focal adhesion, neuroactive ligand-receptor interactions and MAPK signaling were the more heavily enriched pathways for down-regulated genes (Supplemental Table 5).
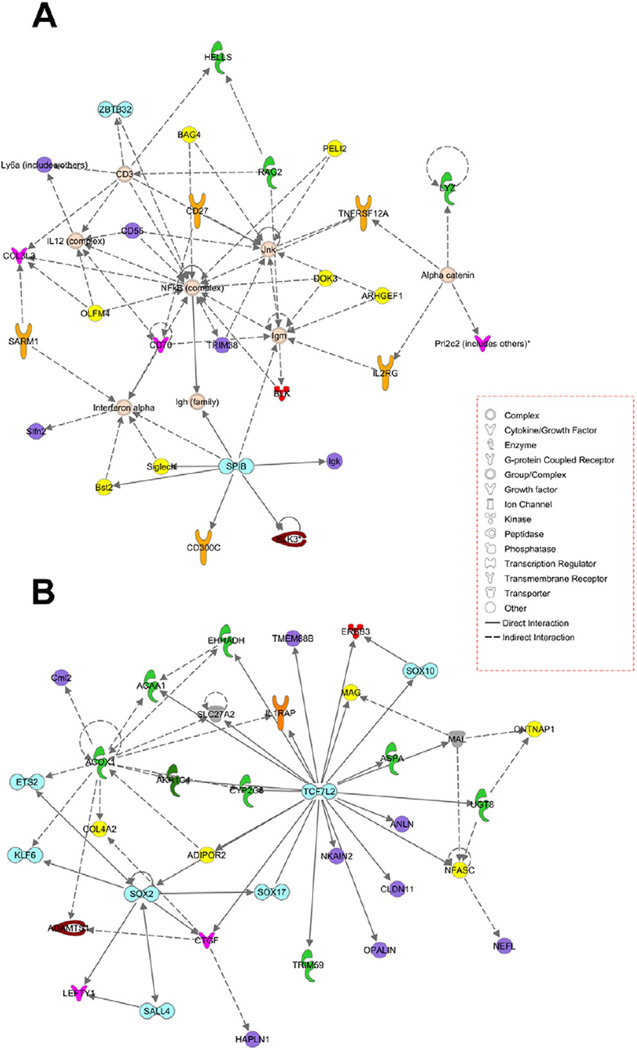
IPA identified cell development, growth, proliferation, and morphology as the top up-regulated molecular and cellular functions and primary immunodeficiency signaling was the top canonical pathway. Cell cycle, cellular development, cell death and survival, and gene expression were down-regulated molecular and cellular functions (Supplemental Table 5). Similarly, embryonic and nervous system development were down-regulated physiological functions. Interactome analysis identified NF-κB and TCF7L2 as the major hubs for up- and down-regulated genes, respectively (Fig. 6). Jnk (c-Jun. N-terminal kinase) was a secondary hub for up-regulated genes.
Fig. 6.
Ingenuity interactome analysis of up-regulated (A) and down-regulated (B) genes in Ciz1−/− mouse cerebellum.
4. Discussion
Similar to human CIZ1 which expresses 7 RefSeq transcripts, mouse Ciz1 expresses 5 RefSeq transcripts, harbors 17 exons, and the first exon is non-coding (Fig. 1). However, Ciz1 is subject to extensive alternative splicing during development and disease states and several cell types have been shown to express numerous additional transcripts (Greaves et al., 2012, Rahman et al., 2007, Rahman et al., 2010). Partial identity at the protein level (65%) suggests that, in comparison to mouse CIZ1, human CIZ1 could have additional or somewhat different cellular roles. Compatible with Northern blot analysis (Warder and Keherly, 2003), data reported by BioGPS (BioGPS.org) and Allen Brain Atlas (www.brain-map.org), we showed that Ciz1 is widely expressed in adult mouse brain and found that cerebellum had the highest expression level among those sensorimotor structures assayed in our study. However, we did not specifically identify and characterize the relative contribution of individual splice variants to the total pool of Ciz1 transcripts during development of the CNS.
Although we were unable to generate irrefutable proof at the protein level, QRT-PCR showed that Ciz1 transcription was eliminated in our gene trap Ciz1−/− mouse model. In another Ciz1-deficient mouse model (Nishibe et al., 2013), Ciz1 expression was only accessed with Northern blotting for which technical details are not provided. Nishibe and colleagues generated their Ciz1 mutant mice by flanking Exon 5 of Ciz1 with loxP sites and crossing floxed mice with CAG—Cre transgenic mice (Nishibe et al., 2013). The Nishibe et al. Ciz1 mutant mice were born at expected Mendelian ratios and no overt abnormalities were identified during a 1.5 year period of observation although the authors do not actually describe behavioral, morphological or pathological phenotyping (Nishibe et al., 2013). However, CIZ1 deficiency rendered cells from mutant mice sensitive to oncogenic transformation. Although we did not pathologically phenotype mice, given the modest quantitative differences in neutrophil and monocyte counts noted between Ciz1−/− mice and their WT littermates, it is possible that CIZ1 deficiency exerts deleterious effects on the maturation of lymphoid and myeloid cells and increases the risk for hematological malignancies in older mice (Nishibe et al., 2013).
Our Ciz1−/− mice generated with gene trap technology were smaller than WT littermates and showed several abnormalities on sensorimotor testing. Ciz1−/− mice were less active in the vertical axis than WT littermates. Overall, female Ciz1−/− mice exhibited more prominent motor deficits than male Ciz1−/− mice, particularly on the rotarod and raised beam task. In contrast, male Ciz1−/− mice were more aggressive than WT littermates in the tube dominance test. Although the cellular underpinnings of these effects were not characterized in our study, it has been shown that CIZ1 co-regulates estrogen receptor α (ERα) by enhancing estrogen transactivation activity by promoting recruitment of the estrogen complex to target gene chromatin (Den Hollander et al., 2006). Moreover, estrogen and other sex hormones are known to exert important effects on cell-cycle checkpoints and other cell cycle proteins (Heckler and Riggins, 2015). In the context of dystonia, it should be noted that most patients with isolated dystonia are females with age of onset in the perimenopausal period (LeDoux, 2012a; LeDoux, 2012b). Comparably, the incidence of breast cancer rises in the postmenopausal period (Jatoi et al., 2008).
Some of the motor aberrations seen in our Ciz1−/− mouse model are similar to those reported in KO, knock-in and transgenic mouse models of isolated or primary dystonia targeting Tor1a and Thap1 (Zhao et al., 2008, Ruiz et al., 2015, Dang et al., 2005, LeDoux, 2011). For example, increased slips on the raised beam task have been described in Tor1a+/− simple KO mice and hMT1 transgenic mice that overexpress mutant ΔE human torsinA (Dang et al., 2005, Song et al., 2012, Zhao et al., 2008). Similar to our Ciz1−/− mice, Thap1C54Y/+ mice and hMT1 transgenic mice show reduced fall latencies on the accelerating rotarod (Ruiz et al., 2015, Song et al., 2012). Directions of change are not consistent across all murine models of dystonia, however. For instance, increased hind stance widths were reported in waddles mice with dystonia but were reduced in our male Ciz1−/− mice (Jiao et al., 2005). Likewise, markedly increased distances traveled were described in Gnal−/− mice surviving to adulthood (Belluscio et al., 1998) whereas the modest effects of Ciz1 elimination on ambulation showed an interaction with gender.
Our study focused on the role of CIZ1 in the sensorimotor CNS and we did not perform pathological phenotyping to search for hematological or soft tissue malignancies (Nishibe et al., 2013). Moreover, we did not exclude the possibility that loss of CIZ1 is associated with quantitative morphological abnormalities of the brain as reported in Tor1a and Thap1 mutant mice (Goodchild et al., 2005, Dang et al., 2005, Zhang et al., 2011, Song et al., 2014). On the other hand, Ciz1−/− mice lived into adulthood, were able to mate successfully and did not exhibit premature death. These data suggest that neurons are able to compensate, in part, for germline deficiency of CIZ1, at least until early adulthood. However, we cannot exclude the possibility that CIZ1 contributes to maturation of one or more cognitive domains or that defective or deficient CIZ1 could cause cognitive decline in older mice (Yates et al., 2015) or plays a role in human Alzheimer's disease (Shen et al., 2016; Dahmcke et al., 2008). It is also possible that conditional KO (cKO) of Ciz1 in adult mice would illicit more robust sensorimotor phenotypes as is the case for Tor1a cKO mice (Liang et al., 2014).
Unlike our Ciz1−/− mice, homozygous KO of Tor1a and Thap1 is lethal in the embryonic or preweanling periods (Dang et al., 2005, LeDoux, 2012a; Goodchild et al., 2005, Tanabe et al., 2012). Approximately 5% of Gnal−/− mice survive to maturity (Belluscio et al., 1998). In contrast, homozygous mutations of other dystonia-associated genes such as CA8/Car8 and HPCA/Hpca are compatible with survival into adulthood (Jiao et al., 2005, Kobayashi et al., 2005). In fact, Hpca−/− mice show no abnormalities in motor activity (Kobayashi et al., 2005) despite the association of severe early-onset dystonia in humans with homozygous and compound heterozygous mutations in HPCA (Charlesworth et al., 2015).
Based on KEGG analysis, it appears that up- and down-regulation of genes in various metabolic pathways served to compensate for CIZ1 deficiency in our mouse model. For example, up-regulation of a nuclear-encoded mitochondrial transferase (Tomm5) and three mitochondrial tRNAs suggest the possibility that CIZ1 plays a role in repair of mitochondrial DNA (mtDNA) as it does for nuclear DNA (nDNA) (Leandro et al., 2015). Down-regulation of several cell surface to nucleus signaling pathways (Supplemental Table 4; TGF-β, MAPK, Toll-receptor, and B-cell) and down-regulation of 33 genes related to cellular development (Supplemental Table 5) suggests that germline deficiency of CIZ1 elicits a cascade of effects that differ from those caused by transient gene silencing (Liu et al., 2015). For illustration, siRNA silencing of Ciz1 in human colorectal cancer cells increases the ratio of cells in the G0/G1 phase and augments apoptosis (Yin et al., 2013).
NF-κB, a major hub for up-regulated genes in Ciz1−/− mice, plays an important role in transcription of DNA in response to cellular stress (Lanzillotta et al., 2015). NF-κB also contributes to the survival and post-mitotic differentiation of neurons (Kaltschmidt and Kaltschmidt, 2015). TCF7L2, the major hub for down-regulated genes, is also a transcription factor. Variants in human TCF7L2 have been associated with Type II diabetes and colorectal cancer (Segditsas and Tomlinson, 2006). In brain, the Wnt/β-catenin pathway, which utilizes TCF family members as effectors, plays a role in development and synaptic plasticity (Wisniewska, 2013). Down-regulation of this pathway in Ciz1−/− mice suggests that CIZ1 may participate in regulation of these same processes.
The role of CIZ1 in post-mitotic neurons of CNS is unclear. The DNA damage response in post-mitotic neurons includes cell-cycle arrest coupled with DNA repair, apoptosis, senescence and differentiation (Benes et al., 2009, Nguyen et al., 2002). However, the cell-cycle machinery, in general, and cell-cycle proteins active at the G1/S checkpoint may also contribute to terminal post-mitotic differentiation of neurons, early neuronal development, epigenetic regulation, signaling (ie., Wnt/β-catenin) and neuronal polarization (Lim and Kaldis, 2013). Cdks, cyclins and other cell-cycle proteins such as CIZ1 appear to have important physiological roles in post-mitotic neurons beyond cell-cycle regulation and repair of DNA double-strand breaks. Many of the up- and down-regulated genes identified by IPA are involved in cellular development and growth, important processes in post-mitotic neurons (Supplemental Table 5) (Herrup, 2013; Nguyen et al., 2002). However, given that our gene expression studies were limited to cerebellum in 10-month-old adult mice, it is quite possible that we failed to identify the effects of CIZ1 deficiency on other molecules and pathways during development and senescence. Moreover, we did not assay the potential differential effects of perturbations such as ionizing radiation on gene-expression profiles in Ciz1−/− and Ciz1+/+ mice.
5. Conclusions
Our data are compatible with the following: (1) Ciz1 gene-trap mice are a valid model of germline CIZ1 deficiency; (2) germline deficiency of CIZ1 is associated with mild motor dysfunction; and (3) up- and down-regulation of genes associated with metabolic pathways, cellular development and cellular growth may compensate, in part, for germline deficiency of CIZ1. Ciz1−/− mice on the C57BL/6J background do not manifest overt dystonia.
Supplementary Material
Acknowledgments
This study was supported by the Neuroscience Institute at the University of Tennessee Health Science Center, Dorothy/Daniel Gerwin Parkinson's Research Fund, and National Institutes of Health grants R01 NS082296 and R01 NS069936.
Footnotes
Appendix A. Supplementary information
Supplementary information to this article can be found online at http://dx.doi.org/10.1016/j.expneurol.2016.05.006.
References
- Ainscough JF, Rahman FA, Sercombe H, Sedo A, Gerlach B, Coverley D. C-terminal domains deliver the DNA replication factor Ciz1 to the nuclear matrix. J. Cell Sci. 2007;120:115–124. doi: 10.1242/jcs.03327. [DOI] [PubMed] [Google Scholar]
- Albanese A, Bhatia K, Bressman SB, Delong MR, Fahn S, Fung VS, Hallett M, Jankovic J, Jinnah HA, Klein C, Lang AE, Mink JW, Teller JK. Phenomenology and classification of dystonia: A consensus update. Mov. Disord. 2013;28:863–873. doi: 10.1002/mds.25475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belluscio L, Gold GH, Nemes A, Axel R. Mice deficient in G(olf) are anosmic. Neuron. 1998;20:69–81. doi: 10.1016/s0896-6273(00)80435-3. [DOI] [PubMed] [Google Scholar]
- Benes FM, Lim B, Subburaju S. Site-specific regulation of cell cycle and DNA repair in post-mitotic GABA cells in schizophrenic versus bipolars. Proc. Natl. Acad. Sci. U. S. A. 2009;106:11731–11736. doi: 10.1073/pnas.0903066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- Charlesworth G, Angelova PR, Bartolome-Robledo F, Ryten M, Trabzuni D, Stamelou M, Abramov AY, Bhatia KP, Wood NW. Mutations in HPCA cause autosomal-recessive primary isolated dystonia. Am. J. Hum. Genet. 2015;96:657–665. doi: 10.1016/j.ajhg.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland NA, Sercombe HE, Ainscough JF, Coverley D. Ciz1 cooperates with cyclin-A-CDK2 to activate mammalian DNA replication in vitro. J. Cell Sci. 2010;123:1108–1115. doi: 10.1242/jcs.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland NA, Sercombe HE, Wilson RH, Coverley D. Cyclin-A-CDK2-mediated phosphorylation of CIZ1 blocks replisome formation and initiation of mammalian DNA replication. J. Cell Sci. 2015;128:1518–1527. doi: 10.1242/jcs.161919. [DOI] [PubMed] [Google Scholar]
- Coverley D, Marr J, Ainscough J. Ciz1 promotes mammalian DNA replication. J. Cell Sci. 2005;118:101–112. doi: 10.1242/jcs.01599. [DOI] [PubMed] [Google Scholar]
- Dahmcke CM, Buchmann-MOLLER S, Jensen NA, Mitchelmore C. Altered splicing in exon 8 of the DNA replication factor CIZ1 affects subnuclear distribution and is associated with Alzheimer's disease. Mol. Cell. Neurosci. 2008;38:589–594. doi: 10.1016/j.mcn.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Dang MT, Yokoi F, Mcnaught KS, Jengelley TA, Jackson T, Li J, Li Y. Generation and characterization of Dyt1 DeltaGAG knock-in mouse as a model for early-onset dystonia. Exp. Neurol. 2005;196:452–463. doi: 10.1016/j.expneurol.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Den Hollander P, Rayala SK, Coverley D, Kumar R. Ciz1, a novel DNA-binding coactivator of the estrogen receptor alpha, confers hypersensitivity to estrogen action. Cancer Res. 2006;66:11021–11029. doi: 10.1158/0008-5472.CAN-06-2336. [DOI] [PubMed] [Google Scholar]
- Dufke C, Hauser AK, Sturm M, Fluhr S, Wachter T, Leube B, Auburger G, Ott T, Bauer P, Gasser T, Grundmann K. Mutations in CIZ1 are not a major cause for dystonia in Germany. Mov. Disord. 2015;30:740–743. doi: 10.1002/mds.26198. [DOI] [PubMed] [Google Scholar]
- Goodchild RE, Kim CE, Dauer WT. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron. 2005;48:923–932. doi: 10.1016/j.neuron.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Greaves EA, Copeland NA, Coverley D, Ainscough JF. Cancer-associated variant expression and interaction of CIZ1 with cyclin A1 in differentiating male germ cells. J. Cell Sci. 2012;125:2466–2477. doi: 10.1242/jcs.101097. [DOI] [PubMed] [Google Scholar]
- Heckler MM, Riggins RB. ERRbeta splice variants differentially regulate cell cycle progression. Cell Cycle. 2015;14:31–45. doi: 10.4161/15384101.2014.972886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrup K. Post-mitotic role of the cell cycle machinery. Curr. Opin. Cell Biol. 2013;25:711–716. doi: 10.1016/j.ceb.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins G, Roper KM, Watson IJ, Blackhall FH, Rom WN, Pass HI, Ainscough JF, Coverley D. Variant Ciz1 is a circulating biomarker for early-stage lung cancer. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E3128–E3135. doi: 10.1073/pnas.1210107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jatoi I, Anderson WF, Rosenberg PS. Qualitative age-interactions in breast cancer: A tale of two diseases? Am. J. Clin. Oncol. 2008;31:504–506. doi: 10.1097/COC.0b013e3181844d1c. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Yan J, Zhao Y, Donahue LR, Beamer WG, Li X, Roe BA, LeDoux MS, Gu W. Carbonic anhydrase-related protein VIII deficiency is associated with a distinctive lifelong gait disorder in waddles mice. Genetics. 2005;171:1239–1246. doi: 10.1534/genetics.105.044487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judex M, Neumann E, Lechner S, Dietmaier W, Ballhorn W, Grifka J, Gay S, Scholmerich J, Kullmann F, Muller-Ladner U. Laser-mediated microdissection facilitates analysis of area-specific gene expression in rheumatoid synovium. Arthritis Rheum. 2003;48:97–102. doi: 10.1002/art.10713. [DOI] [PubMed] [Google Scholar]
- Kaltschmidt B, Kaltschmidt C. NF-KappaB in long-term memory and structural plasticity in the adult mammalian brain. Front. Mol. Neurosci. 2015;8:69. doi: 10.3389/fnmol.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Masaki T, Hori K, Masuo Y, Miyamoto M, Tsubokawa H, Noguchi H, Nomura M, Takamatsu K. Hippocalcin-deficient mice display a defect in cAMP response element-binding protein activation associated with impaired spatial and associative memory. Neuroscience. 2005;133:471–484. doi: 10.1016/j.neuroscience.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Lanzillotta A, Porrini V, Bellucci A, Benarese M, Branca C, Parrella E, Spano PF, Pizzi M. NF-kappaB in innate neuroprotection and age-related neurodegenerative diseases. Front. Neurol. 2015;6:98. doi: 10.3389/fneur.2015.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leandro GS, Sykora P, Bohr VA. The impact of base excision DNA repair in age-related neurodegenerative diseases. Mutat. Res. 2015;776:31–39. doi: 10.1016/j.mrfmmm.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDoux MS. Animal models of dystonia: Lessons from a mutant rat. Neurobiol. Dis. 2011;42:152–161. doi: 10.1016/j.nbd.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDoux MS. Dystonia: Phenomenology. Parkinsonism Relat. Disord. 2012a;18(Suppl. 1):S162–S164. doi: 10.1016/S1353-8020(11)70050-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDoux MS. The genetics of dystonias. Adv. Genet. 2012b;79:35–85. doi: 10.1016/B978-0-12-394395-8.00002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDoux MS, Dauer WT, Warner TT. Emerging common molecular pathways for primary dystonia. Mov. Disord. 2013;28:968–981. doi: 10.1002/mds.25547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang CC, Tanabe LM, Jou S, Chi F, Dauer WT. TorsinA hypofunction causes abnormal twisting movements and sensorimotor circuit neurodegeneration. J. Clin. Invest. 2014;124:3080–3092. doi: 10.1172/JCI72830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140:3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- Liu T, Ren X, Li L, Yin L, Liang K, Yu H, Ren H, Zhou W, Jing H, Kong C. Ciz1 promotes tumorigenicity of prostate carcinoma cells. Front Biosci (Landmark Ed) 2015;20:705–715. doi: 10.2741/4331. [DOI] [PubMed] [Google Scholar]
- Mitsui K, Matsumoto A, Ohtsuka S, Ohtsubo M, Yoshimura A. Cloning and characterization of a novel p21 (Cip1/Waf1)-interacting zinc finger protein, ciz1. Biochem. Biophys. Res. Commun. 1999;264:457–464. doi: 10.1006/bbrc.1999.1516. [DOI] [PubMed] [Google Scholar]
- Nguyen MD, Mushynski WE, Julien JP. Cycling at the interface between neurodevelopment and neurodegeneration. Cell Death Differ. 2002;9:1294–1306. doi: 10.1038/sj.cdd.4401108. [DOI] [PubMed] [Google Scholar]
- Nishibe R, Watanabe W, Ueda T, Yamasaki N, Koller R, Wolff L, Honda Z, Ohtsubo M, Honda H. CIZ1, a p21Cip1/Waf1-interacting protein, functions as a tumor suppressor in vivo. FEBS Lett. 2013;587:1529–1535. doi: 10.1016/j.febslet.2013.03.034. [DOI] [PubMed] [Google Scholar]
- Rahman F, Ainscough JF, Copeland N, Coverley D. Cancer-associated missplicing of exon 4 influences the subnuclear distribution of the DNA replication factor CIZ1. Hum. Mutat. 2007;28:993–1004. doi: 10.1002/humu.20550. [DOI] [PubMed] [Google Scholar]
- Rahman FA, Aziz N, Coverley D. Differential detection of alternatively spliced variants of Ciz1 in normal and cancer cells using a custom exon-junction microarray. BMC Cancer. 2010;10:482. doi: 10.1186/1471-2407-10-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz M, Perez-Garcia G, Ortiz-Virumbrales M, Meneret A, Morant A, Kottwitz J, Fuchs T, Bonet J, Gonzalez-Alegre P, Hof PR, Ozelius LJ, Ehrlich ME. Abnormalities of motor function, transcription and cerebellar structure in mouse models of THAP1 dystonia. Hum. Mol. Genet. 2015;24:7159–7170. doi: 10.1093/hmg/ddv384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25:7531–7537. doi: 10.1038/sj.onc.1210059. [DOI] [PubMed] [Google Scholar]
- Shen X, Chen J, Li J, Kofler J, Herrup K. Neurons in Vulnerable Regions of the Alzheimer's Disease Brain Display Reduced ATM Signaling. eNeuro. 2016:3. doi: 10.1523/ENEURO.0124-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CH, Fan X, Exeter CJ, Hess EJ, Jinnah HA. Functional analysis of dopaminergic systems in a DYT1 knock-in mouse model of dystonia. Neurobiol. Dis. 2012;48:66–78. doi: 10.1016/j.nbd.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CH, Bernhard D, Hess EJ, Jinnah HA. Subtle microstructural changes of the cerebellum in a knock-in mouse model of DYT1 dystonia. Neurobiol. Dis. 2014;62:372–380. doi: 10.1016/j.nbd.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe LM, Martin C, Dauer WT. Genetic background modulates the phenotype of a mouse model of DYT1 dystonia. PLoS One. 2012;7:e32245. doi: 10.1371/journal.pone.0032245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemula SR, Puschmann A, Xiao J, Zhao Y, Rudzinska M, Frei KP, Truong DD, Wszolek ZK, LeDoux MS. Role of Galpha(olf) in familial and sporadic adult-onset primary dystonia. Hum. Mol. Genet. 2013;22:2510–2519. doi: 10.1093/hmg/ddt102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DQ, Wang K, Yan DW, Liu J, Wang B, Li MX, Wang XW, Liu J, Peng ZH, Li GX, Yu ZH. Ciz1 is a novel predictor of survival in human colon cancer. Exp. Biol. Med. (Maywood) 2014;239:862–870. doi: 10.1177/1535370213520113. [DOI] [PubMed] [Google Scholar]
- Warder DE, Keherly MJ. Ciz1, Cip1 interacting zinc finger protein 1 binds the consensus DNA sequence ARYSR(0–2) YYAC. J. Biomed. Sci. 2003;10:406–417. doi: 10.1007/BF02256432. [DOI] [PubMed] [Google Scholar]
- Wisniewska MB. Physiological role of beta-catenin/TCF signaling in neurons of the adult brain. Neurochem. Res. 2013;38:1144–1155. doi: 10.1007/s11064-013-0980-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Lei L, Gu D, Liu H, Wang S. CIZ1 is upregulated in hepatocellular carcinoma and promotes the growth and migration of the cancer cells. Tumour Biol. 2016;37:4735–4742. doi: 10.1007/s13277-015-4309-y. [DOI] [PubMed] [Google Scholar]
- Xiao J, Uitti RJ, Zhao Y, Vemula SR, Perlmutter JS, Wszolek ZK, Maraganore DM, Auburger G, Leube B, Lehnhoff K, LeDoux MS. Mutations in CIZ1 cause adult onset primary cervical dystonia. Ann. Neurol. 2012;71:458–469. doi: 10.1002/ana.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Vemula SR, LeDoux MS. Recent advances in the genetics of dystonia. Curr Neurol Neurosci Rep. 2014;14:462. doi: 10.1007/s11910-014-0462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates SC, Zafar A, Rabai EM, Foxall JB, Nagy S, Morrison KE, Clarke C, Esiri MM, Christie S, Smith AD, Nagy Z. The effects of two polymorphisms on p21cip1 function and their association with Alzheimer's disease in a population of European descent. PLoS One. 2015;10:e0114050. doi: 10.1371/journal.pone.0114050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Wang C, Tang X, Sun H, Shao Q, Yang X, Qu X. CIZ1 regulates the proliferation, cycle distribution and colony formation of RKO human colorectal cancer cells. Mol. Med. Rep. 2013;8:1630–1634. doi: 10.3892/mmr.2013.1716. [DOI] [PubMed] [Google Scholar]
- Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yokoi F, Jin YH, Deandrade MP, Hashimoto K, Standaert DG, Li Y. Altered dendritic morphology of Purkinje cells in Dyt1 DeltaGAG knock-in and purkinje cell-specific Dyt1 conditional knockout mice. PLoS One. 2011;6:e18357. doi: 10.1371/journal.pone.0018357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Wang Y, Dai Y, Wang J, Suo T, Pan H, Liu H, Shen S, Liu H. CIZ1 promoted the growth and migration of gallbladder cancer cells. Tumour Biol. 2015;36:2583–2591. doi: 10.1007/s13277-014-2876-y. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Decuypere M, LeDoux MS. Abnormal motor function and dopamine neurotransmission in DYT1 DeltaGAG transgenic mice. Exp. Neurol. 2008;210:719–730. doi: 10.1016/j.expneurol.2007.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.