Abstract
Barcoding of biological samples is a commonly used strategy to mark or identify individuals within a complex mixture. However, cell barcoding has not yet found wide use in flow cytometry, that would benefit greatly from the ability to analyze pooled experimental samples simultaneously. This is due, in part, to technical and practical limitations of current fluorescent dye-based methods. Here we describe a simple, versatile barcoding strategy that relies on combinations of a single antibody conjugated to different fluorochromes and thus in principle can be integrated into any flow cytometry application. To demonstrate the efficacy of the approach we describe the results of a variety of experiments using live cells as well as fixed and permeabilized cells. The results of these studies show that antibody-based barcoding provides a simple, practical method for identifying cells from individual samples pooled for analysis by flow cytometry that has broad applications in immunological research.
Keywords: Barcoding, phospho-flow, T cells, B cells, flow cytometry, calcium flux
INTRODUCTION
Flow cytometry is a widely used experimental technology with broad applications both for immunological research and for clinical diagnostics including phenotyping cell subsets and identifying changes in cellular activities such as protein expression, cytokine secretion, cell cycle progression, cell-cell interaction and cell death (1–4). Barcoding cells in multiple samples and combining these prior to staining and data acquisition by flow cytometry is an efficient strategy both to achieve uniform staining of pooled individual samples and to improve accuracy by eliminating sample-to-sample variation. Barcoding allows for a relatively short acquisition time by decreasing the sample number and therefore offers a practical way to analyze large numbers of samples. Finally, in principle, when individual samples are barcoded and pooled, the subsequent staining requires less reagent per sample and consequently reduces the cost of analyses.
A current frequently used barcoding strategy, fluorescent cell barcoding (FCB), relies on staining individual samples of cells with dilutions of amine-reactive N-hydroxysuccinimide-derived fluorescent dyes before pooling (5, 6). These dyes stain cells to various degrees based on the dilution of the dye, allowing cells to be distinguished by their fluorescence intensity. This method requires careful optimization of dilutions of dyes, as changes in pH or dye lot may alter the results (5). In addition, solutions of numerous different concentrations of the dyes must be prepared just prior to staining. Based on the unique properties of the cell type or the effects of previous stimulation conditions, further optimization might be required. Since this method does not discriminate between particular cell types, samples may need to be further stained with phenotyping antibodies (Abs), adding another step to the procedure. The dyes used in this method lose their activity relatively quickly upon storage and thus may not be convenient for occasional users. The commonly used fluorochromes, cell barcoding dyes (CBD) 450 and 500 (emissions at 450 nm and 500 nm), are difficult to compensate in flow cytometry because of the close proximity of their emission spectra as well as possible leakage of signal into other channels due to the broad emission range, especially for CBD500. Furthermore, since the barcoding dyes are delivered in a methanol-containing solution, the method is not applicable to real time assays of live cells.
Here, we describe a novel barcoding strategy for use in flow cytometry, based on Abs commonly used for immune cell phenotyping. Cells are stained with the same monoclonal Ab conjugated to different fluorochromes in various combinations. Staining requires no more optimization than is generally necessary for flow cytometry and the fluorochromes can be chosen depending on the availability of reagents as well as the specific needs of the experiment. In addition to fixed and permeabilized cells, live cells can be barcoded allowing for real time functional analysis or short term culturing of the barcoded cells. This strategy can be easily adapted for various flow cytometry applications and offers a convenient alternative to FCB based strategies.
MATERIALS and METHODS
Animals and reagents
C57BL/6 mice were purchased from Jackson Laboratory (Bar Harbor, ME, USA); B10.A and MD4 mice were purchased from Taconic Farms (Hudson, NY, USA). All mice were maintained in NIH animal facilities in compliance with Animal Care and Use Committee standards.
Cells were cultured and stimulated in sterile RF10 media (RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 50 U/ml penicillin, 50 µM streptomycin, 1 mM sodium pyruvate, 2 mM L-glutamine, 0.1 mM non-essential amino acids, 50 µM 2-mercaptoethanol and 10 mM HEPES) (ThermoFisher; Waltham, MA, USA). Samples were stained for flow cytometric analysis using FACS buffer: PBS or HBSS (Lonza; Allendale, NJ, USA) supplemented with 2% FBS, 1% HEPES and 0.09% sodium azide (Sigma-Aldrich; St. Louis, MO, USA). Fix Buffer I (for phospho-flow) or Cytofix/Cytoperm (for intracellular cytokine staining), both purchased from BD Biosciences (Franklin Lakes, NJ, USA), were used as fixation solutions. Calcium flux assays were carried out using either assay buffer (HBSS with calcium and magnesium (ThermoFisher) supplemented with 20mM HEPES and 10mM dextrose (Sigma- Aldrich)) or complete assay buffer (assay buffer supplemented with 2.5mM Probenecid (ThermoFisher)). The Abs used for staining or stimulation purposes are listed in Supplementary Table 1.
Isolation of T and B Lymphocytes from mouse
Animals were euthanized by CO2 asphyxiation followed by cervical dislocation. The superficial cervical, mandibular, superficial/deep axillary, inguinal and mesenteric lymph nodes and the spleen were aseptically removed as described in (7); single cell suspensions were prepared in sterile RF10 media/PBS using a 70 µm cell strainer (BD Biosciences). Cells from spleen were treated with ACK buffer (Lonza) for 5 min on ice to lyse the red blood cells, were washed and resuspended using sterile MACS Buffer (PBS supplemented with 2 mM EDTA and 0.5% BSA) and were passed through a sterile 30 µm cell filter (Partec-Sysmex, Lincolnshire, IL, USA) prior to magnetic separation. To isolate B cells, cell suspensions from spleen were treated with mouse B cell isolation kits according to the manufacturer’s protocol, increasing incubation times at 4 °C to 15 min. Samples were washed using MACS Buffer and applied to LS columns for negative magnetic separation. The initial flow through of purified B cells was passed through the same column a second time to increase purity. T cells were isolated from spleen and lymph nodes using mouse naïve CD4+ T cell isolation kits according to manufacturer’s guidelines. Magnetic cell separation of T cells was carried out using the AutoMACS instrument’s “Depletes” program and the negative fraction, containing purified T cells, was collected. Reagents and tools for magnetic separation were purchased from Miltenyi Biotec (San Diego, CA, USA) unless otherwise stated.
B cell phospho-flow assays
Purified mouse splenic B cells were stained in 2 ml plain RPMI-1640 media containing a 1:250 dilution of Live/Dead Near IR marker (ThermoFisher) for 20 min at 4 °C in the dark to label dead cells prior to stimulation. After two washes with plain RPMI-1640 media, cells were resuspended in RF10 media and 90 µl/well cell suspension was plated into a sterile 96 well round bottom plate (assay plate) (Corning; New York, NY, USA). One row of a separate deep well plate (stimulation plate) (96 wells) (Axygen, Union City, CA, USA) containing either RF10 media only (3 wells) or master mix of 20 µg/ml anti-IgM (F(ab’)2) (3 wells) was aseptically prepared. Both plates were warmed to 37 °C by placing them in a 5% CO2 humidified tissue culture incubator. Stimulation was initiated by rapidly transferring 90 µl/well from the stimulation plate (containing either media or antibody solution) into the assay plate (containing an equal volume of cells) using a multi-channel micropipette yielding 180 µl/well of cell suspension in the assay plate with or without 10 µg/ml anti-IgM antibody. For efficient staining and analysis, 2×105–4×105 cells per well were used. An additional deep well plate (fixation plate) was prepared by adding 1 ml/well Fix Buffer I. This plate was sealed and warmed in the same incubator; at relevant time points (15, 45 or 90 min) cells were mixed by pipetting and 160 µl/well of cell suspension was transferred directly from the assay plate into the fixation plate. For each time point, a total of 18 samples organized in 3 sets of 6 wells (each set containing 3 unstimulated and 3 stimulated samples) were taken. Upon fixing the cells at 37 °C for 10–15 min, cells were centrifuged and resuspended with 200 µl/well FACS buffer and transferred into a 96 well round bottom plate (staining plate) which was kept at 4 °C until all time points were completed.
Cells were then centrifuged, their supernatants removed and the staining plate was placed on ice. Cell pellets were slowly resuspended with 100 µl/well pre-chilled (at −20 °C) Perm Buffer III (BD Biosciences). The plate was covered and incubated on ice for exactly 30 min to permeabilize cells. At the end of permeabilization, 100 µl/well FACS buffer was added into the wells. The plate was centrifuged and supernatants were discarded. This was followed by three additional washes with FACS buffer to completely remove the methanol-containing Perm Buffer III.
Fixed and permeabilized cells were barcoded using mixtures of fluorochrome conjugated barcoding Abs according to the design outlined in Figure 1. Barcoding solutions contained fluorochrome conjugates of barcoding Abs diluted 1/200 in FACS buffer supplemented with 1–2% normal rat and mouse serum (Jackson Immunoresearch, West Grove, PA, USA) and 10 µg/ml anti-CD16/32 to block nonspecific binding of the barcoding and phospho-specific Abs. Cells were incubated with barcoding solutions (50 µl/well in 96 well plate) for 30 min at 4 °C in the dark, followed by four washes of 200 µl/well FACS buffer. Cells with different barcodes were pooled into one well. A small aliquot of pooled cells, stained with only the barcoding Abs, was used as a fluorescence minus one (FMO) control for the subsequent phosphoprotein staining in flow cytometry. Barcoded, pooled cells were stained with 50 µl/well phospho-flow staining mix (prepared by diluting the phospho-specific Abs in FACS buffer according to manufacturer’s guidelines) for 30 min at 4 °C followed by two washes prior to analysis.
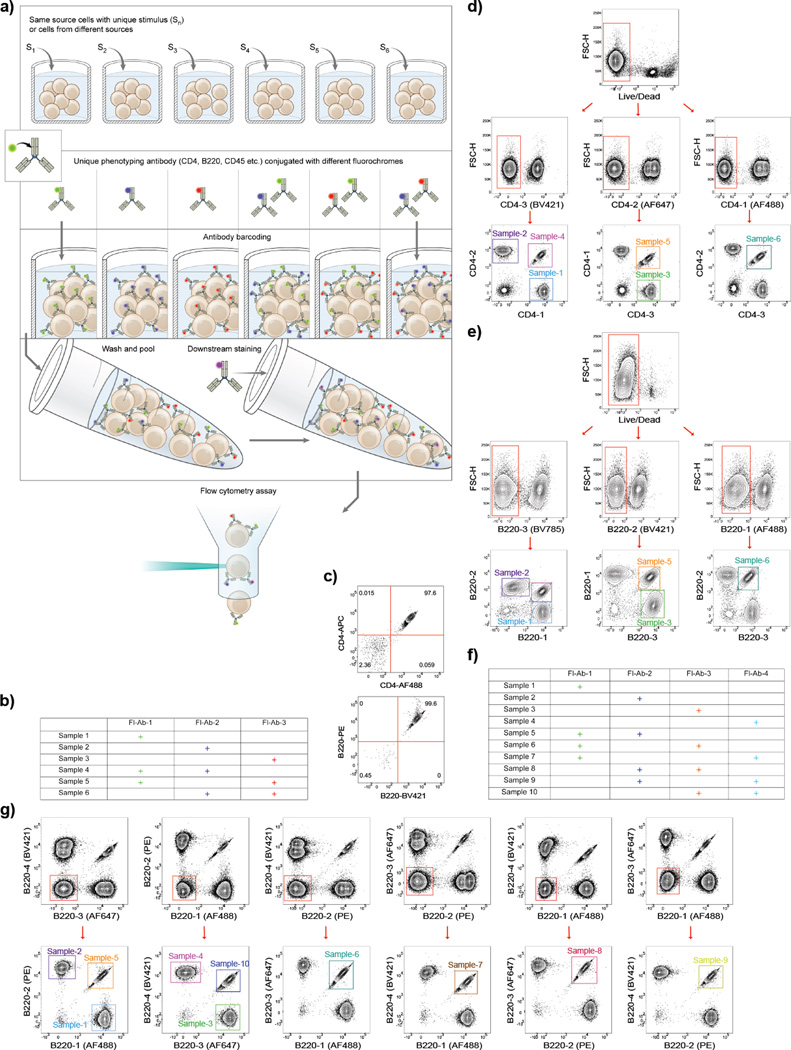
Figure 1.
Ab-barcoding allows identification of T cells and B cells pooled from multiple samples. a) Key steps in barcoding cells using single- or double- stained combinations of three different Fl-Abs for flow cytometry are illustrated. b) Assignment of Fl-Abs to samples to generate six unique barcodes. c) Representative examples of uniformly double stained CD4+ T cells (top) and B cells (bottom) are shown. d–e) Stepwise gating strategy to identify individual populations T cells (d) and B cells (e) in a pool of cells containing six uniquely barcoded samples illustrated in (b) using anti-CD4-Fl-Abs for T cells and anti-B220-Fl-Abs for B cells. Gating steps prior to Live/Dead stage are as shown in (Supplementary Fig. 1). f–g) Representative example of barcoding using four different anti-B220-Fl-Abs in single- or double- stained combinations generating ten unique barcoding patterns as outlined in (f) and demonstrated in (g).
Flow cytometry analysis was carried out using a BD LSR-II cytometer. For each pooled sample, at least 8 × 104 events in the live cell gate were collected. Data were analyzed using FlowJo analysis software (Treestar, Ashland, OR, USA). The geometric mean fluorescence intensity (MFI) of the phosphoprotein signal was calculated for each properly gated sample. The same procedure was repeated for the FMO sample. Using these values, the net MFI was calculated by subtracting the FMO’s MFI from the sample’s MFI. The fold change in phosphorylation was then determined by dividing the net MFI of the stimulated sample by the net MFI of the unstimulated sample for each time point.
B cell calcium flux assays
Six aliquots of pure live splenic B cells (three from wild type and three from MD4 transgenic mice) were barcoded using combinations of three different fluorochrome-conjugated anti-B220 Abs in assay buffer for 30 min at 4 °C followed by four washes. Uniquely barcoded samples as well as an unbarcoded similarly treated samples from wild type mouse were then pooled in to one tube in 4 ml assay buffer and incubated with the loading solution containing Fluo4-AM (calcium indicator dye) and Powerload (to facilitate the entry of Fluo4-AM into the cells) (both purchased from ThermoFisher) for 20 min at room temperature. The final concentrations of Fluo4-AM and Powerload were 1 µM and 25 nM respectively. After one wash, cells were resuspended in 10 ml HBSS and incubated for an additional 20 min at room temperature to allow cells to release excess dye. Cells were then washed once and resuspended in complete assay buffer in FACS tubes. Right before the assay, cells were warmed to 37 °C in a water bath and then applied directly to the cytometer. After a baseline recording for 60 – 75 s, the FACS tube was briefly removed from the cytometer and the first stimulant was added; recording was continued for 4 – 6 min. In some experimental settings, the tube was briefly removed once more and a second stimulant was added which was followed by an additional 4 – 6 min of recording. The entire procedure was carried out in one recording session without pause. The flow rate was kept within the range of 600 – 1000 events/s. Analysis was carried out by plotting median Fluo4 intensity against time.
T cell differentiation assays
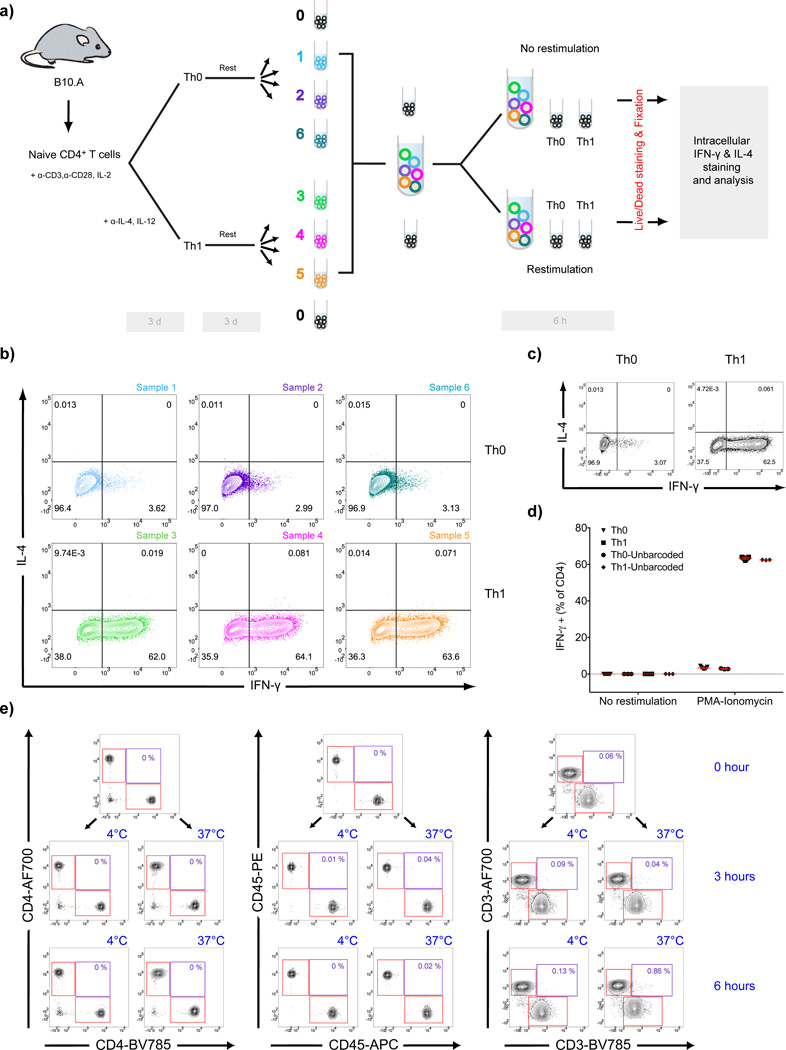
A sterile 24 well tissue culture plate (Corning) was coated with 0.5 ml/well PBS containing 2 µg/ml anti-CD3 and 4 µg/ml anti-CD28 Abs for 2 hours at 37 °C. The plate was washed twice with RF10 media. In each well, 5×105 naïve CD4+ T cells purified from B10.A mice were seeded in 1 ml RF10 media supplemented with either 100 U/ml IL-2 (Peprotech, Rockhill, NJ,USA) only (for Th0 differentiation) or a mixture of 100 U/ml IL-2, 10ng/ml IL-12 (Peprotech) and 10 µg/ml anti-IL-4 antibody (for Th1 differentiation). The plate was incubated at 37 °C in a humidified 5% CO2 incubator for 3 days. Cells were then split 1:2 and transferred into a new plate (resting plate). An equal volume of fresh RF10 media supplemented with 20 U/ml IL-2 was added on top. After 3 days of resting, both Th0 and Th1 differentiated cell suspensions were divided into 3 samples for barcoding with fluorochrome-conjugated anti-CD4 Abs diluted in plain RPMI-1640 media at 4 °C for 20 min. The resulting six uniquely barcoded samples were washed for four times and pooled in RF10 media. For both barcoded and unbarcoded samples restimulation was carried out in parallel by adding cell stimulation cocktail (phorbol 12-myristate 13-acetate (PMA), ionomycin, brefeldin A and monensin) (eBioscience, San Diego, CA, USA) into the growth media. Restimulated and non-restimulated samples were cultured at 37 °C in a humidified 5% CO2 incubator for 6 h. Then cells were stained with Live/Dead marker in FACS buffer followed by fixation, permeabilization and intracellular staining with anti-IL-4 and anti-IFN-γ Abs prior to flow cytometry analysis.
RESULTS
The barcoding strategy described here identifies cells by employing unique combinations of fluorochrome-conjugated Abs (Fl-Abs) and can be applicable to any flow cytometry-based analysis. Theoretically, the barcoding Abs can be of any specificity and can be conjugated to a variety of fluorochromes to barcode various cell types. To demonstrate the efficacy of the method we report the results of several experiments that use this strategy to barcode cell samples for different applications.
Ab-barcoding allows identification of cells pooled from multiple samples
To demonstrate the ability of barcoded samples to be identified once pooled, we barcoded purified mouse T and B cells. Naïve CD4+ T cells were purified from mouse spleen and lymph nodes, CD19+ B cells were purified from mouse spleen. The viability and purity were determined using flow cytometry. T cells were stained with a CD4-specific Ab (anti-CD4) and a Live/Dead marker. B cells were stained with CD19- and B220-specific Abs (anti-CD19 and anti-B220), as well as a Live/Dead marker. The stepwise gating of T and B lymphocytes to gate on single live cells is shown (Supplementary Fig. 1). The cell viability was 95.7% for T cells and 95.4% for B cells. Within the live cell population, T cell purity, defined as the percent of cells that were CD4+, was 98.2% and of this CD4+ population 95% demonstrated the characteristics of naïve T cells (8) defined as CD44lo CD62Lhi (Supplementary Fig. 1a). B cell purity, defined as the percentage of cells that were both CD19+ and B220+, was 99.3% (Supplementary Fig. 1b).
We used the strategy shown in Fig. 1a to barcode six T or B cell populations using combinations of three different Fl-Abs, anti-B220 for B cells and anti-CD4 for T cells. As shown, three of the barcodes require staining with only a single Fl-Ab and three require staining with combinations of Fl-Abs (Fig. 1b). For this strategy to be successful, all cells in the double-labeled samples must be labeled with both Fl-Abs leaving no single-labeled cells. Using two different Fl-Abs to label T cells (AF488 and APC conjugated anti-CD4) and B cells (PE and BV421 conjugated anti-B220), all of the cells were found to be double positive and moreover, the cells appeared on a diagonal in the flow cytometry plot indicating similar binding of both Fl-Abs to the cells (Fig. 1c).
We next demonstrated that six T and B cell samples, barcoded using three different Fl-Abs to create the six barcodes, once pooled, can be identified by flow cytometry. Purified T cells were stained with a Live/Dead marker, divided into six samples and the samples were incubated with the combinations of three different Fl-Abs as shown in Fig. 1b. The samples were pooled and analyzed by flow cytometry. Stepwise gating of the Fl-Ab labeled cells clearly identified the six barcoded populations (Fig. 1d). Briefly, in the live cell population, cells that were negative for one of the three Fl-Abs were gated, and within that gate, subpopulations labeled with one or both of the remaining two Fl-Abs were identified. This gating was repeated for all three Fl-Abs to identify the six barcoded CD4+ T cell populations. By a similar strategy, using three different anti-B220 Fl-Abs six barcoded populations of B cells were identified (Fig. 1e).
In addition, ten B cell samples were barcoded using four different anti-B220-Fl-Abs (Fig. 1f). As shown, this strategy still only required the ability to discriminate single- versus double-positive cells. Using a similar gating strategy to that used to identify six barcoded populations, we were able to readily identify ten different barcoded populations (Fig. 1g)
Creating more than ten barcodes minimally requires the ability to discriminate between quadruple-, triple-, double- and single-labeled cells. A potential problem when using multiple Fl-Abs specific for the same epitope for barcoding is competition between Fl-Abs for the same target epitope resulting in diminished signal intensity. We compared the fluorescence intensity of B cells stained with combinations of five different B220-specific Fl-Abs either alone or in combination with up to four additional Fl-Abs (Fig 2).
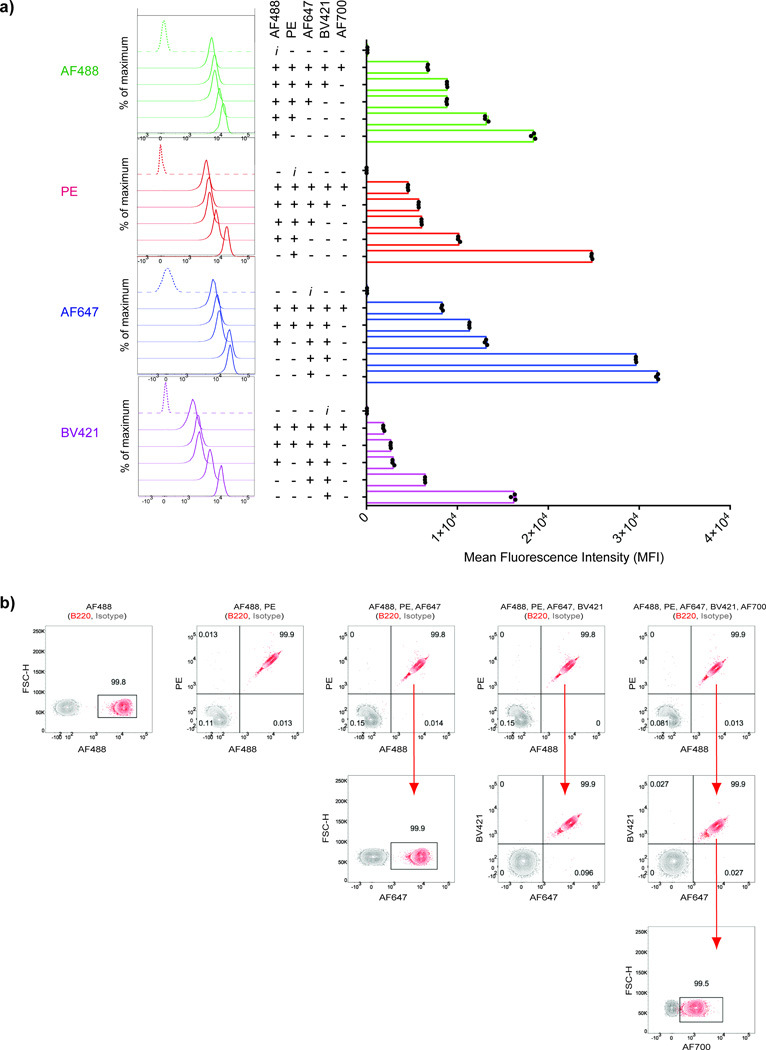
Figure 2.
The decrease in the fluorescence intensity of individual Fl-Ab when combined with multiple other Fl-Abs does not interfere with the identification of barcoded populations. a) anti-B220-Fl-Abs conjugated to either AF488, PE, AF647, BV421 or AF700 and their isotype-matched controls were used to stain purified B cell populations in multiple combinations. The histograms (left) and the MFI values (right) for each fluorochrome are given in comparison with the fluorescently-labeled isotype control Ab. Individual values of triplicate observations are shown as black dots on the MFI column graph. b) B cell samples were stained using the combinations of anti-B220-Fl-Abs (red contour plots) or appropriate fluorescently-labeled isotype control Ab (grey contour plots) and analyzed by flow cytometry. Contour plots are superimposed to determine the boundaries of positive gates. Numbers refer to the percentages of cells positively stained with anti-B220-Fl-Abs for each flow cytometry plot. Results are representative of three independent observations.
Purified B cells were divided into samples that were barcoded using five different Fl-Abs or their fluorochrome conjugated isotype control Abs (Fig. 2a). The concentrations of the five different Fl-Abs used for staining were determined empirically. We first tested all Fl-Abs between 1:100 to 1:400 dilutions from their stock solutions as purchased from the manufacturers and observed no significant difference in brightness (data not shown). As a result we used a 1:200 dilution for all Fl-Abs corresponding to final concentrations of 2.5 µg/mL for AF488, AF700 and AF647; 1 µg/mL for PE and 0.4 µg/mL for BV421. The effect of the addition of up to four different Fl-Abs to a single Fl-Ab was determined (Fig. 2a). As expected, in each case, with the addition of each of the other four Fl-Abs, the fluorescence intensity achieved with the initial Fl-Ab alone dropped to varying degrees. However, all combinations of all Fl-Abs resulted in significantly higher fluorescence intensity as compared to that of the Fl-isotype control Ab. We next demonstrated that cells barcoded using up to five different Fl-Abs could be discriminated and that in all cases cells were uniformly labeled with the Fl-Abs in the barcode mixture. We analyzed the cells pooled from the barcoded samples in the top panel of Fig. 2a. The gating strategy is shown in which the flow cytometry plots of the matched Fl-isotype control-Abs were overlaid to precisely determine the boundaries of the positive population gates (Fig 2b). As shown, it was possible to distinguish all five barcoded populations; for each barcode that required more than one Fl-Ab, the Fl-Abs uniformly stained all cells.
Taken together these results demonstrate that multiple Fl-Abs can be combined to effectively increase the number of possible barcodes, using the same Abs. Hypothetically, using all possible combinations of five Fl-Abs, 32 unique barcodes can be generated. However, achieving 32 barcodes depends on the abundance of the target epitope and the capacity of the flow cytometer to accommodate all the different colors. Therefore pilot trials are recommended for the cell type of interest before designing the combinations.
Barcoding allows efficient analysis of the signaling events following B cell receptor stimulation using phospho-flow
We next determined if barcoding could be applied to an analysis of the functional properties of activated cells in vitro and chose to analyze the phosphorylation of four kinases in naïve mouse splenic B cells following stimulation with anti-IgM. Purified mouse splenic B cells were first stained with Live/Dead marker and then divided into three sets of six samples for each time point (Fig. 3a). In each set, three of the samples were stimulated with soluble anti-IgM (10 µg/mL) and the other three were left untreated. At each time point (15, 45 and 90 min after the addition of anti-IgM), individual samples within each set were fixed, permeabilized, and barcoded using three different anti-B220 Fl-Abs (Fig. 3a). Barcoded samples were pooled as shown and stained with fluorescently labeled Abs specific for phospho-Syk, phospho-Btk, phospho-p38 and phospho-Akt (Fig. 3a). Barcodes were selected to ensure that both single Fl-Ab barcodes and barcodes that required two Fl-Abs were used to barcode unstimulated B cells (barcodes 1,2,6) and anti-IgM-treated B cells (barcodes 3,4,5).
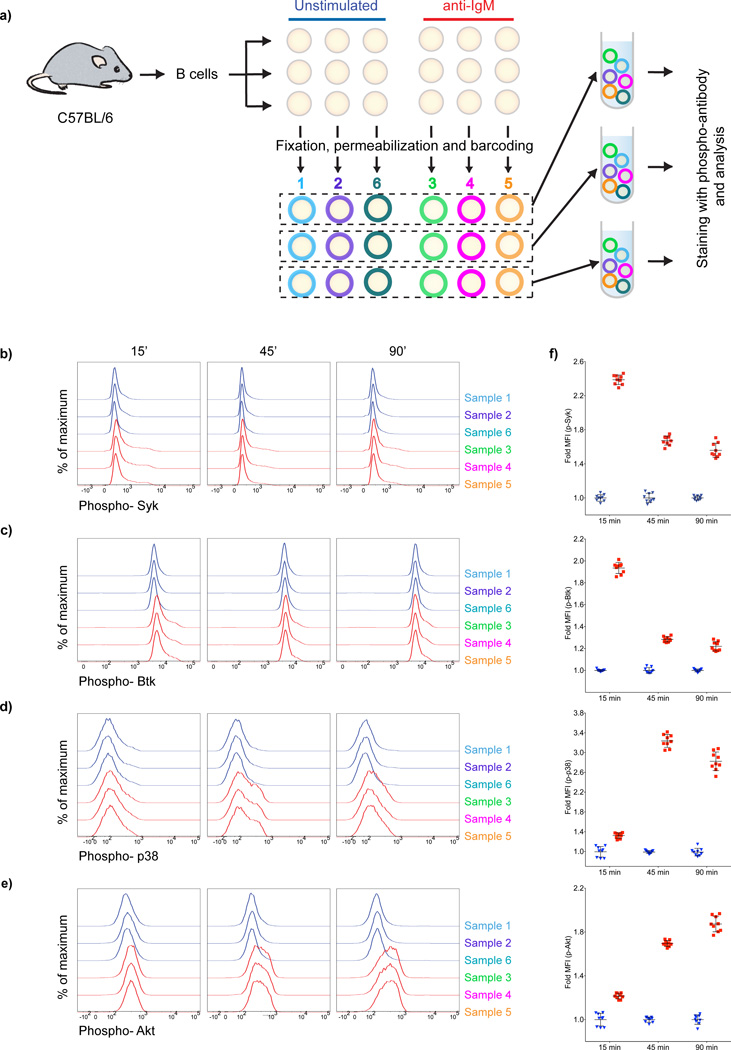
Figure 3.
Ab-based barcoding is effective in standardizing experiments using phospho-flow. (a) Purified mouse splenic B cells were divided into three sets of six samples each for each of three time points that were either unstimulated or stimulated with 10 µg/ml anti-IgM. Fifteen, 45 and 90 min later samples were fixed, permeabilized barcoded and pooled as shown. Pooled samples were stained with specific Abs and analyzed in flow cytometry. (b–e) Time-dependent changes in phosphorylation of Syk (b), Btk (c), p38 (d) and Akt (e) are shown as histogram overlays for both unstimulated cells (blue histograms corresponding to samples 1,2,6) and anti-IgM stimulated cells (red histograms corresponding to samples 3,4,5). f) Fold MFI with time after anti-IgM stimulation (red squares) and unstimulated controls (blue triangles). Each symbol represent one sample of the barcoded replicates. Error bars indicate the standard deviation. Results are representative of more than three independent experiments.
Pooled samples containing triplicates of uniquely barcoded stimulated and unstimulated cells were analyzed in flow cytometry and barcoded cells were discriminated using the gating strategy described in Fig. 1 (Supplementary Fig. 1c). For each phospho-kinase stain, the flow profiles for one set of all six of the barcoded samples (three stimulated with anti-IgM, three unstimulated) are given (Fig. 3b–e) as are the fold MFI for all three triplicate samples (9 individual samples) (Fig. 3f). For each phosphokinase, stimulated cells uniformly showed increased fluorescence intensity compared to the unstimulated cells (Fig. 3b–e). Moreover, we were able to clearly demonstrate the change in phosphorylation dynamics for each kinase (Fig. 3f). As expected, the proximal BCR signaling kinases, Syk and Btk, showed increased phosphorylation at the early time point, 15 min, but phosphorylation was decreased in the 45 and 90 min time points. In contrast, the distal signaling kinases Akt and p38 showed an opposite pattern with little phosphorylation at 15 min but increased phosphorylation at 45 and 90 min. These results are consistent with previously published results on the BCR-mediated phosphorylation dynamics of these kinases as observed in various settings using methods other than phospho-flow (9–11). Taken together, our results provide evidence that barcoding did not alter the subsequent intracellular staining for phosphokinases and thus appears to be a robust method for standardizing the detection of phosphoproteins in multiple separate samples.
Live cell barcoding is an efficient tool for standardizing sample to sample variation in real time functional assays
Lymphocytes respond to a variety of stimuli with an influx of Ca2+ ions into the cytoplasm (12). This physiological event provides the basis for flow cytometry assays, commonly used for analyzing early signaling dynamics as well as determining pathological changes (13). Most of these assays require cells to be loaded with a Ca2+ sensitive dye in buffers lacking essential nutrients and sequential analysis of the cells’ response to stimuli requiring incubation at different temperatures for extended durations of time before analysis (14–16). These assays include an initial base line measurement before the stimulus is added requiring that an aliquot of cells are aspirated by the flow cytometer (16, 17). Differences in the aspirated volume for the baseline measurement can create variability in the concentration of the stimulating agent that is subsequently added to the remaining cell suspension. Thus, standardizing sample-to-sample variations and minimizing nonspecific background effects are often challenging. Here, using purified splenic mouse B cells, we tested our barcoding strategy as an improved approach for overcoming the shortcomings of the current Ca2+ flux assays.
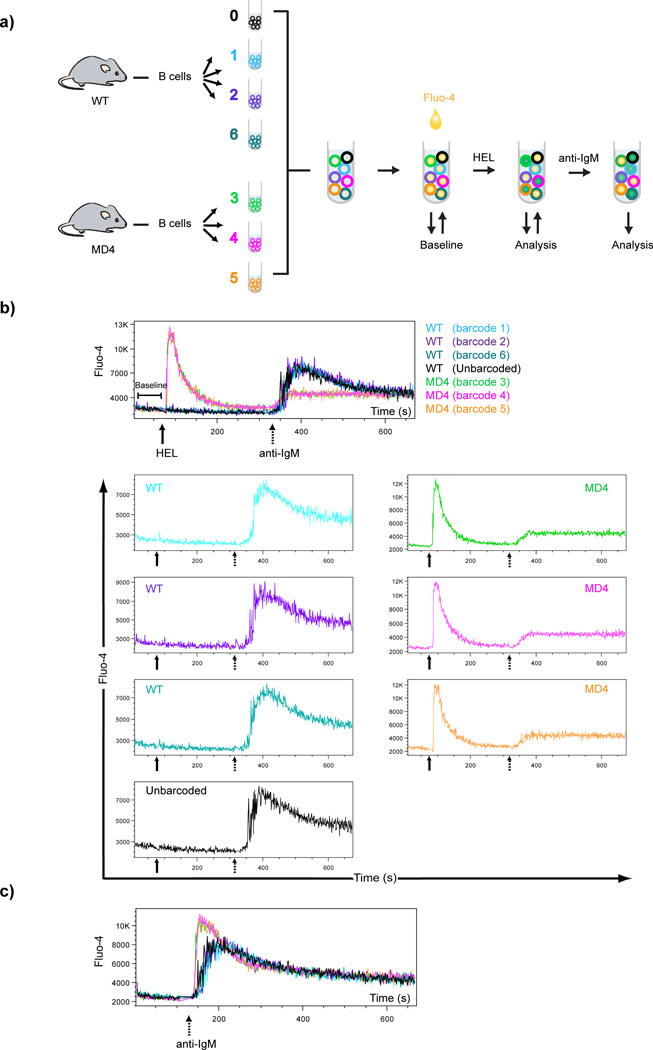
In B cells, stimulation through the BCR triggers a signaling cascade which in turn leads to PLCγ2-mediated rapid Ca2+ uptake into the cytoplasm (18). BCRs can be stimulated either via crosslinking with anti-IgM or through engagement with the antigen for which the BCR is specific. We used B cells from two different mouse strains, namely C57BL/6 (WT) as a source of polyclonal B cells and MD4, a BCR transgenic mouse strain that contains monoclonal B cells that recognize Hen Egg Lysozyme (HEL) (19). Live B cells from both mice were purified and barcoded using combinations of three different anti-B220-Fl-Abs to create six barcodes. Three barcodes were used to label MD4 B cells and three were used to label WT B cells. One potential problem with barcoding live cells prior to stimulation is the effect of the barcoding Ab on the activation state of the cell. Since B220, the target molecule for barcoding Abs here, is a membrane protein tyrosine phosphatase (20), the barcoding Abs could, in theory, trigger a signaling cascade by crosslinking B220 that would affect the response to anti-IgM. Weak stimulation of B cells was reported for certain clones of anti-B220 Abs but only at high concentrations or in double-crosslinked systems (21). For the anti-B220 Ab clone used here, RA3-6B2, no effect of Ab binding to B cells has been reported. Nonetheless, to control for possible effects of the anti-B220-Fl-Abs on B cells, we included unbarcoded B cells from WT mice in the assay. After pooling all samples, cells were loaded with Fluo-4 (22), a fluorescent dye used for quantification of intracellular Ca2+ levels. The pooled samples were warmed to 37 °C prior to analysis. After a 75 s baseline measurement, soluble HEL protein was added to the cells at a final concentration of approximately 2 µg/ml and flow cytometry measurement of Fluo-4 were taken for 5 min. At the end of 5 min, anti-IgM was added to the remaining cells at a final concentration of approximately 10 µg/ml and flow cytometry analysis of Fluo-4 was carried out for an additional 5 min (Fig 4a). Shown are both the composite flow histograms for all samples as well as individual flow histograms for each sample (Fig. 4b). Cells in all six barcoded samples showed uniform baseline Fluo-4 signals. The HEL-specific MD4 B cells showed a uniform Ca2+ flux following the addition of HEL but no response was observed for WT cells. Conversely, subsequent addition of anti-IgM induced a uniform Ca2+ flux by the barcoded WT B cells but only a weak response was observed in MD4 B cells that responded robustly to HEL earlier. Importantly, no differences between the Ca2+ responses of B220 barcoded and unbarcoded WT B cell samples were observed indicating that B220 Abs did not affect the B cells’ response to anti-IgM (Fig 4b). Finally, in a parallel set of experiments, pooled and barcoded samples from WT and MD4 mice were stimulated directly with anti-IgM and all responded with overlapping calcium responses (Fig. 4c). The variation in the intensity of the maximal Ca2+ response between MD4 and WT B cells is not unexpected for B cells taken from different inbred mouse strains.
Figure 4.
Ab-based barcoding as a means to standardize conditions in real time Ca2+ flux assays. a) Samples of B cells purified from WT C57BL/6 mice (barcodes 1,2,6 and unbarcoded) and HEL transgenic MD4 mice (barcodes 3,4,5) were barcoded, pooled and stained with Fluo4 as shown. After a baseline measurement, pooled sample were stimulated with approximately 2 µg/ml HEL protein and Ca2+ uptake analyzed for 5 min. Cells were then stimulated with approximately 10 µg/ml anti-IgM and Ca2+ uptake and analyzed for 5 min. b) Fluo4 intensity histogram overlays of all samples (top panel) as well as individual histograms (bottom panels) over time are shown. Continuous (HEL) and dotted (IgM) arrows indicate the time points when stimuli were added. c) Histograms of Fluo4 intensity for all samples treated with anti-IgM are shown. Data shown are representative of at least four independent experiments.
Taken together these results demonstrate that barcoding cells provides a means to standardized conditions for Ca2+ flux assays including the sample incubation and stimulation times, as well as the concentrations of stimulants, without interfering with the functional response. Moreover, despite all the post-barcoding handling steps, cells were shown to maintain their unique barcoding patterns throughout the experiment (Supplementary Fig. 1d). Finally, by barcoding cells for the real time Ca2+ assay, the experiment time was dramatically decreased from approximately 90 min for an analysis of seven unbarcoded samples to approximately 15 min for seven barcoded samples.
Barcoded and pooled cell populations can be cultured and stimulated for functional assays
Many assays of cellular function involve culturing cells in vitro for periods of time, for instance to compare the effects of the same stimuli on different cell types in vitro. In such experiments barcoding and pooling cells prior to stimulation may prove useful in minimizing variation between samples. To test whether our Ab-based barcoding strategy can be used in such experiments, we devised a simple T cell differentiation assay. Naïve CD4+ T cells obtained from B10.A mice were stimulated for 3 days using plate bound anti-CD3 and anti-CD28 (Fig. 5a). Half of the cells received soluble IL-2, IL-12 and anti-IL-4 to drive Th1 differentiation and the other half received IL-2 only to facilitate Th0 differentiation. Cells were rested for 3 days in media containing low concentrations of IL-2. Cells from each group were then divided into three samples which were barcoded using three different anti-CD4-Fl-Abs as described in Fig 1, resulting in six samples containing barcoded triplicates of Th0 and Th1 differentiated cells. Cells were pooled and divided into two samples which were either restimulated using PMA-ionomycin and protein transport inhibitors for 6 h or left non-restimulated. In a parallel experiment, an aliquot of unbarcoded cells from the Th1 differentiated cells was cultured separately under the same conditions to assess possible effects of barcoding on the experimental outcome. At the end of the 6 h incubation, cells were stained with Live/Dead marker followed by fixation and permeabilization and intracellular staining for IFN-γ (a marker for Th1 cells) and IL-4 (a marker for Th2 cells).
Figure 5.
Ab-based barcoding is compatible with experiments requiring culturing in vitro to activate cells. a) Naïve CD4+ T cells obtained from B10.A mice were stimulated for 3 days by plate bound anti-CD3 and anti-CD28 in the presence of IL-2 alone to facilitate TH0 differentiation or in the presence of IL-2 plus anti-IL-4 and IL-12 to drive Th1 differentiation. Cells were then rested for an additional 3 days. Th0- and Th1-differentiated cells were divided into three samples that were barcoded, pooled and then either restimulated with cell stimulation cocktail or left unstimulated for 6 h. Separate aliquots of unbarcoded Th0- and Th1-differentiated cells were treated similarly. At the end of 6 h, cells were stained with Live/Dead marker, fixed, permeabilized, stained with anti-IL-4 and anti-IFN-γ and analyzed by flow cytometry. b) Representative flow cytometry plots showing the percentages of IFN-γ and/or IL-4 expressing barcoded cells for both Th0- (samples 1,2,6) and Th1- (samples 3,4,5) differentiated, restimulated cells. c) Representative flow cytometry plots showing the percentage of IL-4 and/or IFN-γ producing cells for unbarcoded, restimulated Th0- and Th1-differentiated cells. d) Percentages of IFN-γ+ cells for three sets of barcoded samples (yielding nine samples per condition) and three sets of unbarcoded samples (yielding three samples per condition) for Th0- and Th1-differentiated cells that were restimulated or not. Error bars indicate the standard deviation. e) T cell barcoding antibodies do not detach during short term culturing of live cells. Freshly isolated mouse CD4+ T cells were divided into two groups that were single stained with a different fluorochrome conjugation of the same barcoding antibody. Cells were washed and combined. Flow cytometry plots showing the initial staining of the mixture (0 hour) as well as the levels after incubation at either 4 °C or 37 °C for 3 or 6 hours are shown for CD4 (left panel), CD45 (middle panel) and CD3 (right panel). Single stained populations are shown with red rectangular gates and the double stained populations are shown with purple gates. Percentages refer to the frequency of double stained cells in the mixture. Data shown are representative of three independent experiments.
Among the restimulated samples, Th0-differentiated barcoded cells uniformly stained negative for IFN-γ whereas over 60% of the Th1-differentiated barcoded cells produced IFN-γ (Fig. 5b). Both groups were negative for IL-4 production. IL-4 and IFN-γ staining of unbarcoded Th0- and Th1-differentiated samples showed similar results (Fig. 5c,d) verifying that the Fl-Abs used for barcoding did not affect the outcome of the experiment. Intracellular staining of cytokines in non-restimulated samples was uniformly low in both groups as shown in (Supplementary Fig 2a). Furthermore, for both restimulated and non-restimulated T cells, barcoding Abs remained on the cell surface throughout culturing and post-harvest procedures yielding clearly distinguishable individual populations (Supplementary Fig 2b–c).
The main concern in culturing barcoded samples for long durations in vitro is the theoretical possibility that barcoding Abs may detach and re-attach to other cells, altering the barcoding. To rule out this possibility, we tested the stability of three different barcoding antibodies on CD4+ T cells, namely anti-CD45, anti- CD4 and anti-CD3, under different incubation conditions. For this purpose, two groups of cells each stained with a different single fluorochrome conjugate of the Abs were washed and pooled. Immediately after pooling, the samples were analyzed by flow cytometry. Then, one aliquot of the pooled cells was incubated at 37 °C and a second aliquot was kept at 4 °C. The samples were assayed by flow cytometry to assess the detachment and re-attachment of the barcoding antibodies after 3 h and 6 h incubations. None of the three barcoding Abs generated a double stained population in any of the incubation conditions, ruling out the possibility of Abs detaching and re-attaching to other cells under these assay conditions (Fig. 5e). These results show that barcoding can be integrated in assays that require several hours of culturing.
Ab-based barcoding can be applied to whole spleen cell preparations for phenotype analysis
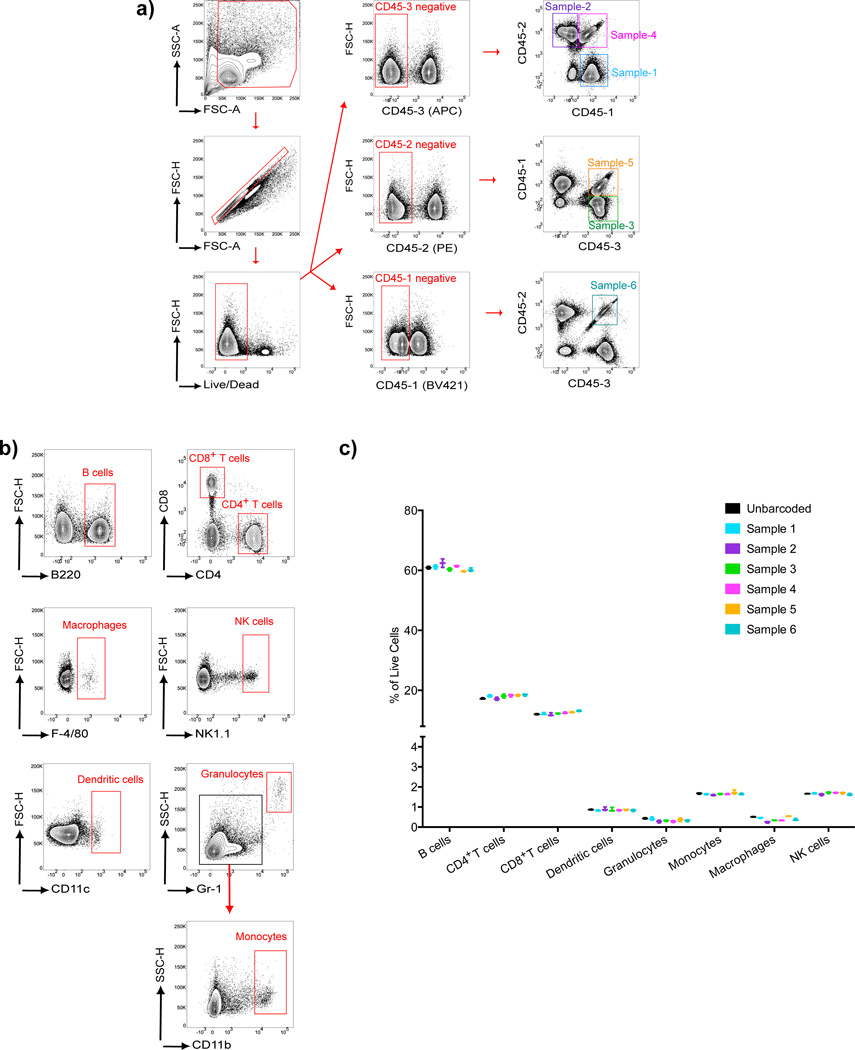
A common experimental design in which the barcoding method described here may be applicable is the analysis of several different cell samples for standard immune-cell phenotyping. To demonstrate the efficacy of barcoding for this application, we barcoded six spleen cell populations in triplicate using three different anti-CD45-Fl-Abs. The three groups of six uniquely barcoded samples were pooled and the three pooled samples were stained with Abs specific for: B220 (B cells); CD8 or CD4 (T cells); CD11c (dendritic cells); Gr-1 (granulocytes, monocytes); CD11b (monocytes); F4/80 (macrophages) and NK1.1 (NK cells) (Fig 6).
Figure 6.
Ab-based barcoding applied to immune cell phenotyping. Splenocytes harvested from C57BL/6 mice were divided into six samples that were barcoded using three different anti-CD45-Fl-Abs and then pooled. The pooled samples and a separate unbarcoded sample of splenocytes were stained with immune cell phenotyping Abs specific for: B220, CD4, CD8, F4/80, NK1.1, CD11c, Gr1 or CD11b. a) Flow cytometry plots showing the gating strategy for the identification of the six uniquely barcoded cell samples. b) Gating strategy used for identification of B cells, T cells, macrophages, NK cells, dendritic cells, granulocytes and monocytes within the mixed splenocyte population. c) Cell subsets were quantified as the percentage of live cells within each barcoded sample and the unbarcoded sample and values obtained from three parallel sets of barcodes and three unbarcoded samples are given. Error bars show the range between the minimum and maximum of observed values. Results are representative of three independent experiments.
As shown, the combinations of the three different anti-CD45-Fl-Abs allowed for the clear identification of the cells from the six different samples by flow cytometry (Fig 6a). The identification of eight different immune cell populations using the phenotyping Abs, adapted from a published strategy (23) is shown (Fig. 6b). The results of this analysis are summarized (Fig. 6c). An example of the identification of these eight immune cell phenotypes in the six barcoded samples and in a spleen cell population that was not barcoded is given (Supplementary Fig. 3a–d) showing the percentage of all live cells that were classified as B cells, CD4+ T cells, CD8+ T cells, DCs, granulocytes, monocytes, macrophages and NK cells. The results show a remarkable consistency of the phenotyping of the spleen cells in the six barcoded samples. Moreover, with respect to the percentages of individual cell subsets, the barcoded samples were indistinguishable from the spleen cell sample that was not barcoded. Thus, the barcoding method described here appears to be a highly robust approach that allows for immune cell phenotyping of different cell samples.
DISCUSSION
Here we describe a novel Ab-based barcoding method for use in flow cytometry. We demonstrated the efficacy of the method using three different Fl-Abs specific for CD4, B220, or CD45. Evidence for the feasibility of barcoding with combinations of up to five different Fl-Abs was also demonstrated in a pilot system. In principle, Abs of any specificity can be used for barcoding and the number of barcodes is limited only by the selection of fluorochromes and the flow cytometry instrumentation used to detect and resolve the fluorochromes. For example, theoretically using 10 different fluorochrome conjugated Abs, it should be possible to barcode 1024 samples. Many potential barcoding Abs are available in multiple fluorochrome conjugates from several manufacturers, therefore offering convenience and flexibility. Although current fluorescent dye-based barcoding techniques (5, 6) allow for the barcoding of up to 16 samples using multiple dilutions of two fluorochromes, barcoding this number of samples is often not practical due to both technical and practical limitations of the method. Having 16 populations in one single flow cytometry plot results in populations that are very close to, if not overlapping, each other. The dye-based method is even less suitable when different stimulation conditions or mixed cell populations generate variations in cell size, dye uptake, or base level fluorescence (6). Taken together, these variations make it difficult to estimate the exact location of the populations in the flow cytometry plot. Our Ab-based barcoding method eliminates this issue by using only one dilution of the Fl-Ab, along with our stepwise gating strategy, which places no more than three populations (two single-stained and one double-stained) on a single flow cytometry plot. Additionally, since the fluorescent dye-based barcoding strategy does not include a phenotyping Ab, an additional fluorochrome and a surface staining step must be included for identification purposes.
The use of Abs for barcoding purposes has been demonstrated previously for applications other than flow cytometry (24–27). Most of those strategies use Abs labeled with unique isotopes or DNA labels for mass spectrometry- or PCR-based analyses, requiring specialized reagents, equipment and deconvolution techniques and they are not compatible with either real time assays on live cells or multicolor flow cytometric analyses.
The Ab-based barcoding method described here is an ideal strategy for efficient use in flow cytometry that can be adapted to a wide range of experimental designs including but not limited to assays using fixed and permeabilized cells, mixed cell populations, live cells for real time assays or short term culturing. The only caveats to this method are those that must be considered when any Fl-Abs are used for flow cytometry (non-specific binding, Fc-receptor binding, etc). It is also possible that for any given barcoding Ab, detachment and re-attachment during culturing in vitro may limit the application of this method. Although we demonstrated that three different barcoding Abs did not significantly detach and re-attach during up to 6 h incubation in vitro at 37°C, it is possible that low affinity Abs or increased acidity in the culture media may result in the detachment of the barcoding Abs. Consequently, we recommend testing any desired barcoding Abs in preliminary assays using the experimental culture conditions. In conclusion, our strategy offers a simple, versatile and reliable barcoding method for flow cytometry applications.
Supplementary Material
Acknowledgments
This study was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases.
We are grateful to Ann Kim for optimizing the B cell isolation protocol, Mirna Peña for maintenance of the animals, Alan Hoofring for generating the illustration in Fig 1a, Nyamekye Obeng-Adjei for helpful comments on the manuscript.
Footnotes
The authors declare that there are no conflicts of interest.
Author Contributions
M.A. conceived the project. M.A., B.A. designed the experiments. M.A., B.A. carried out the experiments. P.M., A.H.H. purified cells and assisted in the experiments. M.A., B.A., S.K.P. analyzed the data. B.A. made the figures. M.A., S.K.P. wrote the manuscript. P.M., E.M.S. edited the manuscript.
References
- 1.Laerum OD, Farsund T. Clinical application of flow cytometry: a review. Cytometry. 1981;2:1–13. doi: 10.1002/cyto.990020102. [DOI] [PubMed] [Google Scholar]
- 2.Shapiro HM. Flow cytometric estimation of DNA and RNA content in intact cells stained with Hoechst 33342 and pyronin Y. Cytometry. 1981;2:143–150. doi: 10.1002/cyto.990020302. [DOI] [PubMed] [Google Scholar]
- 3.Shapiro HM. Flow cytometric probes of early events in cell activation. Cytometry. 1981;1:301–312. doi: 10.1002/cyto.990010502. [DOI] [PubMed] [Google Scholar]
- 4.Christensen ME, Jansen ES, Sanchez W, Waterhouse NJ. Flow cytometry based assays for the measurement of apoptosis-associated mitochondrial membrane depolarisation and cytochrome c release. Methods. 2013;61:138–145. doi: 10.1016/j.ymeth.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 5.Krutzik PO, Clutter MR, Trejo A, Nolan GP. Fluorescent cell barcoding for multiplex flow cytometry. Curr Protoc Cytom. 2011;Chapter 6(Unit 6):31. doi: 10.1002/0471142956.cy0631s55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods. 2006;3:361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- 7.Reeves JP, Reeves PA. Removal of lymphoid organs. Curr Protoc Immunol. 2001;Chapter 1(Unit 1):9. doi: 10.1002/0471142735.im0109s01. [DOI] [PubMed] [Google Scholar]
- 8.Gerberick GF, Cruse LW, Miller CM, Sikorski EE, Ridder GM. Selective modulation of T cell memory markers CD62L and CD44 on murine draining lymph node cells following allergen and irritant treatment. Toxicol Appl Pharmacol. 1997;146:1–10. doi: 10.1006/taap.1997.8218. [DOI] [PubMed] [Google Scholar]
- 9.Pogue SL, Kurosaki T, Bolen J, Herbst R. B cell antigen receptor-induced activation of Akt promotes B cell survival and is dependent on Syk kinase. J Immunol. 2000;165:1300–1306. doi: 10.4049/jimmunol.165.3.1300. [DOI] [PubMed] [Google Scholar]
- 10.Chaturvedi A, Dorward D, Pierce SK. The B cell receptor governs the subcellular location of Toll-like receptor 9 leading to hyperresponses to DNA-containing antigens. Immunity. 2008;28:799–809. doi: 10.1016/j.immuni.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaturvedi A, Martz R, Dorward D, Waisberg M, Pierce SK. Endocytosed BCRs sequentially regulate MAPK and Akt signaling pathways from intracellular compartments. Nat Immunol. 2011;12:1119–1126. doi: 10.1038/ni.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feske S. Calcium signalling in lymphocyte activation and disease. Nat Rev Immunol. 2007;7:690–702. doi: 10.1038/nri2152. [DOI] [PubMed] [Google Scholar]
- 13.Burchiel SW, Edwards BS, Kuckuck FW, Lauer FT, Prossnitz ER, Ransom JT, Sklar LA. Analysis of free intracellular calcium by flow cytometry: multiparameter and pharmacologic applications. Methods. 2000;21:221–230. doi: 10.1006/meth.2000.1002. [DOI] [PubMed] [Google Scholar]
- 14.June CH, Moore JS. Measurement of intracellular ions by flow cytometry. Curr Protoc Immunol. 2004;Chapter 5(Unit 5):5. doi: 10.1002/0471142735.im0505s64. [DOI] [PubMed] [Google Scholar]
- 15.do Ceu Monteiro M, Sansonetty F, Goncalves MJ, O'Connor JE. Flow cytometric kinetic assay of calcium mobilization in whole blood platelets using Fluo-3 and CD41. Cytometry. 1999;35:302–310. [PubMed] [Google Scholar]
- 16.Bailey S, Macardle PJ. A flow cytometric comparison of Indo-1 to fluo-3 and Fura Red excited with low power lasers for detecting Ca(2+) flux. J Immunol Methods. 2006;311:220–225. doi: 10.1016/j.jim.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Desmeules P, Dufour M, Fernandes MJ. A rapid flow cytometry assay for the assessment of calcium mobilization in human neutrophils in a small volume of lysed whole-blood. J Immunol Methods. 2009;340:154–157. doi: 10.1016/j.jim.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Scharenberg AM, Humphries LA, Rawlings DJ. Calcium signalling and cell-fate choice in B cells. Nat Rev Immunol. 2007;7:778–789. doi: 10.1038/nri2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodnow CC, Crosbie J, Adelstein S, Lavoie TB, Smith-Gill SJ, Brink RA, Pritchard-Briscoe H, Wotherspoon JS, Loblay RH, Raphael K, et al. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 1988;334:676–682. doi: 10.1038/334676a0. [DOI] [PubMed] [Google Scholar]
- 20.Ledbetter JA, Tonks NK, Fischer EH, Clark EA. CD45 regulates signal transduction and lymphocyte activation by specific association with receptor molecules on T or B cells. Proc Natl Acad Sci U S A. 1988;85:8628–8632. doi: 10.1073/pnas.85.22.8628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.George A, Rath S, Shroff KE, Wang M, Durdik JM. Ligation of CD45 on B cells can facilitate production of secondary Ig isotypes. J Immunol. 1994;152:1014–1021. [PubMed] [Google Scholar]
- 22.Gee KR, Brown KA, Chen WN, Bishop-Stewart J, Gray D, Johnson I. Chemical and physiological characterization of fluo-4 Ca(2+)-indicator dyes. Cell Calcium. 2000;27:97–106. doi: 10.1054/ceca.1999.0095. [DOI] [PubMed] [Google Scholar]
- 23.Akkaya M, Aknin ML, Akkaya B, Barclay AN. Dissection of agonistic and blocking effects of CD200 receptor antibodies. PLoS One. 2013;8:e63325. doi: 10.1371/journal.pone.0063325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ullal AV, Peterson V, Agasti SS, Tuang S, Juric D, Castro CM, Weissleder R. Cancer cell profiling by barcoding allows multiplexed protein analysis in fine-needle aspirates. Sci Transl Med. 2014;6:219ra219. doi: 10.1126/scitranslmed.3007361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai L, Ong R, Li J, Albani S. A CD45-based barcoding approach to multiplex mass-cytometry (CyTOF) Cytometry A. 2015;87:369–374. doi: 10.1002/cyto.a.22640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agasti SS, Liong M, Peterson VM, Lee H, Weissleder R. Photocleavable DNA barcode-antibody conjugates allow sensitive and multiplexed protein analysis in single cells. J Am Chem Soc. 2012;134:18499–18502. doi: 10.1021/ja307689w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mei HE, Leipold MD, Schulz AR, Chester C, Maecker HT. Barcoding of live human peripheral blood mononuclear cells for multiplexed mass cytometry. J Immunol. 2015;194:2022–2031. doi: 10.4049/jimmunol.1402661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.