Abstract
Retroviral infection requires integration of the viral genome into the host genome. Recombinant integrase proteins may be purified following bacterial expression. A bulk biochemical assay of integrase function relies on the conversion of supercoiled plasmids to linear or relaxed circles. Single molecule molecular tweezer assays of integrase also evaluate the conversion of supercoiled DNA to nicked and broken species. A bacterial nuclease that co-purifies with retroviral integrase may affect the quantitation of integration activity in bulk or single molecule assays. During purification of retroviral integrase from bacteria, fractions may be screened for contaminating nuclease activity. In order to efficiently separate the nuclease from integrase, the binding affinities of each protein must differ. We find that a co-purifying nuclease may be efficiently separated from integrase based on heparin affinity, but not ionic affinity.
Keywords: Retrovirus, integrase, protein purification
Retroviruses must integrate a copy of their viral genome to the host genome to continue the viral life cycle. Integration is performed by the viral enzyme integrase (Coffin, Hughes, and Varmus, 1997). Recombinant integrase (IN) from many retroviruses may be purified following expression in bacteria (Jonsson, Donzella, and Roth, 1993; Marczinovits et al., 1992; Roth, Tanese, and Goff, 1988; Sherman and Fyfe, 1990; Terry et al., 1988; Valkov et al., 2009). After reverse transcription of the viral genomic RNA into a cDNA, retroviral IN has two catalytic activities in vivo. First, IN cleaves two nucleotides from each end of the viral cDNA resulting in recessed 3′ hydroxyl groups, termed 3′ processing. Second, IN joins each 3′ hydroxyl of the viral cDNA to the host genomic DNA in a single step transesterification reaction, called strand transfer. The ultimate product of integration in vivo is the viral genome stably inserted to the host genome.
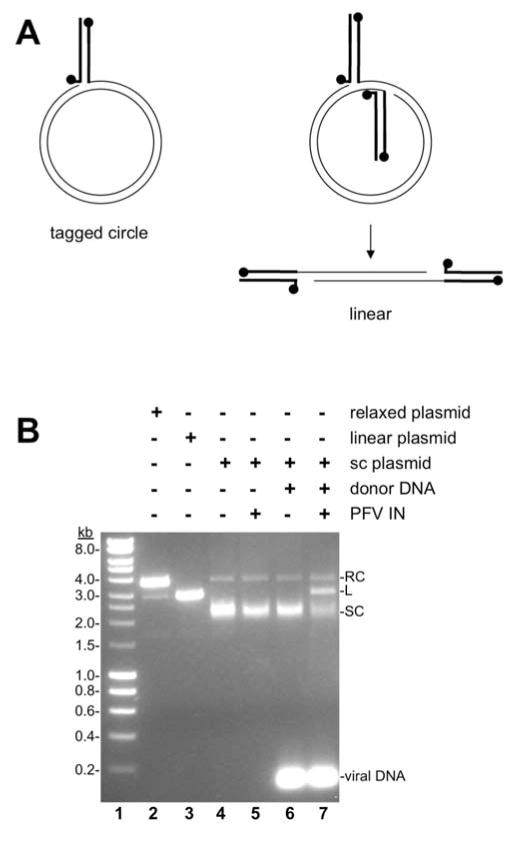
The preferences for retroviral integration sites in vivo, including chromatin features and subtle sequence preference, appear to be unique to each retrovirus (Holman and Coffin, 2005; Shun et al., 2007; Wu et al., 2005). These preferences continue to be explored and defined by assays in vivo and in vitro (Bennett et al., 2014; Hacker et al., 2006; Kang et al., 2006; Serrao et al., 2015; Taganov et al., 2004; Vora and Grandgenett, 1995). Bulk biochemical assays for integration activity in vitro utilize purified recombinant IN with two viral oligomer donor DNAs and a target supercoiled plasmid (Cherepanov, 2007; Li and Craigie, 2005; Li et al., 2006; Sinha and Grandgenett, 2005; Valkov et al., 2009) (Fig. 1). Integration assay products are separated by agarose gel electrophoresis, which readily separates the relaxed circle, linear, and supercoiled species of a plasmid (Fig. 1B, lanes 2–4). IN mediated strand transfer of both viral DNA ends to the target plasmid DNA results in a linear product (Fig. 1A). Retroviral integration assays in vitro may also result in a non-physiologically relevant product consisting of a single viral DNA end joined to the target, termed half site integration (Goodarzi et al., 1995; Vora and Grandgenett, 1995). The half site integration product is a relaxed circle with a single viral DNA oligomer, a tagged circle. Quantitation of linear and relaxed products indicates concerted and half site integration, respectively.
Fig. 1. PFV integration assay.
A. Cartoon of a bulk biochemical retroviral integration assay. Short oligomers of PFV viral DNA ends are shown as bold lines with balls indicating the 5′ ends. Target DNA is shown as thin lines. Right, A supercoiled plasmid target DNA is relaxed by half site integration with a single viral donor DNA yielding a tagged circle. Half site integration is not physiologically relevant. Left, Alternatively, the supercoiled plasmid may be linearized by concerted integration with two viral donor DNA oligonucleotides. Concerted integration is the physiologically relevant integration reaction. B. Agarose gel electrophoresis separates the products of a PFV integration assay. The table at the top of the gel image indicates the addition of DNA forms and PFV IN. A supercoiled plasmid was treated with single strand DNA nicking restriction nuclease Nt.BspQ1 which produces relaxed circles and minimal linear products (lane 2; RC, relaxed circle), linearized with BamHI (lane 3; L, linear), or untreated (lane 4; SC, supercoiled). Addition of PFV IN (lane 5) or viral donor DNA (lane 6) does not change the supercoiled plasmid. However, addition of PFV IN and viral donor DNA results in linear concerted integration products (lane 7).
The prototype foamy virus integrase (PFV IN) has become a model for studying integrase interactions with target DNA. PFV IN is sensitive to clinically relevant HIV-1 IN strand transfer inhibitors (Valkov et al., 2009). It was the first full length integrase protein to be crystallized with viral and target DNAs (Hare et al.; Maertens, Hare, and Cherepanov, 2010). A recent cryo-EM structure illuminates the binding of a tetramer of PFV IN to a mononucleosome (Maskell et al., 2015). Finally, the first single molecule observations of retroviral IN complexes searching target DNA in real time utilized PFV IN (Jones et al., 2016). This protein has become arguably the most biophysically well-characterized retroviral IN.
DNA nucleases with non-specific nicking activity may also relax or linearize a supercoiled plasmid target DNA. The gel mobilities of plasmids relaxed or linearized by a co-purifying nuclease are indistinguishable from half site or concerted integration products, respectively. Furthermore, supercoiled plasmid DNA is a preferred target for PFV IN compared to relaxed circles or linear DNA (Jones et al., 2016). A co-purifying nuclease that converts the plasmid target to relaxed or linearized DNAs will reduce the concentration of preferred supercoiled plasmid and alter the dynamics of the reaction. Thus a co-purifying bacterial nuclease may confound accurate quantitation of retroviral integration assay products in vitro.
During purification of recombinant PFV IN from E. coli strain BL21/DE3 Rosetta cells (Novagen), we noticed a non-specific nuclease activity. We used several chromatography affinity resins to characterize the contaminating bacterial nuclease that co-purifies with PFV IN. Size exclusion chromatography may be used to remove possible co-purifying bacterial nucleases (Hare et al., 2010). However, the volume that may be loaded to size exclusion columns is typically limited, while affinity chromatography has no load volume limitations. There have been no previous reports of the binding affinities of the nuclease that co-purifies with retroviral IN. The differences in binding affinities of PFV IN and the nuclease suggest heparin affinity purification as an alternative strategy to size exclusion chromatography.
All retroviral INs possess inherent endonuclease activity for 3′ processing and utilize a catalytic DDE motif (Coffin et al., 1997; Kulkosky et al., 1992; Sherman and Fyfe, 1990). We purified a catalytically inactive mutant PFV IN(D128N) to analyze and differentiate the binding affinities of IN and the co-purifying nuclease. PFV IN(D128N) and the nuclease have differing affinity for heparin sepharose, and Mono-S cation resin, but not Mono-Q anion resin. Purification and nuclease activity assays were performed three times with equivalent results; representative gels are shown here.
For purification from bacterial expression systems, retroviral IN expression constructs commonly include a hexa-histidine tag for initial purification with nickel resin (Cherepanov et al., 1999; Valkov et al., 2009; Villanueva et al., 2003). PFV IN(D128N) with an amino-terminal hexa-histidine tag was expressed in the Rosetta derivative of BL21(DE3) pLysS cells (Novagen). The culture was grown to OD600 0.9–1.0 and induced with 0.25 mM IPTG and 50 μM ZnCl2 (all chemicals from Sigma-Aldrich) at 25°C for 4 hours (Valkov et al.). The bacterial cells were sonicated in 50 mM Tris-HCl, pH 7.5, 500 mM NaCl, and protease inhibitor PMSF. The lysate was centrifuged at 120,000 g for 1 hour at 4°C. The supernatant was separated by Ni-NTA Superflow resin (Qiagen) with a linear gradient of 20–200 mM imidazole, pH 8.0. Fractions were analyzed by PAGE stained with Coomassie Brilliant Blue. Fractions containing PFV IN(D128N) were combined and supplemented with 10 mM DTT and 0.1 mM EDTA. Tags, such as the hexa-histidine tag, may impair the ability of retroviral IN to form a functional multimer. A PreScission site between the tag and PFV IN allowed the hexa-histidine tag to be removed by digestion with human rhinovirus (HRV) 3C protease (Novagen) overnight at 4°C.
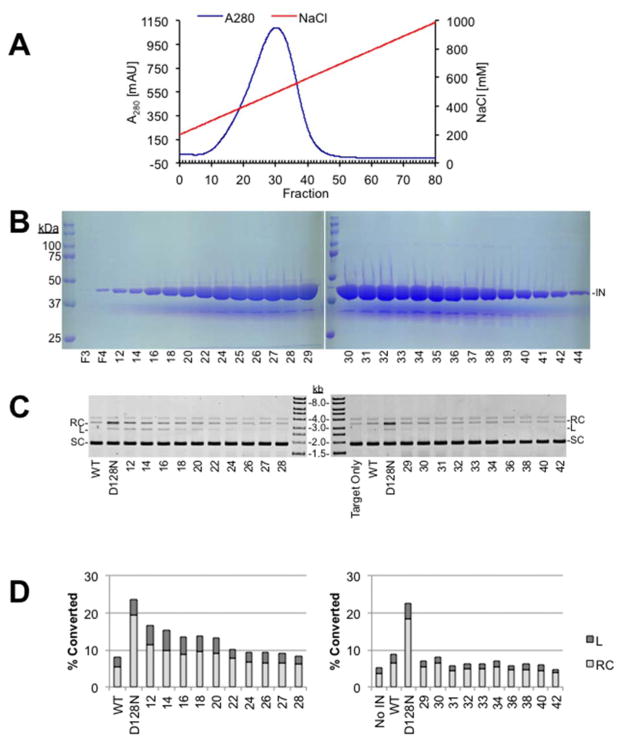
Recombinant retroviral IN is commonly fractionated by heparin sepharose affinity resin with a salt gradient following nickel chromatography (Cherepanov et al., 1999; Valkov et al., 2009; Villanueva et al., 2003). The digestion reaction was diluted with 3 volumes 50 mM Tris-HCl, pH 7.5, 10 mM DTT, and 0.1 mM EDTA to reduce the salt concentration. Digested and diluted PFV IN(D128N) was bound to heparin sepharose resin in 50 mM Tris, pH 7.5, 10 mM DTT, 0.1 mM EDTA and eluted with a linear gradient of 0.2–1 M NaCl (Fig. 2A). Heparin fractions were analyzed by PAGE for the presence of IN protein (Fig. 2B) and by agarose gel electrophoresis for nuclease activity (Fig. 2C and D). Nuclease assays combined 3 μl of heparin fraction, 50 ng supercoiled plasmid pMP2, 10 mM HEPES, pH 7.5, 110 mM NaCl, 5 mM MgSO4, 4 μM ZnCl2, and 10 mM DTT in a final volume of 15 μl. No viral donor DNA was included in the nuclease assays. The reactions were incubated at 37°C for 90 minutes. Proteinase K and SDS were added to stop the reactions and digest proteins. DNA species were evaluated by agarose gel electrophoresis. Ethidium bromide stained gels were scanned with a Typhoon 9410 variable mode imager (GE Healthcare Life Sciences). Bands in each lane were quantified with ImageQuant TL software. The volume values of the relaxed circle, linear, and supercoiled bands were added to yield the total DNA volume in each lane. The percentage of the total DNA volume converted to relaxed circle and linear products is shown as “% Converted”. Reactions that did not include IN protein (No IN) displayed 4.4 (±0.7)% of the total DNA as relaxed circles or linear products. As a control wild type PFV IN was purified with Ni-NTA Superflow followed by heparin sepharose. The heparin sepharose fractions were analyzed by the endonuclease assay and an integrase assay. Fractions that were negative for endonuclease activity and positive for integrase activity were combined, dialyzed, and stored at −80°C. This nuclease free preparation of wild type PFV IN routinely showed 8.8 (±0.9)% of the DNA converted to products. In contrast, a previous purification of PFV IN(D128N) that combined all of the heparin sepharose fractions with PFV IN(D128N) protein present and did not exclude fractions with nuclease activity displayed 24.4 (±2.1)% conversion to relaxed circles and linear products.
Fig. 2. Heparin sepharose chromatography of PFV IN(D128N).
A. Chromatography profile of PFV IN(D128N) fractionation with heparin sepharose. Blue line indicates the A280. Red line indicates the gradient of NaCl. B. Coomassie stained PAGE analysis of selected fractions following heparin sepharose chromatography of PFV IN(D128N), 44 kDa (IN). F3 is the void volume and F4 is the wash volume. Fraction numbers are indicated at the bottom of the gel images. C. Nuclease assay of heparin sepharose fractions separated by agarose, stained with ethidium bromide, and imaged with a Typhoon laser scanner. Relaxed circles (RC), linear (L), and supercoiled (SC) plasmid are indicated. Nuclease free PFV IN wild type (WT) is included as a negative control for nuclease activity. A previous purification of PFV IN(D128N) that did not exclude the co-purifying nuclease is included as a positive control for nuclease activity (D128N). D. Quantitation of the nuclease assay. Relative relaxation or linearization of the total plasmid associated with each heparin sepharose fraction is shown.
The heparin sepharose fractions with the peak nuclease activity did not correlate with the fractions of peak PFV IN(D128N) protein (Fig. 2). The highest concentration of PFV IN(D128N) protein was in fractions 29–32. The co-purifying nuclease was not readily visualized by Coomassie stain, which could indicate that it is at a low concentration or has a molecular weight less than 20 kDa (Fig. 2B). However, the nuclease assays indicated higher activity in fractions 12–22 and less activity in fractions 24–42 (Fig. 2C and D). Lower nuclease activity fractions 24–42 were combined and further analyzed for cation and anion affinity.
Half of the low nuclease PFV IN(D128N) heparin fractions 24–42 were analyzed by Mono Q chromatography, a strong anion resin. The column was run in 50 mM Tris, pH 7.5, 10 mM DTT, 0.1 mM EDTA, and a linear gradient of 0.1–1 M NaCl. PFV IN(D128N) does not bind to this resin and passed through the column. No OD280 peaks indicating protein were observed throughout the salt gradient. Several fractions were tested for nuclease activity, but had no detectable nuclease. This suggested the contaminating nuclease does not bind to Mono Q and passed through the column similar to PFV IN. Coomassie stained PAGE showed the PFV IN(D128N) in the void volume. Neither protein showed affinity for Mono Q anion resin.
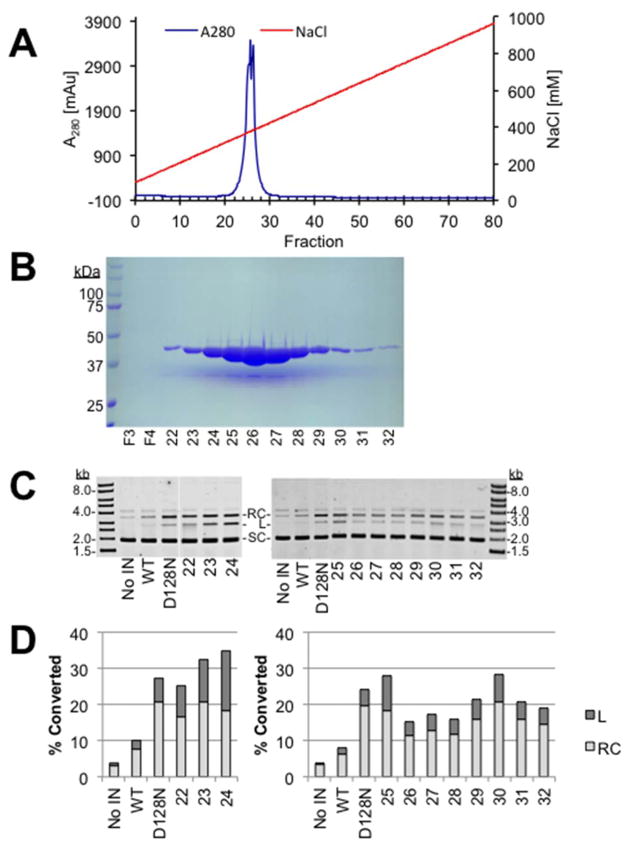
The remaining half of the low nuclease PFV IN(D128N) heparin sepharose fractions 24–42 were also analyzed by Mono S chromatography, a strong cation resin. This column was run in the same buffers as the Mono Q column. PFV IN(D128N) eluted in a sharp peak from the Mono S resin largely in fractions 25–27 (Fig. 3A and B). The co-purifying nuclease also bound to the Mono S resin with peak activity in fraction 24 (Fig. 3C and D). There was little separation of PFV IN(D128N) and the co-purifying nuclease by Mono-S chromatography. The near overlap of the PFV IN(D128N) protein peak and the nuclease activity peak suggests that altering the elution gradient would not effectively resolve the two proteins.
Fig. 3. Mono-S cation chromatography of PFV IN(D128N).
A. Chromatography profile of PFV IN(D128N) fractionation with Mono-S anion resin. Blue line indicates the A280. Red line indicates the gradient of NaCl. B. Coomassie stained PAGE analysis of selected fractions following Mono-S chromatography of PFV IN(D128N), 44 kD (IN). F3 is the void volume and F4 is the wash volume. Fraction numbers are indicated at the bottom of the gel images. C. Nuclease assay of Mono-S fractions separated by agarose, stained with ethidium bromide, and imaged with a Typhoon laser scanner. Relaxed circles (RC), linear (L), and supercoiled (SC) plasmid are indicated. Wild type PFV IN without nuclease (WT) and a previous preparation of PFV IN(D128N) with co-purifying nuclease (D128N) are included as controls. D. Quantitation of the nuclease assay. Relative relaxation or linearization of the total plasmid associated with each Mono-S fraction is shown.
Results of retroviral integration assays in vitro may be confused by a co-purifying bacterial nuclease. We have found that the co-purifying nuclease was most effectively separated from PFV IN by fractionation with heparin sepharose resin. Fractionation by Mono-S cation resin revealed that the peak of PFV IN protein and the peak of nuclease activity are distinguishable, but not efficiently separated. Mono-Q anion resin did not bind either PFV IN or the co-purifying nuclease. When size exclusion chromatography is unavailable or impractical, heparin sepharose chromatography combined with nuclease assays may efficiently isolate PFV IN without co-purifying nuclease activity.
Highlights.
A bacterial DNA nuclease co-purifies with recombinant retroviral integrase.
Integrase assays may be confounded by the contaminating nuclease.
The nuclease and integrase have differing affinity for heparin sepharose.
Acknowledgments
This work was support by NIH AI099854 to KEY.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bennett GR, Peters R, Wang XH, Hanne J, Sobol RW, Bundschuh R, Fishel R, Yoder KE. Repair of oxidative DNA base damage in the host genome influences the HIV integration site sequence preference. PloS one. 2014;9:e103164. doi: 10.1371/journal.pone.0103164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 2007;35:113–24. doi: 10.1093/nar/gkl885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Surratt D, Toelen J, Pluymers W, Griffith J, De Clercq E, Debyser Z. Activity of recombinant HIV-1 integrase on mini-HIV DNA. Nucleic Acids Res. 1999;27:2202–10. doi: 10.1093/nar/27.10.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1997. [PubMed] [Google Scholar]
- Goodarzi G, Im GJ, Brackmann K, Grandgenett D. Concerted integration of retrovirus-like DNA by human immunodeficiency virus type 1 integrase. Journal of virology. 1995;69:6090–7. doi: 10.1128/jvi.69.10.6090-6097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker CV, Vink CA, Wardell TW, Lee S, Treasure P, Kingsman SM, Mitrophanous KA, Miskin JE. The integration profile of EIAV-based vectors. Mol Ther. 2006;14:536–45. doi: 10.1016/j.ymthe.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232–6. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman AG, Coffin JM. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6103–7. doi: 10.1073/pnas.0501646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N, Lopez MA, Jr, Hanne J, Peake M, Lee JB, Fishel R, Yoder KE. Retroviral Intasomes Search a Target DNA by 1D-Diffusion Rarely Resulting in Integration. Nat Commun. 2016 doi: 10.1038/ncomms11409. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson CB, Donzella GA, Roth MJ. Characterization of the forward and reverse integration reactions of the Moloney murine leukemia virus integrase protein purified from Escherichia coli. J Biol Chem. 1993;268:1462–9. [PubMed] [Google Scholar]
- Kang Y, Moressi CJ, Scheetz TE, Xie L, Tran DT, Casavant TL, Ak P, Benham CJ, Davidson BL, McCray PB., Jr Integration site choice of a feline immunodeficiency virus vector. Journal of virology. 2006;80:8820–3. doi: 10.1128/JVI.00719-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkosky J, Jones KS, Katz RA, Mack JP, Skalka AM. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–8. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Craigie R. Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. J Biol Chem. 2005;280:29334–9. doi: 10.1074/jbc.M505367200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Mizuuchi M, Burke TR, Jr, Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. The EMBO journal. 2006;25:1295–304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens GN, Hare S, Cherepanov P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature. 2010;468:326–9. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczinovits I, Molnar J, Soki J, Fodor I. Overexpression and purification of enzymatically active recombinant integrase protein of Rous sarcoma virus. Virus genes. 1992;6:301–6. doi: 10.1007/BF01702568. [DOI] [PubMed] [Google Scholar]
- Maskell DP, Renault L, Serrao E, Lesbats P, Matadeen R, Hare S, Lindemann D, Engelman AN, Costa A, Cherepanov P. Structural basis for retroviral integration into nucleosomes. Nature. 2015;523:366–9. doi: 10.1038/nature14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MJ, Tanese N, Goff SP. Gene product of Moloney murine leukemia virus required for proviral integration is a DNA-binding protein. J Mol Biol. 1988;203:131–9. doi: 10.1016/0022-2836(88)90097-6. [DOI] [PubMed] [Google Scholar]
- Serrao E, Ballandras-Colas A, Cherepanov P, Maertens GN, Engelman AN. Key determinants of target DNA recognition by retroviral intasomes. Retrovirology. 2015;12:39. doi: 10.1186/s12977-015-0167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman PA, Fyfe JA. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:5119–23. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–78. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Grandgenett DP. Recombinant human immunodeficiency virus type 1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegration complexes from virus-infected cells. Journal of virology. 2005;79:8208–16. doi: 10.1128/JVI.79.13.8208-8216.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov KD, Cuesta I, Daniel R, Cirillo LA, Katz RA, Zaret KS, Skalka AM. Integrase-specific enhancement and suppression of retroviral DNA integration by compacted chromatin structure in vitro. Journal of virology. 2004;78:5848–55. doi: 10.1128/JVI.78.11.5848-5855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry R, Soltis DA, Katzman M, Cobrinik D, Leis J, Skalka AM. Properties of avian sarcoma-leukosis virus pp32-related pol-endonucleases produced in Escherichia coli. Journal of virology. 1988;62:2358–65. doi: 10.1128/jvi.62.7.2358-2365.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkov E, Gupta SS, Hare S, Helander A, Roversi P, McClure M, Cherepanov P. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 2009;37:243–55. doi: 10.1093/nar/gkn938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva RA, Jonsson CB, Jones J, Georgiadis MM, Roth MJ. Differential multimerization of Moloney murine leukemia virus integrase purified under nondenaturing conditions. Virology. 2003;316:146–60. doi: 10.1016/s0042-6822(03)00559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vora AC, Grandgenett DP. Assembly and catalytic properties of retrovirus integrase-DNA complexes capable of efficiently performing concerted integration. Journal of virology. 1995;69:7483–8. doi: 10.1128/jvi.69.12.7483-7488.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B, Burgess SM, Munroe DJ. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. Journal of virology. 2005;79:5211–4. doi: 10.1128/JVI.79.8.5211-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]