Abstract
Objective
Articular cartilage harbors chondrogenic progenitor cells (CPCs), a population that responds chemotactically to cell death. Because this behavior is reminiscent of macrophages, we hypothesized that CPCs have macrophage-like capabilities for scavenging cell and tissue debris through phagocytosis.
Design
CPCs, chondrocytes, synoviocytes, and macrophages were cultured with fluorophore-labeled chondrocyte debris for 3, 6, 12, or 24 hours. Debris internalization was quantified by confocal microscopy and flow cytometry. Confocal microscopy was also used to test CPCs and chondrocytes for uptake of fluorophore-labeled fibronectin fragments (Fn-fs), a form of extracellular matrix debris. Lysosome activity and mass in CPCs and chondrocytes were measured using fluorescent probes. The relative expression of phagocytosis-related genes and proteins was evaluated by polymerase chain reaction (PCR) and immunoblotting, respectively. Pulse-chase experiments were performed to determine if the debris internalized by CPCs and chondrocytes was cleared, and if clearance was affected by a cathepsin B inhibitor.
Results
More macrophages, synoviocytes, and CPCs internalized cell debris than chondrocytes at all time points. While uptake remained flat in chondrocytes at ~10%, in the other cell types it peaked at more than 60% after 12 – 24 hours. Relative to chondrocytes, CPCs showed significantly higher rates of Fn-fs engulfment, greater lysosome activity and mass, and over-expressed phagocytosis-related genes and proteins. Pulse-chase experiments revealed time- and cathepsin B- dependent clearance of cell debris in CPCs, but not in chondrocytes.
Conclusions
CPCs phagocytized cell and matrix debris much more efficiently than chondrocytes, supporting the hypothesis that they play a macrophage-like role in injured cartilage.
Keywords: Chondrogenic Progenitor Cells, Phagocytosis, Chondrocytes, Synoviocytes, Macrophages
Introduction
The long-held belief that articular cartilage harbors only one cell type, the chondrocyte, has given way in the face of evidence for a second distinct type, the chondrogenic progenitor cell (CPC), which has been identified in healthy, injured, and osteoarthritic cartilage [1–6]. Under normal circumstances CPCs are thought to represent less than 1% of cartilage cells. However, our previous work documented the emergence and proliferation of CPCs on cartilage surfaces after a mechanical injury, which leads to several fold increases in their numbers in and near the sites of tissue damage and chondrocyte death [6]. These cells were found to migrate to injury sites along alarmin and chemokine gradients, a chemotactic behavior that chondrocytes do not perform. However, like superficial chondrocytes, CPCs express relatively high levels of lubricin, a boundary lubricant that protects cartilage from mechanical damage [6, 7]. Migrating CPCs are morphologically distinguished from non-motile chondrocytes by their elongated and sometimes dendritic shape, which contrasts sharply with the typically spherical chondrocytes. CPCs express some, but not all of the markers used to identify MSCs. The overall pattern of CPC gene expression revealed by microarray analysis is more akin to synovial fibroblasts and mesenchymal stem cells than chondrocytes [8]. Unlike pluripotent MSCs, CPCs are limited in their ability to form different tissues; however, as their name implies, CPCs reliably generate hyaline cartilage under chondrogenic conditions [9].
Phagocytosis is described as the engulfment and destruction of extracellular objects and substances by cells. Engagement of cell-surface receptors by extracellular ligands leads to internalization via phagosomes and endosomes, followed by lysosomal degradation, a process that depends on proteases, which include cathepsin B, the most abundant cathepsin in lysosomes [10]. Phagocytosis plays a prominent role in the immune response to tissue damage, where it contributes to the clearance of pro-inflammatory necrotic tissue and cell remnants, a vital tissue repair function supported mainly by macrophages. Cell-death-related ligands serve as chemokines that lure circulating monocytes to wounds, where they become mobile macrophages [11, 12]. While many different cell types, including synoviocytes, are capable of chemotaxis and phagocytosis, macrophages are specialists that relocate and scavenge debris with particular alacrity. This occurs regularly throughout vascularized tissues, but the peculiar anatomy of synovial joints may thwart the ability of macrophages to reach damaged cartilage in large numbers, in that most chondral lesions are physically remote from the synovium, where sentinel macrophages and synoviocytes are stationed and blood vessels offer an ongoing supply of circulating monocytes. Supporting this conclusion are data indicating that synovial inflammation in joints with isolated chondral injuries is relatively mild [13].
The observations cited above point to a potential deficiency in debris clearance as a limitation to spontaneous cartilage repair. Chondrocytes isolated from osteoarthritic patients show some phagocytic activity [14], but their ability to clear cell debris at injury sites is weakened by their unresponsiveness to injury-related chemokines [6]. The lack of other extrinsic scavenging mechanisms led us to consider whether CPCs could provide the required phagocytic activities. As an initial test of this hypothesis we used flow cytometry and confocal microscopy to compare CPCs to chondrocytes with respect to the uptake of fluorescently-labeled cell and extracellular matrix debris. Fn-fs fragments were chosen to represent matrix debris despite the fact that fibronectin is a minor component of the cartilage matrix (<1%). This was based on observations that Fn-fs drive local chondrolysis in injured cartilage, which makes them a high-priority target for wound-site clearance [15]. Chondrocytes were isolated from full-thickness cartilage (including the superficial, transitional, and deep zones), or from the top third of the matrix (including the superficial and upper transitional zones). Macrophages and synoviocytes, which are known to be phagocytic, were included in the cell debris uptake studies. Lysosomal activity in CPCs and chondrocytes was measured by flow cytometric analysis of intracellular cathepsin activity. A cathepsin B inhibitor was used to assess the role of lysosomal proteases in the clearance of DiO-labeled cell debris. The relative expression of phagocytosis markers in CPCs and chondrocytes was evaluated by PCR and immunoblotting.
Method
Cartilage Harvest and Culture
Fresh articular cartilage was harvested from bovine tibial plateaus of healthy stifle joints (Bud’s Meat, Riverside, IA). The cartilage was then cultured in standard media (1:1 mixture of DMEM and F12, supplemented with 10% fetal bovine serum, 50 μg/ml L-ascorbate, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2.5 μg/ml Fungizone).
Cell Isolation and Culture
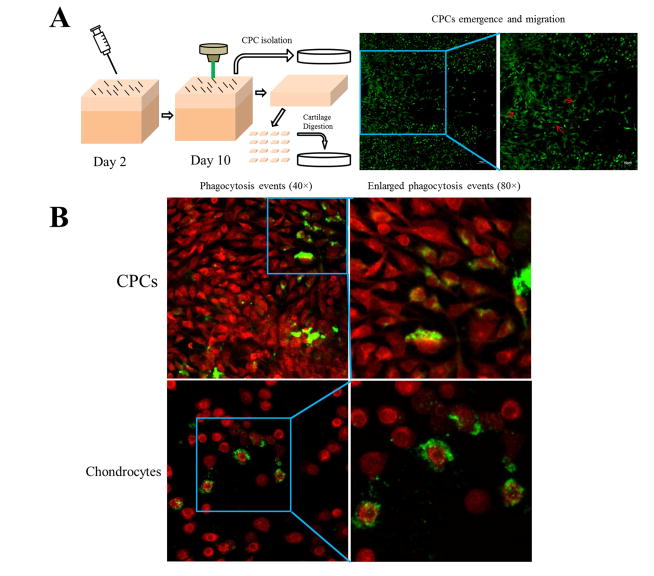
CPC detection and isolation was performed as previously described (Fig 1A) [6]. Briefly, a sterile 18 G needle was dragged on the cartilage surface of each explant to create multiple matrix tears and induce chondrocyte death. After 10-days in culture, the emergence of CPCs on the injured cartilage surface was then confirmed. Briefly, explants were washed with Hanks’ Balanced Salt Solution and stained with 1 μM Calcein Green-AM (Invitrogen, Grand Island, NY) for 30 mins, followed by confocal microscopy detection. For CPC isolation, each explant surface was treated with 0.25% Trypsin-EDTA (Gibco, Grand Island, NY) for 10 mins to isolate CPCs, culture media was added to end trypsinization, and cell suspension was then centrifuged at 300 G for 10 mins. Cells were resuspended and seeded in multiple 35 mm dishes. After isolation of CPCs, the underlying cartilage tissue was shaved off the subchondral bone, minced into smaller pieces, and digested in 0.03% collagenase/protease (dissolved in culture media) for 16 hrs. The digestion media was then centrifuged (300 G for 10 mins) and resuspended, cells were seeded into multiple 35 mm dishes. To determine if phagocytic cells were enriched in the superficial zone, chondrocytes from the upper 1/3 cartilage were separated from the bottom 2/3 prior to cell isolation using a customized fixture [9]. All cells were allowed to adapt to culture conditions for at least two days before measuring phagocytic activities.
Figure 1.
Cell isolation and internalization of cell debris by CPCs and chondrocytes. A) Schematic representation of the procedures for CPC and chondrocytes isolation and confocal image showing CPCs migrating on the cartilage surface post injury (arrows). B) Confocal images show green labeled cell debris with cells counter-stained with Calcein Red-Orange. The majority of the green-labeled debris in the CPC culture was internalized (upper panels), whereas in chondrocytes the label was surface-bound (lower panels).
Synovium tissue was obtained from fresh bovine knee joint, then minced to smaller pieces to attach on culture dishes. The culture media was added onto the synovium tissue drop by drop after couple hours of dry attachment. Small amount of culture media was replenished on the following day until the synovium tissue was no longer attached. Synoviocytes were collected for experiments at passage 1.
The mouse macrophage cell line (RAW 264.7) was a generous gift from Dr. Wendy Maury (The University of Iowa). Macrophages were cultured in macrophage-specific culture media (DMEM, supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2.5 μg/ml Fungizone).
Generation of DiO-labeled Cell debris
The lipophilic fluorescent tracer 3-octadecyl-2-[3-(3-octadecyl-2(3H)-benzoxazolylidene)-1-propenyl]-, perchlorate (DiO) was used to label lipids and lipophilic proteins in freshly isolated chondrocytes [14]. Cells were resuspended at 106 cells/ml in culture media, and DiO solution (Molecular Probes, Eugene, OR) was then added at 5 μl/ml. After 20 minutes of incubation at 37 °C, the mixture was centrifuged (1500 rpm for 5 mins) and resuspended in 2 ml culture media (around 1×107 cells), 10 freeze-thaw cycles (liquid nitrogen and 37 °C water bath for 20 mins, alternatively) were applied to generate the DiO-labeled cell debris.
Generation of FITC-labeled Fibronectin Fragments (Fn-fs)
Human fibronectin fragments (Fn-fs) mixture, containing Fn-fs of 29 kDa, 40 – 60 kDa and 120 – 160 kDa, (generated by Dr. Gene Homandberg) was labeled by fluorescein isothiocyanate (FITC). Briefly, Fn-fs solution was dialyzed against 0.1 M NaHCO3 for 6 hours, then incubated with FITC stock solution (3 mg/ml) at a ratio of 1:170 (Fn-fs : FITC) by mass for 2 hrs at room temperature. FITC conjugated Fn-fs solution was then dialyzed exhaustively against 1× PBS to remove excess materials.
Detection and Quantification of Phagocytosis+ Cells
After 1 day in culture, DiO-labeled cell debris (100 μl) and FITC-labeled Fn-fs (10 μg) were added to each dish (35 mm). The cells were then cultured for various time periods (3 hrs, 6 hrs, 12 hrs, and 24 hrs). For microscopy analysis, cells were washed with Hanks’ Balanced Salt Solution and stained with 1 μM Calcein Red-Orange (Invitrogen, Grand Island, NY) for 30 mins. For flow cytometry analysis, cells were trypsinized after washing in Hanks solution, and then suspended in 1 ml culture media in 5 ml Falcon Polystyrene Tubes (BD Bioscience, San Jose, CA). Hoechst 33258 (BD Biosciences, San Jose, CA) was added to each sample to a concentration of 4 μg/ml to label dead cells.
Measurement and Quantification of Lysosome Activity
After 1 day in culture, CPCs and chondrocytes were subjected to DiO-labeled cell debris (100 μl) for 12 hrs, then stained with cathepsin B substrate and Hoechst 33342 provided in VIVAprobe™ Lysosome Assay Kit (VIVA Bioscience, UK) following the manufacturer’s instruction for microscopy analysis. After staining, cell suspensions were prepared and assessed by flow cytometry as described above.
Pulse-Chase Experiment of Cell Debris Degrading Time Evaluation
CPCs and chondrocytes were seeded in dishes (35 mm) and incubated with DiO-labeled cell debris (100 μl) with or without 4 μM Z-Phe-Gly-NHO-Bz-pMe, a cathepsin B inhibitor (Calbiochem, Billerica, MA) for 12 hrs. The cells were then washed with Hanks solution 3 times and were cultured for an additional 12 hrs or 24 hrs with or without cathepsin B inhibitor. The cells were then isolated and processed for flow cytometry analysis.
Real-time Polymerase Chain Reaction Analysis
50 ng RNA was reverse transcribed to complimentary DNA using TaqMan reverse transcription reagents (Applied Biosystem, Grand Island, NY). qPCR reactions were performed with SYBR Green reagents (Applied Biosystem, Grand Island, NY) and custom specific primers (Integrated DNA Technologies, Coralville, IA), including CD 68 (Forward: GGA GTA ATG GTT CCC AGC CC, Reverse: CTG CAG TGG ATC CTG CTT GA), CD 14 (Forward: ACC ACC CTC AGT CTC CGT AA, Reverse: GCC GAG ACT GGG ATT GTC AG), GULP1 (Forward: TGG ATG CAT ACT CCC GAA GC, Reverse: AGC TGG CAA TTG TGT TGA ACT), LAMP1 (Forward: GTG AAG AAT GGC AAC GGG AC, Reverse: TTA TTC TGG GGC CCA CTC CT). All qPCR experiments were performed in triplicate. Each gene expression level was normalized to β-actin. The fold change was calculated by the 2−ΔΔCt method.
Western Blot Analysis for Lysosome Activity
Lysosomal-associated membrane protein 1 (LAMP1), a 90–120 kDa protein that is indicator of lysosome mass, CD 68, CD14, and GULP1 were analyzed by western blot, primary CPCs and chondrocytes with or without cell debris (100 μl), were seeded in 6-well plates. Western blot procedures were essentially the same as previously described [16]. Briefly, cells were lysed in cold lysis buffer and total protein concentration was determined with the BCA Protein Assay kit (Thermo Fisher Scientific, Rockford, IL). Proteins were denatured with 2× sample buffer, a total of 5 μg proteins from each lysis were resolved in 12.5% SDS gels and blotted onto nitrocellulose membranes, after blocking with 5% non-fat milk for 1 hr, the blots were incubated at 4 °C overnight with LAMP1, CD 68, CD 14, GULP1, and β-actin antibody respectively. The blots were incubated with horseradish peroxidase-conjugated anti-mouse IgG for LAMP1, CD 68, CD 14, and anti-rabbit IgG for GULP1 and β-actin in 5% BSA in TBST for 1 hr at room temperature, followed by reaction with Super Signal West Dura Chemiluminescent Substrate (Thermo Fisher Scientific, Rockford, IL). Chemiluminescence signals were detected with Kodak Biomax Xar Film (Sigma Aldrich, Rochester, NY).
Statistical Analysis
Data were reported as means with 95% confidence interval. Statistical analyses were conducted with IBM SPSS (Version 23) and GraphPad Prism 6. All data were assumed to be independent. For phagocytosis+ cell quantification, CPCs and chondrocytes (whole thickness) were isolated from three independent bovine explants, synovium tissue and chondrocytes (superficial zone) were isolated from another three independent bovine explants, and macrophages were from three batches of the Raw 264.7 cell line. Two-sided two-sample t-test was applied to assess the difference between CPC and chondrocytes (from whole thickness and superficial zone, respectively) at each time point (3 hrs, 6 hrs, 12 hrs, and 24 hrs). For lysosome activity assessment and pulse-chase experiments, CPCs and chondrocytes of each group were isolated from three independent bovine explants and two-sided two-sample t-test was performed to test difference between CPCs and chondrocytes, as well as two groups of CPCs (12 hrs versus 12 hrs with cathepsin B inhibitor and 24 hrs versus 24 hrs with cathepsin B inhibitor). For gene expression analysis, CPCs and chondrocytes were isolated from three independent bovine explants and each data point was the mean of three technical repeats. Two-sided two-sample t-test was used to evaluate gene expression difference between CPCs and chondrocytes. A two-tailed P-value < 0.05 was considered statistically significant.
Results
Phagocytosis Activity Comparison between CPCs and Chondrocytes
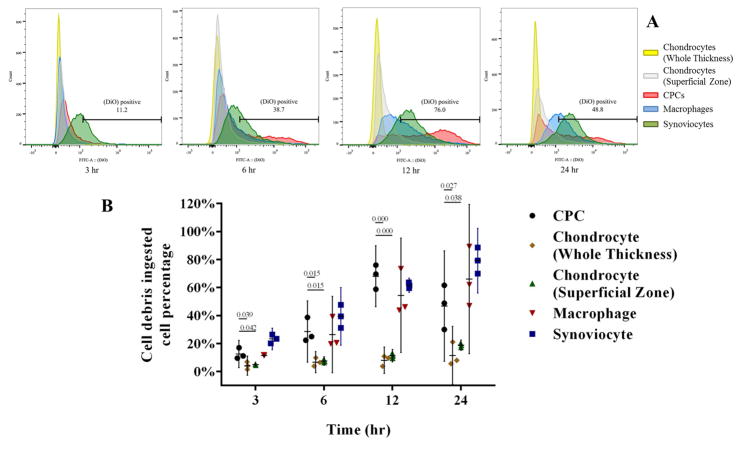
Confocal microscopy analysis showed CPCs vigorously emerged and migrated to the injured site on cartilage surface (Fig 1A). For samples added with cell debris, it showed that both CPCs and chondrocytes were labeled with DiO; however, most of the label in CPCs was intracellular, whereas most chondrocyte labeling was on the cell surface (Fig 1B). A similar phenomenon was observed FITC labeled Fn-fs, which were internalized by CPCs, but not by chondrocytes (Fig 2A). Flow cytometry analysis quantitatively confirmed that the percentage DiO+ CPCs was significantly higher than chondrocytes at each time point (3 hrs, 6 hrs, 12 hrs, and 24 hrs) (Fig 3B). In addition, DiO+ CPCs increased dramatically (12.6% for 3 hrs, 28.6% for 6 hrs) and peaked at 12 hrs (68.1%) (Fig 3B). Notably, CPCs were in the same level with macrophages at first two time points (DiO+ macrophages were 11.5%, 26.4% for 3 hrs, 6 hrs, respectively), and surpassed macrophages (54.4%) at 12 hrs, macrophages (66.0%) reversed over CPCs (46.8%) at 24 hrs, while DiO+ chondrocytes (from whole thickness) augmented gradually (4.2%, 6.7%, 8.1%, and 11.6% for each time point, respectively), superficial zone chondrocytes were slightly higher than chondrocytes from whole thickness (4.9%, 7.6%, 11.3% and 18.8% for each time point, respectively), but no significant zone-related differences were observed. DiO+ synoviocytes showed similar increasing pattern with macrophages throughout the whole time course (Fig 3B).
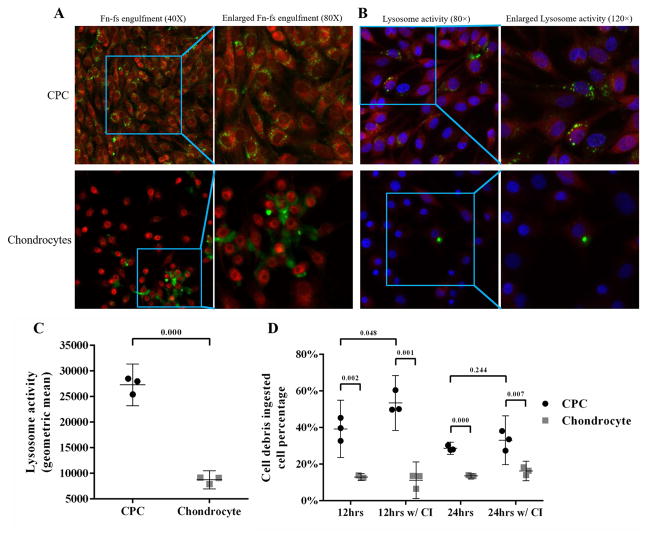
Figure 2.
Fn-fs engulfment and lysosome activity in CPCs and chondrocytes. A) Confocal imaging revealed that FITC-labeled Fn-fs (green) were engulfed by CPCs, to a much greater extent than chondrocytes. B) Confocal images show lysosome activity in CPCs and chondrocytes incubated with labeled cell debris. In the panels on the right, the fluorescent lysosome activity probe (red) and cell debris (green) was imaged in cells that were counter stained with a nuclear probe (blue). Both the green and red probes stained CPCs much more intensely than chondrocytes. C) Flow cytometric analysis showed lysosome activities in CPCs were significantly higher than chondrocytes. (n = 3 per group). D) DiO+ CPC percentages are significantly higher than chondrocytes at 12 hrs and 24 hrs with or without cathepsin B inhibitor, DiO+ CPC percentage was significantly elevated with cathepsin B inhibitor at 12 hrs, and was slightly elevated at 24 hrs. No change was observed in chondrocytes in any situation (n = 3 per group, CI: cathepsin B inhibitor). Data are the individual values (shapes) with the corresponding mean ± 95% confidence interval (range). The numbers above the bars indicate P values for differences between groups.
Figure 3.
Flow cytometric analysis of phagocytosis and lysosome activity in CPCs and chondrocytes. A) Representative comparisons among CPCs, chondrocytes (Whole thickness), chondrocytes (Superficial/transitional zones), synoviocytes and macrophages. CPCs were comparable to macrophages and synoviocytes, which showed a significantly higher percentage of DiO+ than chondrocytes at every time point. The number in each figure represents DiO+ CPC percentage. (DiO+ cell percentage of other 4 cell types not shown). B) DiO+ cell percentages increased over time for all cell types and were higher in CPCs, synoviocytes, and macrophage populations than in chondrocyte populations isolated from full-thickness cartilage or from the upper zones (n = 3 per group per time point). The differences between CPCs and chondrocytes were significant at all time points. Data are the individual values (shapes) with the corresponding mean ± 95% confidence interval (range). The numbers above the bars indicate P values for differences between groups.
Lysosome Activity Comparison between CPCs and Chondrocytes
CPCs and chondrocytes were stained to assess cathepsin B activity, a marker of lysosomal activity. Confocal imaging revealed that almost all the cytoplasm of CPCs was red-fluorescent, while in chondrocytes, red fluorescence was significantly dimmer in the cytoplasm (Fig 2B). This indicated greater per cell lysosome activity in CPCs (Green fluorescence indicates cell debris). This impression was confirmed by flow cytometric analysis, which showed an average of 3.1-fold higher signal intensity in CPCs versus chondrocytes (Fig 2C).
Cathepsin B Inhibitor Effect on Cell Debris Degradation of CPCs and Chondrocytes
The pulse-chase experiment revealed that the percentage of CPCs with internalized debris decreased after removing the debris from 39.2% at 12 hrs post-removal to 28.7% at 24 hrs post-removal, while chondrocytes maintained virtually the same level at the two time points (12.9% at 12 hrs and 13.5% at 24 hrs). The percentage of positive CPCs was significantly higher at both time points in the presence of cathepsin B inhibitor (53.4% at 12 hrs at 33.0% 24 hrs. In contrast, cathepsin inhibition had minimal effects on chondrocyte debris ingestion percentage remained at similar (11.2% positive at 12 hrs and 16.2% positive at 24 hrs) (Fig 2D).
Quantitative Real-Time PCR Analysis of Phagocytosis Markers
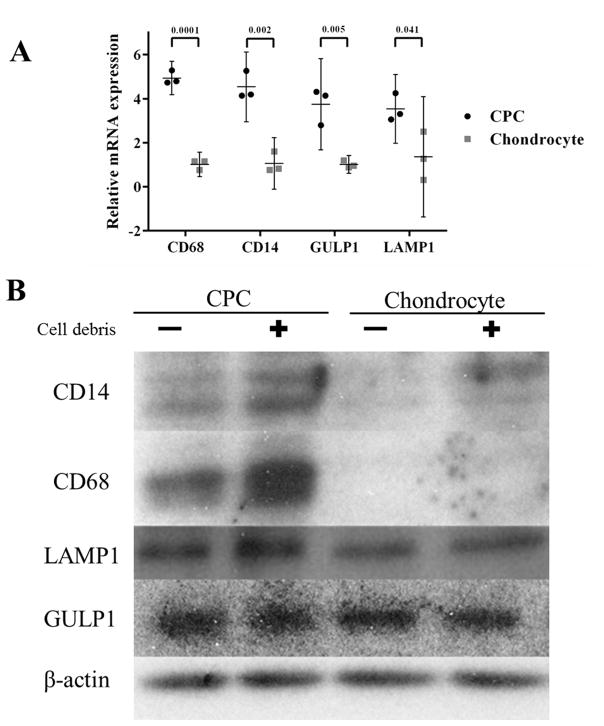
Gene expression analysis exhibited significantly higher expression of all phagocytosis markers in CPCs versus chondrocytes (4.9 fold, 4.5 fold, 3.8 fold and 3.5 fold for CD 68, CD 14, GULP1 and LAMP1, respectively) (Fig 4A).
Figure 4.
Expression of phagocytosis markers in CPCs and chondrocytes. A) real-time PCR showed that CPCs significantly over-expressed in all four phagocytosis markers relative to chondrocytes (n = 3 per group). Data are the individual values (shapes) with the corresponding mean ± 95% confidence interval (range). The numbers above the bars indicate P values for statistical differences between groups. B) Western blots showed higher CD14, CD68, and LAMP1 protein expression in CPCs than in chondrocytes. Incubation with cell debris induced expression in CPCs, but not in chondrocytes. A 4th marker, GULP, was expressed at similar levels in CPCs and chondrocytes. β-actin expression was served as loading control.
Western Blot Analysis of Lysosome Marker
Compared to chondrocytes, CPCs lysate samples showed significantly increased band signals indicating increased CD 14, CD 68, and LAMP1 expression. In addition, cell debris stimulated significantly higher expression of these three phagocytosis markers in CPCs, but not in chondrocytes; however, there was no significant difference between CPCs and chondrocytes with respect to GULP1 expression (Fig 4B).
Discussion
CPCs internalized cell debris significantly more avidly than chondrocytes. The percentage of CPCs with intracellular debris increased by nearly 6-fold, from 12% at 3 hours, to 68% at 12 hours. Similar time-related increases in the uptake of debris were observed in synoviocytes and macrophages but not in chondrocytes. Debris internalization by chondrocytes from the top 1/3 of the cartilage matrix did not differ significantly from chondrocytes from full-thickness cartilage. Relative to chondrocytes, CPCs over-expressed markers associated with phagocytosis at both the RNA and protein levels, which was consistent with relative enhancement of lysosomal mass and cathepsin activity in CPCs.
CPCs were also superior to chondrocytes in internalizing Fn-fs, a form of ECM debris that induces the chondrolytic activities of chondrocytes with a potency that rivals interleukin-1 beta [17]. Much of the fibronectin present in cartilage is concentrated in pericellular matrices, where proteolytic fragments are likely to exert direct biologic effects via integrin and toll-like receptors [18]. We showed previously in an osteochondral explant model that Fn-fs levels were elevated in cartilage under the same impact-injury circumstances that provoked CPC activation and chemotaxis [15]. Taken together, these data support the hypothesis that fibronectin proteolysis poses a unique threat to matrix stability, and that the clearance of Fn-fs by CPCs may be biologically significant.
The protein markers chosen for analysis here have been used to identify a variety of cells with phagocytic activity, including cells of the monocyte-macrophage lineage [19–22]. To our knowledge their expression has not been described in mesenchymal progenitor cells. Although we found that the expression of most phagocytosis-related markers was up-regulated in CPCs versus chondrocytes, GULP1 stands out as an exception in that expression was similar in the two cell types. The reason for this is unclear, but the fact that GULP1 expression was not inducible by cell debris in CPCs or chondrocytes is consistent with the absence of apoptotic bodies in freeze-thawed cell extracts, which contained factors released from necrotic cells rather than apoptotic cells.
The results of our cell debris uptake assays should be interpreted with some caution. First, it is conceivable that debris that was not labeled by the lipophilic DiO probe (e.g. non-membrane – associated proteins, mitochondrial DNA etc.) was taken up at quite different rates by the two cell types, indicating greater divergence in overall phagocytic performance than was apparent in our experiments. A second caveat concerns the freeze-thaw method used to kill cells, which does not generate the same mixture of necrotic and apoptotic cell debris encountered by CPCs or macrophages in mechanically-injured cartilage. This may be significant, as apoptotic bodies are subject to macrophage engulfment and have been shown to moderate the inflammatory response that drives chemotaxis and phagocytosis. Thus, if the reaction of CPCs to apoptotic body engulfment is similar to that of macrophages, we might expect weaker phagocytic activity in vivo than indicated by our purely necrotic cell debris-based assay. In addition, although the mouse-derived macrophage cell line used in this study is a standard model, the phagocytic behavior of the cells may not be fully representative of primary macrophages. Although debris appeared to bind to chondrocytes surfaces, we cannot rule out the possibility that extracellular matrix produced by chondrocytes hindered debris from binding to phagocytic receptors. Finally, we assessed the statistical difference between CPCs and chondrocytes as independent observations since these two cell population contain intrinsically distinct properties even though each batch of CPCs and chondrocytes were isolated from same bovine explants. Thus we conducted two-sided two-sample t-test to evaluate the difference between these two cell types.
In vascularized tissues, inflammatory cells of hematopoietic origin are drawn to injured tissues by alarmins contained in debris released from necrotic and apoptotic cells. Acting through innate immune receptors, alarmins stimulate immunocyte chemotaxis and phagocytosis, and cytokine release. The ensuing localized inflammatory reaction is geared towards sterilizing and debriding wounds, and only when cell and tissue debris are cleared does inflammation subside [23]. How this unfolds in injured cartilage where immunocytes are relatively scarce is still unclear, but our findings to date suggest that CPCs can provide the same debris-clearing functions as macrophages. This conclusion is consistent with our previous data, which showed that alarmin-activated CPCs over-expressed pro-inflammatory chemokines and cytokines including IL8 and CXCL12 [8], indicating the potential for a CPC-initiated proinflammatory cascade similar to that initiated by immunocytes in wound sites. Notably, since CPCs also possess phagocytic capabilities that ultimately quench the alarmin signaling driving inflammation, like macrophages they may be empowered to complete the inflammatory cycle and make way for tissue regeneration. However, additional experiments are needed to characterize the fate of the debris internalized by CPCs before we can conclude that they are fully capable of debris clearance in this setting.
CPCs are not the only cells originating in the joint that are capable of scavenging cell debris: Synoviocytes, mesenchymal stem cells, and dendritic cells show phagocytic activity and are likely to be involved in healing responses in some joint tissues [12, 24–29]. However, in contrast to CPCs these cells must migrate long distances across healthy cartilage to reach a focal cartilage lesion, a process that may be hindered by the anti-adhesive lubricin coating on cartilage surfaces. This problem is obviated in the case of CPCs, which are recruited locally from nearby cartilage and only need to travel over short distances on damaged articular surfaces that are typically depleted of lubricin. The advantages of proximity and good traction would appear to have important consequences for healing cartilage lesions, as CPCs residing near experimental cartilage defects easily migrate into and populate scaffolds, where they can be induced to regenerate hyaline cartilage in situ [30]. Even if this process never occurs spontaneously in vivo, there is clearly some intrinsic repair potential available to exploit therapeutically.
Acknowledgments
We sincerely thank Barbara Laughlin and John Bierman for the acquisition of the cow stifles and osteochondral explants. The flow cytometry data presented herein were obtained at the Flow Cytometry Facility, which is a Carver College of Medicine/Holden Comprehensive Cancer Center core research facility at the University of Iowa. The Facility is funded through user fees and the generous financial support of the Carver College of Medicine, Holden Comprehensive Cancer Center, and Iowa City Veterans Administration Medical Center.
Funding source
This work was supported by the National Institutes of Health (CORT NIH P50 AR055533), by the Department of Defense (W81XWH-10-1-0702a), and by the University of Iowa Department of Orthopaedics and Rehabilitation.
Footnotes
Author contributions
Conception and design: Zhou, Zheng, Martin
Analysis and interpretation of the data: Zhou, Zheng, Martin
Drafting of the article: Zhou, Martin, Buckwalter
Critical revision of the article for important intellectual content: Martin, Buckwalter
Final approval of the article: Zhou, Zheng, Buckwalter, Martin
Statistical expertise: Zhou
Collection and assembly of data: Zhou
Zhou (cheng-zhou@uiowa.edu) and Martin (james-martin@uiowa.edu) take responsibility for the integrity of the work as a whole, from inception to finished article.
Conflict of interest
The authors have no conflicts of interest to declare regarding this study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Cheng Zhou, Email: cheng-zhou@uiowa.edu.
Hongjun Zheng, Email: zhengh@wudosis.wustl.edu.
Joseph A. Buckwalter, Email: joseph-buckwalter@uiowa.edu.
James A. Martin, Email: james-martin@uiowa.edu.
References
- 1.Williams R, Khan IM, Richardson K, Nelson L, McCarthy HE, Analbelsi T, et al. Identification and clonal characterisation of a progenitor cell sub-population in normal human articular cartilage. PLoS One. 2010;5:e13246. doi: 10.1371/journal.pone.0013246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koelling S, Kruegel J, Irmer M, Path JR, Sadowski B, Miro X, et al. Migratory chondrogenic progenitor cells from repair tissue during the later stages of human osteoarthritis. Cell Stem Cell. 2009;4:324–335. doi: 10.1016/j.stem.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 3.Dowthwaite GP, Bishop JC, Redman SN, Khan IM, Rooney P, Evans DJ, et al. The surface of articular cartilage contains a progenitor cell population. J Cell Sci. 2004;117:889–897. doi: 10.1242/jcs.00912. [DOI] [PubMed] [Google Scholar]
- 4.Alsalameh S, Amin R, Gemba T, Lotz M. Identification of mesenchymal progenitor cells in normal and osteoarthritic human articular cartilage. Arthritis Rheum. 2004;50:1522–1532. doi: 10.1002/art.20269. [DOI] [PubMed] [Google Scholar]
- 5.Hattori S, Oxford C, Reddi AH. Identification of superficial zone articular chondrocyte stem/progenitor cells. Biochem Biophys Res Commun. 2007;358:99–103. doi: 10.1016/j.bbrc.2007.04.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seol D, McCabe DJ, Choe H, Zheng H, Yu Y, Jang K, et al. Chondrogenic progenitor cells respond to cartilage injury. Arthritis Rheum. 2012;64:3626–3637. doi: 10.1002/art.34613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seol D, Choe H, Ramakrishnan PS, Jang K, Kurriger GL, Zheng H, et al. Organ culture stability of the intervertebral disc: rat versus rabbit. J Orthop Res. 2013;31:838–846. doi: 10.1002/jor.22285. [DOI] [PubMed] [Google Scholar]
- 8.Zhou C, Zheng H, Seol D, Yu Y, Martin JA. Gene expression profiles reveal that chondrogenic progenitor cells and synovial cells are closely related. J Orthop Res. 2014;32:981–988. doi: 10.1002/jor.22641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu Y, Zheng H, Buckwalter JA, Martin JA. Single cell sorting identifies progenitor cell population from full thickness bovine articular cartilage. Osteoarthritis Cartilage. 2014;22:1318–1326. doi: 10.1016/j.joca.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarantis H, Grinstein S. Subversion of phagocytosis for pathogen survival. Cell Host Microbe. 2012;12:419–431. doi: 10.1016/j.chom.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Yang J, Zhang L, Yu C, Yang XF, Wang H. Monocyte and macrophage differentiation: circulation inflammatory monocyte as biomarker for inflammatory diseases. Biomark Res. 2014;2:1. doi: 10.1186/2050-7771-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartok B, Firestein GS. Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol Rev. 2010;233:233–255. doi: 10.1111/j.0105-2896.2009.00859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bondeson J, Blom AB, Wainwright S, Hughes C, Caterson B, van den Berg WB. The role of synovial macrophages and macrophage-produced mediators in driving inflammatory and destructive responses in osteoarthritis. Arthritis Rheum. 2010;62:647–657. doi: 10.1002/art.27290. [DOI] [PubMed] [Google Scholar]
- 14.Jiao K, Zhang J, Zhang M, Wei Y, Wu Y, Qiu ZY, et al. The identification of CD163 expressing phagocytic chondrocytes in joint cartilage and its novel scavenger role in cartilage degradation. PLoS One. 2013;8:e53312. doi: 10.1371/journal.pone.0053312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding L, Guo D, Homandberg GA, Buckwalter JA, Martin JA. A single blunt impact on cartilage promotes fibronectin fragmentation and upregulates cartilage degrading stromelysin-1/matrix metalloproteinase-3 in a bovine ex vivo model. J Orthop Res. 2014;32:811–818. doi: 10.1002/jor.22610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang KW, Ding L, Seol D, Lim TH, Buckwalter JA, Martin JA. Low-intensity pulsed ultrasound promotes chondrogenic progenitor cell migration via focal adhesion kinase pathway. Ultrasound Med Biol. 2014;40:1177–1186. doi: 10.1016/j.ultrasmedbio.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Homandberg GA, Ummadi V, Kang H. High molecular weight hyaluronan promotes repair of IL-1 beta-damaged cartilage explants from both young and old bovines. Osteoarthritis Cartilage. 2003;11:177–186. doi: 10.1016/s1063-4584(02)00371-0. [DOI] [PubMed] [Google Scholar]
- 18.Hwang HS, Park SJ, Cheon EJ, Lee MH, Kim HA. Fibronectin fragment-induced expression of matrix metalloproteinases is mediated by MyD88-dependent TLR-2 signaling pathway in human chondrocytes. Arthritis Res Ther. 2015;17:320. doi: 10.1186/s13075-015-0833-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devitt A, Moffatt OD, Raykundalia C, Capra JD, Simmons DL, Gregory CD. Human CD14 mediates recognition and phagocytosis of apoptotic cells. Nature. 1998;392:505–509. doi: 10.1038/33169. [DOI] [PubMed] [Google Scholar]
- 20.Travaglione S, Falzano L, Fabbri A, Stringaro A, Fais S, Fiorentini C. Epithelial cells and expression of the phagocytic marker CD68: scavenging of apoptotic bodies following Rho activation. Toxicol In Vitro. 2002;16:405–411. doi: 10.1016/s0887-2333(02)00028-0. [DOI] [PubMed] [Google Scholar]
- 21.Huynh KK, Eskelinen EL, Scott CC, Malevanets A, Saftig P, Grinstein S. LAMP proteins are required for fusion of lysosomes with phagosomes. EMBO J. 2007;26:313–324. doi: 10.1038/sj.emboj.7601511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sullivan CS, Scheib JL, Ma Z, Dang RP, Schafer JM, Hickman FE, et al. The adaptor protein GULP promotes Jedi-1-mediated phagocytosis through a clathrin-dependent mechanism. Mol Biol Cell. 2014;25:1925–1936. doi: 10.1091/mbc.E13-11-0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kraus S, Arber N. Inflammation and colorectal cancer. Curr Opin Pharmacol. 2009;9:405–410. doi: 10.1016/j.coph.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 24.Tso GH, Law HK, Tu W, Chan GC, Lau YL. Phagocytosis of apoptotic cells modulates mesenchymal stem cells osteogenic differentiation to enhance IL-17 and RANKL expression on CD4+ T cells. Stem Cells. 2010;28:939–954. doi: 10.1002/stem.406. [DOI] [PubMed] [Google Scholar]
- 25.Hinds KA, Hill JM, Shapiro EM, Laukkanen MO, Silva AC, Combs CA, et al. Highly efficient endosomal labeling of progenitor and stem cells with large magnetic particles allows magnetic resonance imaging of single cells. Blood. 2003;102:867–872. doi: 10.1182/blood-2002-12-3669. [DOI] [PubMed] [Google Scholar]
- 26.Schwachula A, Riemann D, Kehlen A, Langner J. Characterization of the immunophenotype and functional properties of fibroblast-like synoviocytes in comparison to skin fibroblasts and umbilical vein endothelial cells. Immunobiology. 1994;190:67–92. doi: 10.1016/S0171-2985(11)80284-6. [DOI] [PubMed] [Google Scholar]
- 27.Savina A, Amigorena S. Phagocytosis and antigen presentation in dendritic cells. Immunol Rev. 2007;219:143–156. doi: 10.1111/j.1600-065X.2007.00552.x. [DOI] [PubMed] [Google Scholar]
- 28.Nagl M, Kacani L, Mullauer B, Lemberger EM, Stoiber H, Sprinzl GM, et al. Phagocytosis and killing of bacteria by professional phagocytes and dendritic cells. Clin Diagn Lab Immunol. 2002;9:1165–1168. doi: 10.1128/CDLI.9.6.1165-1168.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Albert ML, Pearce SF, Francisco LM, Sauter B, Roy P, Silverstein RL, et al. Immature dendritic cells phagocytose apoptotic cells via alphavbeta5 and CD36, and cross-present antigens to cytotoxic T lymphocytes. J Exp Med. 1998;188:1359–1368. doi: 10.1084/jem.188.7.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu Y, Brouillette MJ, Seol D, Zheng H, Buckwalter JA, Martin JA. Functional full-thickness articular cartilage repair by rhSDF-1alpha loaded fibrin/ha hydrogel network via chondrogenic progenitor cells homing. Arthritis Rheumatol. 2015 doi: 10.1002/art.39049. [DOI] [PMC free article] [PubMed] [Google Scholar]