Abstract
DNA methylation is an important epigenetic modification that can have profound and widespread effects on gene expression and on cellular fate and function. Recent work has shown that DNA methylation plays critical roles in hematopoietic development and hematopoietic disease. DNA methyltransferases (DNMTs) and Ten-eleven translocation (TET) enzymes are required for adding and removing methyl “marks” from DNA, respectively, and both sets of gene have been shown to be necessary for proper formation and maintenance of hematopoietic stem cells and for differentiation of downstream hematopoietic lineages during development. DNA methylation and demethylation enzymes have also been implicated in hematopoietic disorders such as acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS). Here, we review some of the recent literature regarding the role of DNA methylation in hematopoietic health and disease.
DNA methylation and hematopoiesis
In vertebrates, hematopoiesis takes place in distinct waves. Primitive hematopoiesis occurs early during embryonic development, and is responsible for generating primitive erythroid and myeloid blood cells. This is followed by a wave of definitive-derived erythroid and myeloid lineage-only blood cells that emerge from early erytho-myeloid progenitor cells (EMPs). Finally, definitive hematopoietic stem cells (HSCs) responsible for generating erythroid, myeloid and lymphoid lineages emerge (1–3). HSCs are specified from hemogenic endothelium in the floor of the dorsal aorta and are responsible for generating all blood cell types throughout adulthood (4, 5). Both the essential process and the major signaling molecules responsible for correct specification of HSCs are highly conserved amongst vertebrates (6, 7). Differentiation of HSCs from hemogenic endothelium and emergence of the different blood lineages from HSCs is highly complex and tightly regulated. Cell-type intrinsic transcription factors downstream from key signaling pathways regulate hematopoiesis. For HSCs, the runt related transcription factor Runx1 plays a critical role in their differentiation from hemogenic endothelium (8). In recent years it has become evident that in addition to signaling networks and transcription factors, epigenetic regulation also plays a crucial role during hematopoiesis. Epigenetic mechanisms such as histone modifications, chromatin remodeling proteins, DNA modifications (methylation and hydroxyl methylation), and small RNAs have all been shown to influence gene expression during development and adult life. During development, differentiation of HSCs and their downstream blood lineages is characterized by changes in both gene expression and the epigenetic landscape, especially DNA methylation (9). Changes in DNA methylation also influence gene expression patterns during adult hematopoiesis, and are often associated with aberrant specification of blood cells and hematologic pathologies. High throughput RNA sequencing and whole genome bisulfite sequencing techniques have revealed some of these changes, and have led to a number of proposed roles for DNA methylation in blood cell differentiation from HSCs in normal development and disease (10, 11). In this review, we focus on recent findings highlighting novel roles for DNA methylation and DNA methylation machinery during normal and pathological hematopoiesis and hematopoietic development.
DNA methylation and de-methylation machinery and its function
In eukaryotes, DNA methylation is carried out by a family of proteins called DNA methyltransferases (DNMTs). DNMTs methylate cytosine residues in CG dinucleotides. Localized regions of DNA containing high densities of CG dinucleotides are called CpG islands (CGIs), and these are often targeted for methylation by DNMTs (Figure 1). CGI methylation is carried out by two classes of DNMT enzymes, maintenance DNMTs and de novo DNMTs. Maintenance DNMTs are responsible for maintaining already-placed methyl “marks” during DNA replication. In vertebrates, DNMT1 is the maintenance DNA methyltransferase that adds methyl groups to copy methyl marks to newly synthesized DNA strand during cell division (12). De novo DNA methyltransferases, in contrast, add new methyl marks to DNA. Unlike the single maintenance DNA methyltransferase, vertebrates have a number of different de novo methyltransferases that are expressed in different cell types and tissues at different times, and that apparently target different but overlapping sets of CGIs (13). DNMT3A and DNMT3B are examples of two de novo class DNA methyltransferases that add new DNA methylation patterns as new cell fates are specified (14). As discussed further below, DNMT3A and DNMT3B have both been implicated in hematopoiesis and HSC biology. DNA methylation is reversible, and there are additional “Ten-eleven-translocation” (TET) enzymes important for removal of DNMT-mediated methyl marks are removed by a set of. Three vertebrate TET proteins, TET1, TET2, and TET3 oxidize methyl cytosine to 5-hydroxymethyl cytosine and other downstream oxidized forms of methyl cytosine and eventually removal of the marks entirely (15, 16). Hydroxymethyl marks are actively removed by TET proteins and DNA repair machinery (17), although a portion of the hydroxylmethyl marks are also removed passively through cell division (18). Methyl marks can also be lost passively through failure of the maintenance enzyme DNMT1 to duplicate these marks on newly synthesized DNA strands during cell division-associated DNA replication. Together, DNMTs and TET proteins help to maintain a correct balance between DNA methylation and demethylation during normal development.
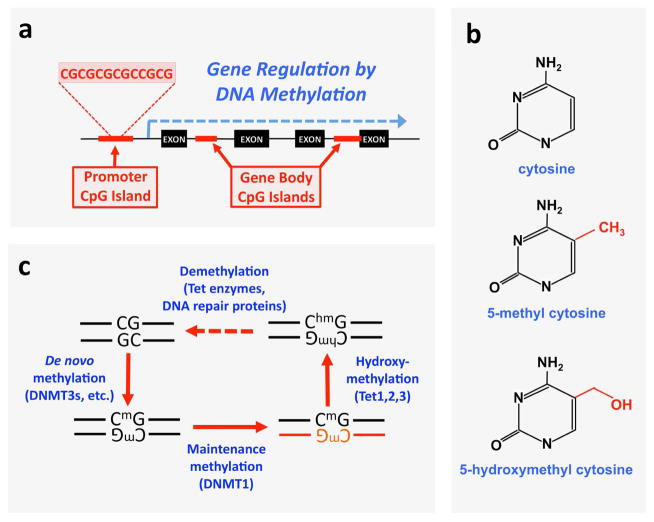
Figure 1. DNA methylation and demethylation processes.
a, Schematic showing the relative positions of the CpG islands associated with a gene locus. Promoter CpG islands are found in upstream regulatory regions, and gene body CpG islands are present anywhere between the start and stop codon. Promoter CpG islands are involved in gene silencing, while recent evidence suggests that gene body CpG islands promote gene activation, b, Chemical structure of cytosine, 5-methyl cytosine, and 5-hydroxymethyl cytosine. c, Methylation and demethylation is driven by distinct sets of enzymes in vivo. Newly established methyl “marks” are added by de novo methyltransferases, including the Dnmt3 family. The maintenance methyltransferase Dnmt1 ensures persistence of these marks in replicating DNA by copying the methyl marks onto to newly synthesized strands (red) during mitosis. Tet proteins hydroxylate methyl groups on cytosine, with subsequent removal of the hydroxymethyl tag by Tet and other DNA repair enzymes leading back to unmethylated DNA.
A large body of evidence has documented an important role for DNA methylation in regulating gene expression (reviewed extensively in (19, 20). In mammals, X-chromosome inactivation is initiated by expression of Xist, a non-coding RNA. This is followed by changes to the chromatin and acquisition of silencing histone marks. Finally, the inactivated gene loci are methylated to assure complete silencing of the X-chromosome (21–23). Inhibition of the methylation in the final step can lead to incomplete silencing of certain loci, suggesting DNA methylation to the promoter and enhancer regions plays a crucial role in assuring complete silencing of the transcription (24, 25). Similar to X-chromosome inactivation, DNA methylation is also involved in genomic imprinting, where one copy of the gene is kept silenced, often by methylating the promoter and enhancer regions. This is done to achieve expression from only the maternal or paternal copy, which plays crucial roles in cell differentiation and embryonic development (26). In mammals, genomic imprinting was first observed for Igf2, Igf2r and H19 gene loci (27–29). Subsequently several more genes were discovered whose regulation was dependent on DNA methylation based genomic imprinting (26, 30). The role of promoter enhancer methylation and suppression of gene expression was further supported by analysis of Dnmt1 knock out mice. Dnmt1 null mice showed aberrant expression of imprinted and X- chromosome linked genes (31).
In recent years, DNA methylation has also been implicated in activation of transcription. Dnmt3b has been shown to preferentially bind and methylate gene body regions of actively transcribed genes in mouse ES cells (32). Preferential gene body methylation of actively expressing genes by Dnmt3b has also been observed in human colon cancer cell lines (33). These results suggest that Dnmt3b-mediated gene body DNA methylation has a positive role on gene expression, and it may be acting to maintain open chromatin structure for active transcription rather than act to silence. The discovery that gene body DNA methylation is strongly associated with gene activation has opened new avenues for future research to understand the role of DNA methylation in gene expression and identify novel gene body-specific regions controlling this activation.
Members of the DNMT family have also been shown to have non-DNA methylation related activities. Mammalian DNMT1, DNMT3L, DNMT3A and DNMT3B methylate DNA, but DNMT2 preferentially binds and methylates tRNAs and potentially other RNA substrates (34), although the functional consequences of this are not well understood. However, it is evident that the DNMT-mediated modifications to both DNA and RNA, and potentially other substrates, can influence gene expression (35, 36).
DNA methylation in normal hematopoiesis and malignancies
As noted above, members of the DNMT3 family of de novo DNA methyltransferases have been shown to play important roles in hematopoiesis. There are two DNMT3 family members in mammals, DNMT3A and DNMT3B. In mice, DNMT3a has been shown to play a role in adult HSC differentiation (11). Deletion of DNMT3a leads to hypomethylation of specific gene loci but hypermethylation of other loci, and the overall level of DNA methylation is relatively unchanged in the Dnmt3a deficient HSCs. However, these changes in methylation and concomitant aberrant gene expression lead to gradual overproliferation and blockage in downstream lineage differentiation. Combined loss of DNMT3a and DNMT3b leads to more severe defects in HSC proliferation and differentiation (11, 37). Specification of HSC begins during embryogenesis in the hemogenic endothelium, with activation of the essential but transiently expressed transcription factor Runx1. Studies carried out using Runx1 deleted animals show that this gene plays a crucial and highly conserved role in embryonic specification of HSCs (8, 38). However, once HSCs are specified Runx1 is downregulated and its function becomes dispensable (39). A recent zebrafish study from our laboratory has shown that DNMT3-dependent DNA methylation promotes continued HSC specification and identity in the absence of Runx1 via its role in maintaining expression of a key downstream transcription factor in HSC (40). Zebrafish have six DNMT3 paralogs, dnmt3aa, dnmt3ab, dnmt3ba, dnmt3bb.1, dnmt3bb.2 and dnmt3bb.3. Zebrafish dnmt3bb.1, the closest homolog to mammalian DNMT3b, is expressed specifically in developing HSC in the trunk, and it is required downstream from Runx1 to maintain HSC fate (Figure 2). Runx1 induces expression of dnmt3bb.1 in developing HSCs, and loss of dnmt3bb.1 leads to hypomethylation of gene body CGIs in the key downstream HSC transcription factor cmyb and reduction in cmyb expression, forcing HSCs to undergo apoptosis. Endothelial-specific overexpression of dnmt3bb.1 is sufficient to rescue cmyb-expressing HSCs in runx1-null embryos. Furthermore, overexpression of dnmt3bb.1 in zebrafish early blastomeres specifically induces cmyb as well as downstream myeloid, lymphoid, and erythroid gene expression (40). HSC rescue in runx-1 deficient animals, and specific expression of hematopoietic genes in “naïve” blastomere cells that normally do not exhibit hematopoietic gene expression, suggests that dnmt3bb.1 expression is both necessary and sufficient to promote hematopoietic gene expression. The induction of downstream hematopoietic genes appears to be indirect, however, via dnmt3bb.1’s role in promoting cmyb expression. Mutants in cmyb have hematopoietic defects that mirror those of dnmt3bb.1 (41), expression of cmyb in the early blastula results in a similar induction of downstream hematopoietic lineage gene expression, and cmyb knockdown prevents dnmt3bb.1-promoted downstream hematopoietic gene expression in blastulae (40). Together, these results show that the de novo DNA methyltransferase dnmt3bb.1 plays an essential role in maintaining HSC specified during early embryogenesis by promoting the expression of a key HSC transcription factor, cmyb. Like zebrafish dnmt3bb.1, mouse DNMT3b is also expressed by developing HSCs in the dorsal aorta during definitive hematopoiesis (42), and further studies should help shed light on the conserved function of these genes in vertebrates. As might be expected to ensure perdurance of “marks” placed in the DNA of HSC by DNMT3 family members, the maintenance DNA methyltransferase DNMT1 is also required for hematopoiesis. Loss of DNMT1 in adult HSC leads to defects in self-renewal, bone marrow niche retention, and the ability to generate proper downstream blood lineages (43). Similarly, loss of zebrafish dnmt1 also causes defects in HSC development during embryogenesis, at least in part via its effects on the cebpa gene (44).
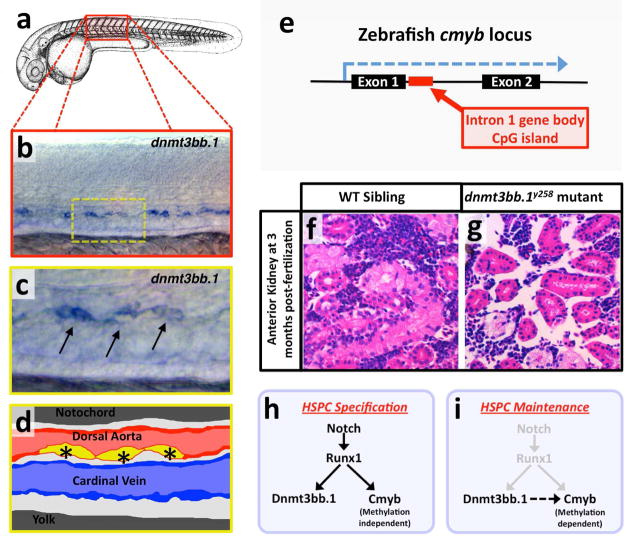
Figure 2. Dnmt3bb.1 is an essential epigenetic factor in HSC fate maintenance.
a, Camera lucida drawing of a zebrafish embryo with a red box noting the approximate region of the trunk shown in the in situ hybridization image in panel b. b,c Whole mount in situ hybridization of zebrafish trunk probed for dnmt3bb.1, showing expression in the developing HSCs (arrows in panel c). Yellow inset box in panel b indicates the magnified area shown in panel c. d, diagram corresponding to panel c showing dorsal aorta (red), cardinal vein (blue), and dnmt-positive HSCs in the floor of the dorsal aorta (yellow, with asterisks). e, Schematic showing the zebrafish cmyb locus and the position of the intron 1 gene body CpG island. f,g, Images through the anterior kidneys (adult hematopoietic organ in zebrafish equivalent to the bone marrow in higher vertebrates) of wild type sibling (f) and dnmt3bb.1y258 mutant (g) adult zebrafish, stained with hematoxylin/eosin, revealing a profound loss of hematopoietic tissue in mutant animals. h, Notch-Runx1 signaling controls specification of HSPCs in the ventral wall of the dorsal aorta during early embryogenesis. This signaling pathway initiates HSPC expression of both the key transcription factor Cmyb and the epigenetic regulator Dnmt3bb.1. i, As development proceeds Runx1 expression is down-regulated and continued maintenance of active Cmyb expression in HSPC depends on Dnmt3bb.1–mediated Cmyb gene body DNA methylation, ensuring maintenance of HSPC cell fate during the Runx1-independent phase. Images are from Gore et al. (2016).
DNMT3 family members have also been shown to play important roles in hematopoietic malignancies. DNMT3a has been very strongly linked to leukemia (45, 46). Somatic DNMT3a mutations are found in 30% and 8% patients of acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) respectively. In AML patients, a wide variety of mutations are seen in the methyltransferase domain of DNMT3a that result in both truncated proteins and missense variants, including a specific mutation in residue R882 that has been shown to reduce the catalytic activity of the mutant protein (47). AML patients carrying the R882 mutation in DNMT3a show no significant difference in overall levels of DNA methylation, but show hypomethylation of specific gene loci (45), although the changes in methylation that have been identified have thus far not strongly correlated with aberrant expression of genes linked to these differentially methylated regions. Additional whole genome bisulfite sequencing and RNA sequencing studies together with functional studies in model organisms will likely be required to further analyze R882 versus wild type DNMT3a mediated methylation and gene expression changes, and their potential impact on hematological malignancies. It is important to note that HSCs from DNMT3a deficient mice do not develop spontaneous AML like disorders (11), although in irradiated mice DNMT3a deficient transplanted HSCs develop a wide spectrum of hematopoietic malignancies (48). These observations suggest secondary mutations and abnormal DNA methylation due to inefficient DNMT3a activity may contribute together to malignant transformation. In line with this notion, mutations in Npm1, Flt3 and Idh1 co-exist with high frequency in DNMT3a mutant AML (49), although it is not clear how these mutations interact with DNMT3a to contribute to malignancy. Although methylation of specific genes such as p15, e-cadherin, and others has been used for prognostic prediction of AML and MDS (50), a strong correlation has not been identified between DNMT3a mutation and methylation of these loci in AML patients. Aberrant DNA methylation and mutations in DNMT3a are also commonly seen in different types of B and T cell lymphomas (51). Whole genome bisulfite sequencing of Burkitt lymphoma, a B cell-type lymphoma, shows global DNA hypomethylation compared to normal cells. These changes in DNA methylation are strongly correlated with down regulation of NF-kB and Jak/Stat signaling pathways (52). Together, these and other observations suggest that in addition to playing crucial roles in the normal development of HSCs, aberrant activity of DNA methyltransferases and acquisition of secondary mutations may contribute significantly to convert these cells to malignancy. In a handful of cases, asymptomatic carriers for somatic mutations in Dnmt3a have been reported, suggesting acquisition of secondary mutations or clonal amplification of mutation-carrying HSCs may be important contributing factors to hematopoietic malignancies (53, 54).
DNA demethylation in normal hematopoiesis and malignancies
The process of removing methyl groups from cytosine residues in DNA is performed by a special class of proteins called Ten-eleven translocation (TET) enzymes. These proteins oxidize the methyl group in methylcytosine to convert it to hydroxymethyl cytosine, which subsequently undergoes further oxidization with eventual removal of the methyl tag (15, 16). TET protein-mediated epigenetic changes to the genome also influence hematopoietic development. Interestingly, TET1 was first discovered in mixed lineage leukemia patients, as a fusion gene with histone methyltransferase (55). Less than 1% of AML patients have mutations in TET1, however (56), suggesting mutations in Tet1 coding regions are not generally responsible for hematological disorders. However, TET1 has been shown to be a direct target of MLL-fusion proteins and is significantly up-regulated in MLL-rearranged leukemia, leading to a global increase of 5-hydroxymethylcytosine level (57). Loss of Tet1 in HSCs leads to biased differentiation towards B cells. Tet1 deficient B cells gradually accumulate high levels of DNA damage and that presumably contributes to their malignancies (58). It is unclear how Tet proteins directly regulate HSC gene expression and downstream cell lineage specification.
TET2 is more frequently mutated in myeloid diseases, with mutations found at a frequency of approximately 7–20% in AML and up to 50% in chronic myelomonocytic leukemia (59, 60). HSCs from Tet2 mutation-carrying CMML patients show significantly higher global DNA methylation levels but reduced hydroxymethylation (61). Mutations in the Tet2 coding sequence are most common in AML and CMML. Missense mutations in the catalytic domain of Tet2 protein are thought to cause impaired methyl cytosine oxidation, leading to aberrant gene expression and defective hematopoiesis (61–63). Tet2 mutations are also frequently seen in different lymphoid malignancies at a very high frequency of 20–76% (64, 65). Similarly, bone marrow-derived cells from mice carrying deletions in the Tet2 locus show significant reduction in hydroxymethylation (61, 66, 67). Aged Tet2 null mice showed a wide array of hematological, especially myeloid malignancies, and bone marrow-derived HSC proliferation is significantly increased in Tet2 null mice, although not all strains of mice carrying tet2 deficiency develop malignant phenotypes (64). Tet2-deficient HSCs show skewed hematopoietic differentiation programs with increased monocytic and myeloblastic lineage differentiation (64, 67). Interestingly, the HSC overproliferation defects seen in Tet2 knock out mice are similar to those in DNMT3a knock out mice, suggesting that a correct balance between DNA methylation and demethylation is important for controlling HSC proliferation. In the zebrafish, mutation of tet2 alone does not result in phenotypes similar to Tet2 mouse knockouts (68, 69), although combined loss of tet2 and tet3 activity leads to defective hematopoiesis, suggesting that tet3 function compensates for loss of tet2 (69). Zebrafish embryos deficient for tet2/3 activity show defective HSC emergence from the hemogenic endothelium, leading to death of the “stalled” HSC progenitors and subsequent reduction in downstream blood lineages (69). Combined deficiency of Tet2 and Tet3 results in significantly greater reduction in levels of hydroxymethylation compared to individual deficiencies, further supporting the notion that the two genes have partially redundant function (70). Like Tet1, mutations in Tet3 are very rare in hematopoietic malignancies. Conditional loss of Tet3 in mice HSCs does not lead to significant decrease in myeloid, lymphoid and erythroid populations (70), suggesting a less critical role for this gene compared to Tet1 and Tet2. Together, these results identify Tet genes as major players in HSC development, though they may have different functions in different organisms.
Conclusions and future directions
Recent data suggests that a carefully regulated balance between Dnmt mediated methylation and Tet mediated hydroxymethylation of cytosine residues is crucial for normal hematopoietic development. It has also become clear that aberrant changes to this key epigenetic process contribute significantly to the development of hematological malignancies. In the future, technological advances in genome sequencing and analysis should help to further elucidate how these epigenetic changes target specific gene loci, and how specific epigenetic modifications result in altered hematopoietic gene expression. Single cell sequencing methods will facilitate analysis of cell populations found in hematopoietic malignancies, helping to reveal clonal diversity and associated changes to gene expression. Together with genomic analysis, additional studies on DNA methylation and demethylation machinery in model organisms will also be required to elucidate the precise molecular mechanisms regulating DNA methylation and demethylation during development and organogenesis. Although there is not always a precise gene-to-gene correspondence in function between different vertebrate model organisms or between model organisms and humans (as noted for the Tet genes, above), the overall functions of the DNMT and TET gene families appear to be conserved, although additional studies will be needed to parse out the common and diverse roles for these genes in development and gene regulation in different species.
The mechanisms directing specific Dnmt and Tet proteins to distinct sets of target sites in the genome are still largely unknown. Association with cell type specific transcription factors or other co-factors may play a role. Dnmts and Tet proteins may also be guided to specific sites via other types of initiating epigenetic modifications to local chromatin regions (as discussed above for X-chromosome inactivation). Dnmts and Tet proteins have been reported to interact with a variety of other proteins, including transcription factors and histone modifying enzymes (71–74). Recent findings showing that Dnmt3b methylation is preferentially targeted to gene body regions compared to promoters suggests that DNA methylation enzymes also have preferences when it comes to targeting to different regions of genes (32, 33). Gene body methylation by Dnmt3b appears to act as a positive regulator for gene expression, suggesting methylation in this context helps to maintain active chromatin structure accessible for RNA polymerase, although further work will be needed to explore mechanisms of gene activation by gene body methylation. In the future, more comprehensive knowledge of the interactions that direct DNA methylation and demethylation machinery to specific sites in the genome and the functional consequences for expression of specific genes should help us to better understand how mutant or aberrantly expressed Dnmt and Tet proteins lead to hematopoietic malignancy.
DNA methylation is an important epigenetic mechanism regulating gene expression
DNA Methyltransferases (DNMTs) add repressing or activating methyl “marks” to DNA
Ten-Eleven Translocation (TET) enzymes help to remove methyl marks from DNA
DNMT and TET genes play key roles during both normal and pathologic hematopoiesis
Acknowledgments
The Weinstein lab is supported by the intramural program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development. We would like to apologize to authors whose work we could not cite due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bertrand JY, et al. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development. 2007;134:4147–4156. doi: 10.1242/dev.012385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frame JM, McGrath KE, Palis J. Erythro-myeloid progenitors: “definitive” hematopoiesis in the conceptus prior to the emergence of hematopoietic stem cells. Blood Cells Mol Dis. 2013;51:220–225. doi: 10.1016/j.bcmd.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoder MC. Inducing definitive hematopoiesis in a dish. Nat Biotechnol. 2014;32:539–541. doi: 10.1038/nbt.2929. [DOI] [PubMed] [Google Scholar]
- 4.Hirschi KK. Hemogenic endothelium during development and beyond. Blood. 2012;119:4823–4827. doi: 10.1182/blood-2011-12-353466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mikkola HK, Orkin SH. The journey of developing hematopoietic stem cells. Development. 2006;133:3733–3744. doi: 10.1242/dev.02568. [DOI] [PubMed] [Google Scholar]
- 6.Paik EJ, Zon LI. Hematopoietic development in the zebrafish. Int J Dev Biol. 2010;54:1127–1137. doi: 10.1387/ijdb.093042ep. [DOI] [PubMed] [Google Scholar]
- 7.Kim AD, Stachura DL, Traver D. Cell signaling pathways involved in hematopoietic stem cell specification. Exp Cell Res. 2014;329:227–233. doi: 10.1016/j.yexcr.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cedar H, Bergman Y. Epigenetics of haematopoietic cell development. Nat Rev Immunol. 2011;11:478–488. doi: 10.1038/nri2991. [DOI] [PubMed] [Google Scholar]
- 10.Jeong M, et al. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nature genetics. 2014;46:17–23. doi: 10.1038/ng.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Challen GA, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nature genetics. 2012;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schermelleh L, et al. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic acids research. 2007;35:4301–4312. doi: 10.1093/nar/gkm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hattori N, et al. Preference of DNA methyltransferases for CpG islands in mouse embryonic stem cells. Genome research. 2004;14:1733–1740. doi: 10.1101/gr.2431504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lei H, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 15.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seisenberger S, et al. Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Philos Trans R Soc Lond B Biol Sci. 2013;368:20110330. doi: 10.1098/rstb.2011.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes & development. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nature reviews Genetics. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 21.Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto I, Heard E. Lessons from comparative analysis of X-chromosome inactivation in mammals. Chromosome Res. 2009;17:659–669. doi: 10.1007/s10577-009-9057-7. [DOI] [PubMed] [Google Scholar]
- 23.Patrat C, et al. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5198–5203. doi: 10.1073/pnas.0810683106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sado T, et al. X inactivation in the mouse embryo deficient for Dnmt1: distinct effect of hypomethylation on imprinted and random X inactivation. Developmental biology. 2000;225:294–303. doi: 10.1006/dbio.2000.9823. [DOI] [PubMed] [Google Scholar]
- 25.Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153:773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartolomei MS, Ferguson-Smith AC. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barlow DP, Stoger R, Herrmann BG, Saito K, Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991;349:84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- 28.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 29.DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64:849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- 30.Plasschaert RN, Bartolomei MS. Genomic imprinting in development, growth, behavior and stem cells. Development. 2014;141:1805–1813. doi: 10.1242/dev.101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 32.Baubec T, et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015;520:243–247. doi: 10.1038/nature14176. [DOI] [PubMed] [Google Scholar]
- 33.Yang X, et al. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaefer M, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes & development. 2010;24:1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu F, et al. Molecular and enzymatic profiles of mammalian DNA methyltransferases: structures and targets for drugs. Curr Med Chem. 2010;17:4052–4071. doi: 10.2174/092986710793205372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chembiochem. 2011;12:206–222. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- 37.Challen GA, et al. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell stem cell. 2014;15:350–364. doi: 10.1016/j.stem.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464:112–115. doi: 10.1038/nature08761. [DOI] [PubMed] [Google Scholar]
- 39.Liakhovitskaia A, et al. Restoration of Runx1 expression in the Tie2 cell compartment rescues definitive hematopoietic stem cells and extends life of Runx1 knockout animals until birth. Stem Cells. 2009;27:1616–1624. doi: 10.1002/stem.71. [DOI] [PubMed] [Google Scholar]
- 40.Gore AV, et al. Epigenetic regulation of hematopoiesis by DNA methylation. Elife. 2016;5 doi: 10.7554/eLife.11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sood R, et al. Development of multilineage adult hematopoiesis in the zebrafish with a runx1 truncation mutation. Blood. 2010;115:2806–2809. doi: 10.1182/blood-2009-08-236729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe D, Suetake I, Tajima S, Hanaoka K. Expression of Dnmt3b in mouse hematopoietic progenitor cells and spermatogonia at specific stages. Gene expression patterns : GEP. 2004;5:43–49. doi: 10.1016/j.modgep.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 43.Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell stem cell. 2009;5:442–449. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, et al. DNA methyltransferase 1 functions through C/ebpa to maintain hematopoietic stem and progenitor cells in zebrafish. J Hematol Oncol. 2015;8:15. doi: 10.1186/s13045-015-0115-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. The New England journal of medicine. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan XJ, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nature genetics. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 47.Yamashita Y, et al. Array-based genomic resequencing of human leukemia. Oncogene. 2010;29:3723–3731. doi: 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- 48.Mayle A, et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood. 2015;125:629–638. doi: 10.1182/blood-2014-08-594648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamaguchi S, et al. IDH1 and IDH2 mutations confer an adverse effect in patients with acute myeloid leukemia lacking the NPM1 mutation. Eur J Haematol. 2014;92:471–477. doi: 10.1111/ejh.12271. [DOI] [PubMed] [Google Scholar]
- 50.Shen L, et al. DNA methylation predicts survival and response to therapy in patients with myelodysplastic syndromes. J Clin Oncol. 2010;28:605–613. doi: 10.1200/JCO.2009.23.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Couronne L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. The New England journal of medicine. 2012;366:95–96. doi: 10.1056/NEJMc1111708. [DOI] [PubMed] [Google Scholar]
- 52.Kretzmer H, et al. DNA methylome analysis in Burkitt and follicular lymphomas identifies differentially methylated regions linked to somatic mutation and transcriptional control. Nature genetics. 2015;47:1316–1325. doi: 10.1038/ng.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hahn CN, et al. A tale of two siblings: two cases of AML arising from a single pre-leukemic DNMT3A mutant clone. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2015;29:2101–2104. doi: 10.1038/leu.2015.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Genovese G, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. The New England journal of medicine. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ono R, et al. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23) Cancer Res. 2002;62:4075–4080. [PubMed] [Google Scholar]
- 56.N. Cancer Genome Atlas Research. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. The New England journal of medicine. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang H, et al. TET1 plays an essential oncogenic role in MLL-rearranged leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:11994–11999. doi: 10.1073/pnas.1310656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cimmino L, et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nature immunology. 2015;16:653–662. doi: 10.1038/ni.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lorsbach RB, et al. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2003;17:637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 60.Langemeijer SM, Aslanyan MG, Jansen JH. TET proteins in malignant hematopoiesis. Cell Cycle. 2009;8:4044–4048. doi: 10.4161/cc.8.24.10239. [DOI] [PubMed] [Google Scholar]
- 61.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang YI, et al. Evaluation of allelic strength of human TET2 mutations and cooperation between Tet2 knockdown and oncogenic Nras mutation. Br J Haematol. 2014;166:461–465. doi: 10.1111/bjh.12871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pronier E, et al. Inhibition of TET2-mediated conversion of 5-methylcytosine to 5-hydroxymethylcytosine disturbs erythroid and granulomonocytic differentiation of human hematopoietic progenitors. Blood. 2011;118:2551–2555. doi: 10.1182/blood-2010-12-324707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quivoron C, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 65.Lemonnier F, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. 2012;120:1466–1469. doi: 10.1182/blood-2012-02-408542. [DOI] [PubMed] [Google Scholar]
- 66.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moran-Crusio K, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gjini E, et al. A zebrafish model of myelodysplastic syndrome produced through tet2 genomic editing. Mol Cell Biol. 2015;35:789–804. doi: 10.1128/MCB.00971-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li C, et al. Overlapping Requirements for Tet2 and Tet3 in Normal Development and Hematopoietic Stem Cell Emergence. Cell Rep. 2015;12:1133–1143. doi: 10.1016/j.celrep.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ko M, et al. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol Rev. 2015;263:6–21. doi: 10.1111/imr.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robertson KD, et al. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nature genetics. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 72.Macaluso M, Cinti C, Russo G, Russo A, Giordano A. pRb2/p130-E2F4/5-HDAC1-SUV39H1-p300 and pRb2/p130-E2F4/5-HDAC1-SUV39H1-DNMT1 multimolecular complexes mediate the transcription of estrogen receptor-alpha in breast cancer. Oncogene. 2003;22:3511–3517. doi: 10.1038/sj.onc.1206578. [DOI] [PubMed] [Google Scholar]
- 73.Brenner C, et al. Myc represses transcription through recruitment of DNA methyltransferase corepressor. The EMBO journal. 2005;24:336–346. doi: 10.1038/sj.emboj.7600509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lehnertz B, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Current biology : CB. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]